Abstract
We present a method for detecting the presence of Mycoplasma genitalium, Mycoplasma hominis, Ureaplasma parvum, and Ureaplasma urealyticum organisms, which are thought to be associated with nongonococcal urethritis (NGU) and other genitourinary infections, in clinical samples. This method consists of PCR amplification of a part of the 16S rRNA gene followed by 96-well microtiter plate hybridization assay using four species-specific capture probes to detect the targets. To test the efficacy of this method, we applied it to the detection of the four species in the urine of patients with NGU. There were no cross-reactions with other human mycoplasmas or ureaplasmas, and the PCR-microtiter plate hybridization assay detected as few as 10 copies of the 16S rRNA gene of each of the four species. Based on these results, this PCR-microtiter plate hybridization assay can be considered an effective tool for the diagnosis of genitourinary infections with mycoplasmas or ureaplasmas.
Urethritis, an infection often acquired via sexual contact, is categorized into gonococcal urethritis or nongonococcal urethritis (NGU) depending on the presence or absence of Neisseria gonorrhoeae. In NGU, the infection is attributed to the pathogenic role of Chlamydia trachomatis, which has been detected in 30 to 40% of men with NGU (17), or to other pathogens, including mycoplasmas and ureaplasmas, in chlamydia-negative patients.
In our previous study, we reported a method to identify all human-origin mycoplasmas and ureaplasmas using PCR amplification and phylogenetic analysis (22). This method was shown to successfully detect 13 species of mycoplasmas and two species of ureaplasmas. In urinary samples from a cohort of men with NGU, however, we detected only four species of mycoplasma or ureaplasma—Mycoplasma genitalium, Mycoplasma hominis, Ureaplasma parvum, and Ureaplasma urealyticum (22). Among these four species, only M. genitalium was significantly more frequent in men with NGU or chlamydia-negative NGU than in asymptomatic controls. However, the number of subjects examined in the previous study (22) was too small to definitively conclude that the other three species, M. hominis, U. parvum, and U. urealyticum, do not play pathogenic roles in NGU.
Although our phylogeny-based technique has proven useful for identifying mycoplasmas and ureaplasmas in clinical samples, it requires the use of subcloning and sequencing to distinguish among multiple mycoplasmas and/or ureaplasmas in a sample. In the present study, therefore, we employed a DNA probe method to identify the nucleotide sequences of M. genitalium, M. hominis, U. parvum, and U. urealyticum and applied it to a newly developed hybridization-based microtiter plate assay to detect those four species in first-pass urine samples of men with NGU and asymptomatic controls.
MATERIALS AND METHODS
Prototype strains.
The following prototype strains of 15 species of human mycoplasmas and ureaplasmas were used in this study. The following mycoplasmas and U. urealyticum (T-strain 960) were provided by Tsuguo Sasaki, Laboratory of Sterility Assurance, the National Institute of Infectious Diseases, Tokyo, Japan: M. fermentans (PG18), M. genitalium (G37), M. hominis (PG21), M. lipophilum (MaBy), M. orale (CH19299), M. penetrans (GTU-54-6A1), M. pneumoniae (FH), M. primatum (HRC/292), and M. salivarium (PG20). The following mycoplasmas and U. parvum (ATCC 27818) were obtained from the American Type Culture Collection: M. buccale (ATCC 23636), M. faucium (ATCC 25293), M. pirum (ATCC 25960), and M. spermatophilum (ATCC 49695).
Clinical specimens.
The urine samples used in this study were the same as those used in our previous study (22). Briefly, the samples were obtained from male patients with urethritis and from asymptomatic males between July 1999 and July 2000. A diagnosis of urethritis was based on the observation of five or more polymorphonuclear leukocytes per oil-immersion field (×1,000) in a Gram-stained endourethral swab specimen. As described previously (22), DNA extracts derived from urine samples were amplified by PCR and subjected to phylogeny-based identification procedures. The 55 samples thereby identified as mycoplasma or ureaplasma positive (16 were M. genitalium positive, 2 were M. hominis positive, 14 were U. parvum positive, and 23 were U. urealyticum positive) were then subjected to PCR-microtiter plate hybridization assay. In addition, 14 M. hominis-positive and 10 U. parvum-positive urine samples newly identified were used.
DNA extraction.
DNA was extracted as described previously (22). Briefly, the precipitates from 1 ml of urine were harvested by centrifugation at 15,000 × g for 30 min. The precipitates were treated with proteinase K at 55°C for 2 h in digestion buffer, and the DNA was extracted using the phenol-chloroform method. The DNA extracts were first used for phylogenetic analysis, and then the remainders were stored at −70°C for use in the PCR-microtiter hybridization assay.
PCR.
A forward primer, My-ins (5′-GTAATACATAGGTCGCAAGCGTTATC-3′, nucleotides [nt] 520 to 545), and two biotinylated reverse primers, MGSO-2-Bi (5′-biotin-CACCATCTGTCACTCTGTTAACCTC-3′, nt 1036 to 1012) and UGSO-Bi (5′-biotin-CACCACCTGTCATATTGTTAACCTC-3′, nt 1012 to 988), were used to amplify an approximately 520-bp region of the 16S rRNA gene of mycoplasmas and ureaplasmas. The positions of all primers used, except UGSO-Bi, were numbered according to the nucleotide sequence of the M. genitalium G37 (GenBank accession number X77334). The primer sequence of UGSO-Bi was obtained from the U. urealyticum T-strain 960 (GenBank accession number AF073450). A PCR mixture contained 1× PCR buffer [50 mM Tris-HCl (pH 8.3), 10 mM KCl, 5.0 mM (NH4)2SO4, 2.0 mM MgCl2], 0.5 μM My-ins; 0.25 μM (each) MGSO-2-Bi and UGSO-Bi; 0.2 mM (each) dATP, dCTP, and dGTP; 0.6 mM dUTP; 1.25 U of FastStart Taq DNA polymerase (Roche Molecular Biochemicals, Mannheim, Germany); and 10 μl of prepared DNA solution in a total volume of 50 μl. PCR was performed using the GeneAmp PCR System 9600 (Applied Biosystems, Foster City, Calif.) under the following conditions: an initial cycle at 95°C for 10 min, followed by 50 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and elongation at 72°C for 60 s, with a final cycle at 72°C for 7 min. The PCR products were then subjected to hybridization assays. To compare the results of the hybridization assay with those of agarose gel electrophoresis, the remainders of the PCR products were separated in 3% agarose gel, then stained with ethidium bromide (1 μg/ml) to visualize the DNA bands from the PCR products with a UV transilluminator.
Plasmid construction.
The 771-bp DNA fragment including the whole sequence of the PCR product of M. genitalium was amplified using a primer set, My-1S (5′-GAACAGCCACAATGGGACTGAGA-3′, nt 293 to 315) and My-2A (5′-TCACGACACGAGCTGACGACAAC-3′, nt 1063 to 1041). The amplified product was introduced into pT7Blue T-Vector (Novagen Inc., Madison, Wis.) to construct a positive control plasmid as described previously (22). After purification of the plasmid using a QIAprep Spin Miniprep Kit (Qiagen Inc., Hilden, Germany), the amount of DNA was quantified by measuring the optical density (OD) at 260 nm, and the copy number was calculated. Ten copies of the positive control plasmid were used for each batch of PCR. Tris-EDTA buffer containing no DNA was used as a negative control.
Internal control.
To monitor the presence of inhibitors in the PCR, we deleted the M. genitalium-specific capture probe-binding region from the positive control plasmid and constructed an internal control plasmid. Ten copies of the internal control were added to the PCR mixture for each clinical sample tested.
Hybridization.
Amplified products of M. genitalium, M. hominis, U. parvum, and U. urealyticum were detected by hybridization-based microtiter plate assay with species-specific oligonucleotide probes. We designed the capture probes in the species-specific regions demonstrated by the alignment of human mycoplasmas and ureaplasmas (GenBank accession numbers AB069810 to AB069824). Four capture probes—Mgen-P3-Am (5′-TCGGAGCGATCCCTTCGGT-3′) specific for M. genitalium, Mhom-P10-Am (5′-GACACTAGCAAACTAGAGTTAG-3′) specific for M. hominis, Upar-P6-Am (5′-GTCTGCCTGAATGGGTCGGT-3′) specific for U. parvum, and Uure-P4-Am (5′-GGCTCGAACGAGTCGGTGT-3′) specific for U. urealyticum—were 5′ aminated with a six-carbon spacer. We also designed a capture probe to detect the amplified product of the internal control (IC-P4-Am: 5′-CTAGCTGTCGGCTGGAATTC-3′). These probes were each immobilized to a microtiter plate, DNA-BIND 1 × 8 strip well plate (Corning Inc., Corning, N.Y.), as described in the manufacturer's instructions. The amplified products were denatured by boiling for 5 min and were quickly chilled in ice. One hundred microliters of hybridization buffer (5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.02% sodium dodecyl sulfate) was added to the wells of each microtiter plate immobilized by the capture probes. Five microliters of denatured amplicons was then added, and hybridization was carried out at 37°C for 90 min. Following hybridization, the wells were washed twice (soaking for 5 min) with wash buffer I (360 μl/well; 0.2× SSC, 0.1% sodium dodecyl sulfate). One hundred microliters of streptavidin-peroxidase (Roche Molecular Biochemicals) was added per well and allowed to incubate for 15 min at 37°C. Then the wells were washed twice (soaking for 5 min) with wash buffer II (360 μl/well; 0.1% Tween 20 in phosphate-buffered saline). One hundred microliters of BM Blue POD Substrate (Roche Molecular Biochemicals) was added per well and allowed to sit for 10 min at room temperature. The reaction was stopped by the addition of hydrosulfuric acid (100 μl/well), and the OD at 450 nm was measured by the Novapath Microplate Reader (Bio-Rad, Richmond, Calif.). The cutoff OD value was determined to be 0.300 by measuring the negative control value 20 times. The mean value plus 4 standard deviations was 0.256. Samples with an OD value of ≥0.300 were considered positive, and those with a value <0.300 were considered negative.
Sensitivity.
To estimate the sensitivity of the PCR-hybridization assay, the three fragments that included the whole sequence of the PCR product of the 16S rRNA gene of M. hominis, U. parvum, and U. urealyticum, respectively, were amplified and introduced into pT7Blue T-Vector (Novagen) as described under “Plasmid construction” above.
Specificity.
The DNAs prepared from human mycoplasmas and ureaplasmas were subjected to PCR amplification. Nonreacted primers and deoxynucleoside triphosphates were removed from amplified products using a QIAquick PCR Purification kit (Qiagen Inc.), and then the amount of DNA was quantified by measuring the OD at 260 nm. To evaluate the specificity of the capture probes, 300 ng of amplified DNA of each prototype strain was assayed per well.
Detection of mixed DNA.
The four plasmids, respectively, containing the DNA fragments of the four mycoplasma and ureaplasma species were mixed in two combinations—M. genitalium with U. urealyticum, and M. hominis with U. parvum—in each of the following mix ratios: 100 to 0, 99 to 1, 95 to 5, 90 to 10, 70 to 30, 50 to 50, 30 to 70, 10 to 90, 5 to 95, 1 to 99, and 0 to 100. PCR was performed with 105 copies of mixed plasmid per reaction. Amplified products were subjected to both direct sequencing as described previously (22) and microtiter plate hybridization assay, and the ability to detect mixed DNA was compared between the two methods.
RESULTS
Specificity.
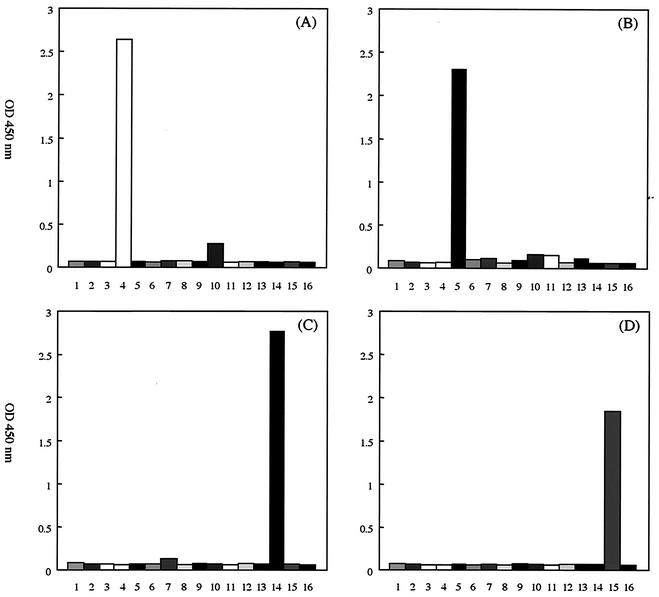
The amplified products of each DNA fragment of human mycoplasma or ureaplasma were observed by agarose gel electrophoresis and staining with ethidium bromide (data not shown). To evaluate the specificity of the capture probes, 300 ng of the amplified DNA of each prototype strain was assayed by the hybridization-based microtiter plate assay. Mhom-P10-Am, Upar-P6-Am, and Uure-P4-Am detected only their targets, and there was no cross-reaction with other human mycoplasmas or ureaplasmas (Fig. 1). Although there was a weak reaction of Mgen-P3-Am with M. pneumoniae, the OD value was below the cutoff (Fig. 1). When PCR-microtiter plate hybridization assay was performed with 1010 copies of M. pneumoniae DNA per reaction, the OD value with Mgen-P3-Am was below the cutoff (data not shown). There was no cross-reaction with a variety of genitourinary tract contagions, such as Chlamydia trachomatis, Neisseria gonorrhoeae, Candida albicans, herpes simplex virus types 1 and 2, or adenovirus types 40 and 41 (data not shown).
FIG. 1.
The species-specificity of the four capture probes. The microtiter plate hybridization assay was performed with 300 ng of amplified DNA derived from 15 prototype strains of human mycoplasmas and ureaplasmas, and the optical density at 450 nm was measured. Column 1, M. buccale; column 2, M. faucium; column 3, M. fermentans; column 4, M. genitalium; column 5, M. hominis; column 6, M. lipophilum; column 7, M. orale; column 8, M. pirum; column 9, M. penetrans; column 10, M. pneumoniae; column 11, M. primatum; column 12, M. salivarium; column 13, M. spermatophilum; column 14, U. parvum; column 15, U. urealyticum; column 16, blank. (A) Mgen-P3-Am; (B) Mhom-P10-Am; (C) Upar-P6-Am; (D) Uure-P4-Am.
Sensitivity.
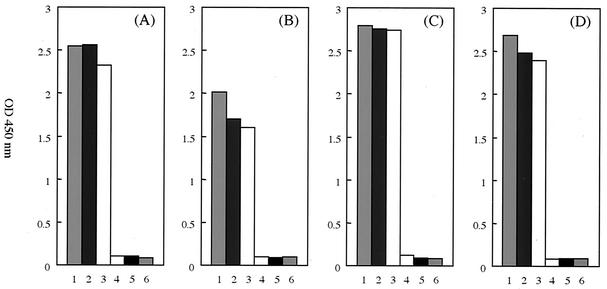
Recombinant plasmids, into which were inserted the partial DNA fragments of M. genitalium, M. hominis, U. parvum, and U. urealyticum, respectively, were diluted with Tris-EDTA buffer. To determine the sensitivity of the PCR-microtiter plate hybridization assay, a series of 10-fold dilutions of the plasmid, ranging from 103 to 10−1 copies per reaction, was used. The sensitivity of this assay was 10 copies of DNA per reaction (Fig. 2). Amplified products were also electrophoretically separated on 3% agarose gels and stained with ethidium bromide, and again, the lower limit of visual detection was 10 copies of DNA per reaction.
FIG. 2.
The sensitivity of four species-specific PCR-microtiter plate hybridizations. To determine the sensitivity, a PCR-microtiter plate hybridization assay was performed using plasmid containing the partial 16S rRNA genes of M. genitalium, M. hominis, U. parvum, and U. urealyticum. Column 1, 103 copies plasmid; column 2, 102 copies plasmid; column 3, 101 copies plasmid; column 4, 100 copies plasmid; column 5, 10−1 copies plasmid; column 6, blank. (A) Mgen-P3-Am; (B) Mhom-P10-Am; (C) Upar-P6-Am; (D) Uure-P4-Am.
Mixed DNA detection.
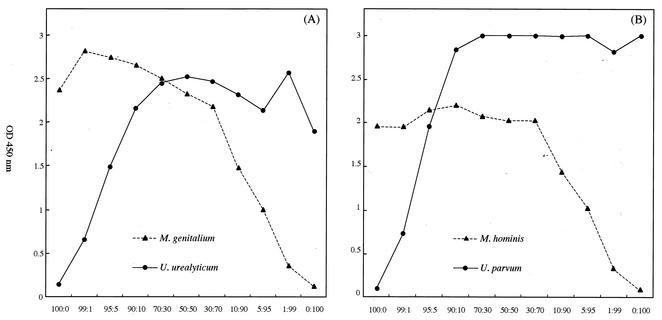
Because DNA is often present in clinical samples in various combinations, we prepared mixed DNA samples of different ratios. Using these samples, we compared the ability to detect mixed DNA between the direct sequencing method and microtiter plate hybridization assay. When the minor population of DNA was more than 30%, it was detected by direct sequencing (data not shown). On the other hand, the hybridization-based assay was able to detect the minor population of DNA even when its content was as low as 1% (Fig. 3).
FIG. 3.
Detection of mixed DNA by PCR-microtiter plate hybridization assay. The plasmids, containing partial DNA fragments of M. genitalium, M. hominis, U. parvum, and U. urealyticum, respectively, were mixed at various ratios: 100 to 0, 99 to 1, 95 to 5, 90 to 10, 70 to 30, 50 to 50, 30 to 70, 10 to 90, 5 to 95, 1 to 99, and 0 to 100. The PCR-microtiter plate hybridization assay was performed with 105 copies mixed plasmid per reaction, and the optical density at 450 nm was measured. Both panels show mixed plasmid: M. genitalium and U. urealyticum (A) and M. hominis and U. parvum (B).
Detection of mycoplasmas and ureaplasmas from urine samples.
As previously described (22), portions of the DNA solutions extracted from urine samples were subjected to the PCR amplification and phylogeny-based identification procedures. In this study, the PCR-microtiter plate hybridization assay was performed anew with each of the remaining DNA solutions. The same species, identified by phylogenetic analysis, were also detected by hybridization-based microtiter plate assay (Table 1). In some of the samples, two species of mycoplasmas or ureaplasmas were detected simultaneously (Table 1). Furthermore, we added and examined 24 urine samples, already identified as M. hominis or U. parvum positive by phylogenetic analysis, obtained from men with urethritis. From 8 and 2 of the 14 M. hominis-positive samples, U. parvum and U. urealyticum were simultaneously detected in addition to M. hominis, respectively, by hybridization assay. With the four remaining M. hominis-positive samples and 10 U. parvum-positive samples, the same results were obtained by both methods.
TABLE 1.
Detection of mycoplasmas and ureaplasmas
| Classification of specimens (no.)a | Detected organism(s)b | No. of specimens from indicated patients
|
||
|---|---|---|---|---|
| GU | NGU | Asymptomatic | ||
| M. genitalium-positive (16) | M. genitalium | 2 | 10 | 1 |
| M. genitalium and U. urealyticum | 3 | |||
| M. hominis-positive (2) | M. hominis | 1 | ||
| M. hominis and U. parvum | 1 | |||
| U. parvum-positive (14) | U. parvum | 1 | 4 | 9 |
| U. urealyticum-positive (23) | U. urealyticum | 6 | 13 | 3 |
| U. urealyticum and U. parvum | 1 | |||
Identified by phylogenetic analysis in our previous study (22).
Detected by microtiter plate hybridization assay in this study.
DISCUSSION
M. genitalium was first isolated in urethral cultures from two men with NGU in 1981 (20). Although M. genitalium has been proposed as a cause of human NGU, the precise role of this mycoplasma in the etiology of NGU has not been established because of the immense difficulty of isolating it from clinical samples. More recently, however, PCR-based assays have facilitated the detection of M. genitalium in clinical samples (8, 13), and a significant association has been demonstrated between M. genitalium and NGU (2, 3, 5-7, 11, 12, 19). In experimentally infected chimpanzees, M. genitalium has been shown to induce symptomatic genital infections with inflammatory and antibody responses, suggesting that M. genitalium may be a pathogen of NGU (21).
In 1999, U. urealyticum biovars 1 and 2 were classified into U. parvum and U. urealyticum, respectively (10). Many reports have suggested a relationship between NGU and ureaplasmas. However, the colonization rates in asymptomatic males make it difficult to reach an unequivocal conclusion with respect to the etiologic role of ureaplasmas (9, 15, 18). Most of these reports have discussed the role of ureaplasmas without discriminating between the two species, U. parvum and U. urealyticum. Recently, Povlsen et al. demonstrated that the prevalence of U. urealyticum among men with NGU was higher than that among men without NGU (14). Therefore, U. urealyticum may play an important role in the etiology of NGU.
M. hominis has been associated with bacterial vaginosis, pelvic inflammatory disease, postpartum fever, and postabortal fever, as well as a number of gynecological infections (4, 16, 18). However, its role in NGU is poorly understood. Other mycoplasmas isolated from the human urogenital tract, such as M. fermentans and M. penetrans, do not seem to be associated with NGU, as discussed in our previous studies (1).
We previously amplified the 16S rRNA gene of mycoplasmas and ureaplasmas from urine samples by PCR and stored the data on the nucleotide sequences of the amplified products (22). Comparison with the nucleotide sequences of mycoplasmas and ureaplasmas of amplified products from clinical samples and those obtained from GenBank demonstrated that there were some species-specific regions in approximately 250 bp, including the V4 and V5 regions of the 16S rRNA gene. In the present study, to detect M. genitalium, M. hominis, U. parvum, and U. urealyticum with high specificity, we designed oligonucleotide probes at the respective sites of the species-specific regions. Hybridization-based microtiter plate assays using three of the four oligonucleotide capture probes—Mhom-P10-Am, Upar-P6-Am and Uure-P4-Am—detected only their respective targets and not other human mycoplasmas or ureaplasmas. A weak reaction of Mgen-P3-Am with M. pneumoniae was observed. However, with the amplified product from 1010 copies of M. pneumoniae DNA per reaction, the OD value was below the cutoff value. Therefore, few possibilities for misidentification were present.
The results of detection of mycoplasmas and ureaplasmas in clinical samples by the hybridization-based microtiter plate assay were compared with those by the direct sequencing method. The two methods performed similarly, although the former method detected more than one strain from each of the samples. Considering the specificity of this assay, it was supposed that the hybridization-based assay showed no cross-reaction and was better able to detect mixed infections than the direct sequence method. This assumption was supported by the results of assays with artificial mixed-infection samples using the plasmids.
For the detection of fastidious bacterial strains, such as M. genitalium, a PCR-based method was extremely useful. The hybridization-based microtiter plate assay in the present study required only 2 to 3 h and cost less than the method that determined the nucleotide sequences of the amplicons. This assay improved the ability to detect mixed infections compared with the direct sequencing method, because there was no need to subclone the amplified products when multiple strains of mycoplasmas or ureaplasmas were present in clinical specimens. We believe the PCR-microtiter plate hybridization assay method described here provides a rapid and effective measure to detect human mycoplasmas and ureaplasmas which is useful for etiological and epidemiological studies of these pathogens.
Acknowledgments
We thank Tsuguo Sasaki, Laboratory of Sterility Assurance, the National Institute of Infectious Diseases, Tokyo, Japan, for providing prototype strains of mycoplasmas and ureaplasmas.
REFERENCES
- 1.Deguchi, T., C. B. Gilroy, and D. Taylor-Robinson. 1996. Failure to detect Mycoplasma fermentans, Mycoplasma penetrans, or Mycoplasma pirum in the urethra of patients with acute nongonococcal urethritis. Eur J. Clin. Microbiol. Infect. Dis. 15:169-171. [DOI] [PubMed] [Google Scholar]
- 2.Deguchi, T., and S. Maeda. 2002. Mycoplasma genitalium: another important pathogen of nongonococcal urethritis. J. Urol. 167:1210-1217. [DOI] [PubMed] [Google Scholar]
- 3.Gambini, D., I. Decleva, L. Lupica, M. Ghislanzoni, M. Cusini, and E. Alessi. 2000. Mycoplasma genitalium in males with nongonococcal urethritis: prevalence and clinical efficacy of eradication. Sex. Transm. Dis. 27:226-229. [DOI] [PubMed] [Google Scholar]
- 4.Hillier, S. L., R. P. Nugent, D. A. Eschenbach, M. A. Krohn, R. S. Gibbs, D. H. Martin, M. F. Cotch, R. Edelman, J. G. Pastorek II, A. V. Rao, et al. 1995. Association between bacterial vaginosis and preterm delivery of a low-birth-weight infant. N. Engl. J. Med. 333:1737-1742. [DOI] [PubMed] [Google Scholar]
- 5.Horner, P. J., C. B. Gilroy, B. J. Thomas, R. O. Naidoo, and D. Taylor-Robinson. 1993. Association of Mycoplasma genitalium with acute non-gonococcal urethritis. Lancet 342:582-585. [DOI] [PubMed] [Google Scholar]
- 6.Jensen, J. S., H. T. Hansen, and K. Lind. 1996. Isolation of Mycoplasma genitalium strains from the male urethra. J. Clin. Microbiol. 34:286-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jensen, J. S., R. Orsum, B. Dohn, S. Uldum, A. M. Worm, and K. Lind. 1993. Mycoplasma genitalium: a cause of male urethritis? Genitourin Med. 69:265-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jensen, J. S., S. A. Uldum, J. Sondergard-Andersen, J. Vuust, and K. Lind. 1991. Polymerase chain reaction for detection of Mycoplasma genitalium in clinical samples. J. Clin. Microbiol. 29:46-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katoh, N. 1985. Significance of Ureaplasma urealyticum and Clostridium difficile in nongonococcal urethritis. Kansenshogaku Zasshi 59:687-700. (In Japanese.) [DOI] [PubMed]
- 10.kong, F., G. James, Z. Ma, S. Gordon, W. Bin, and G. L. Gilbert. 1999. Phylogenetic analysis of Ureaplasma urealyticum-support for the establishment of a new species, Ureaplasma parvum. Int. J. Syst. Bacteriol. 49:1879-1889. [DOI] [PubMed]
- 11.Maeda, S., M. Tamaki, M. Nakano, M. Uno, T. Deguchi, and Y. Kawada. 1998. Detection of Mycoplasma genitalium in patients with urethritis. J. Urol. 159:405-407. [DOI] [PubMed] [Google Scholar]
- 12.Mena, L., X. Wang, T. F. Mroczkowski, and D. H. Martin. 2002. Mycoplasma genitalium infections in asymptomatic men and men with urethritis attending a sexually transmitted diseases clinic in New Orleans. Clin. Infect Dis. 35:1167-1173. [DOI] [PubMed] [Google Scholar]
- 13.Palmer, H. M., C. B. Gilroy, P. M. Furr, and D. Taylor-Robinson. 1991. Development and evaluation of the polymerase chain reaction to detect Mycoplasma genitalium. FEMS Microbiol. Lett. 61:199-203. [DOI] [PubMed] [Google Scholar]
- 14.Povlsen, K., E. Bjornelius, P. Lidbrink, and I. Lind. 2002. Relationship of Ureaplasma urealyticum biovar 2 to nongonococcal urethritis. Eur. J. Clin. Microbiol. Infect. Dis. 21:97-101. [DOI] [PubMed] [Google Scholar]
- 15.Schwartz, M. A., and T. M. Hooton. 1998. Etiology of nongonococcal nonchlamydial urethritis. Dermatol. Clin. 16:727-733, xi. [DOI] [PubMed]
- 16.Shimada, M., T. Kotani, S. Ohtaki, H. Sameshima, T. Ikenoue, T. Sasaki, and T. Kenri. 1999. Clinicobacteriological studies on the nine cases with upper genital tract Mycoplasma hominis infection. Kansenshogaku Zasshi 73:646-651. (In Japanese.) [DOI] [PubMed]
- 17.Taylor-Robinson, D. 1996. The history of nongonococcal urethritis. Thomas Parran Award Lecture. Sex. Transm. Dis. 23:86-91. [DOI] [PubMed] [Google Scholar]
- 18.Taylor-Robinson, D., and P. M. Furr. 1997. Genital mycoplasma infections. Wien. Klin. Wochenschr. 109:578-583. [PubMed] [Google Scholar]
- 19.Totten, P. A., M. A. Schwartz, K. E. Sjostrom, G. E. Kenny, H. H. Handsfield, J. B. Weiss, and W. L. Whittington. 2001. Association of Mycoplasma genitalium with nongonococcal urethritis in heterosexual men. J. Infect. Dis. 183:269-276. [DOI] [PubMed] [Google Scholar]
- 20.Tully, J. G., D. Taylor-Robinson, R. M. Cole, and D. L. Rose. 1981. A newly discovered mycoplasma in the human urogenital tract. Lancet i:1288-1291. [DOI] [PubMed]
- 21.Tully, J. G., D. Taylor-Robinson, D. L. Rose, P. M. Furr, C. E. Graham, and M. F. Barile. 1986. Urogenital challenge of primate species with Mycoplasma genitalium and characteristics of infection induced in chimpanzees. J. Infect. Dis. 153:1046-1054. [DOI] [PubMed] [Google Scholar]
- 22.Yoshida, T., S. Maeda, T. Deguchi, and H. Ishiko. 2002. Phylogeny-based rapid identification of mycoplasmas and ureaplasmas from urethritis patients. J. Clin. Microbiol. 40:105-110. [DOI] [PMC free article] [PubMed] [Google Scholar]