Abstract
In 1997 and 1998, H3N2 influenza A viruses emerged among pigs in North America. Genetic analyses of the H3N2 isolates demonstrated that they had distinctly different genotypes. The most commonly isolated viruses in the United States have a triple-reassortant genotype, with the hemagglutinin, neuraminidase, and PB1 polymerase genes being of human influenza virus origin, the nucleoprotein, matrix, and nonstructural genes being of classical swine influenza virus origin, and the PA and PB2 polymerase genes being of avian influenza virus origin. In contrast, a wholly human H3N2 virus was isolated from a single baby pig in Ontario, Canada, in 1997, but it did not spread within the swine population. Genetic differences between this wholly human virus and the triple-reassortant viruses may affect their replication efficiencies in pigs. In the present study we compared the pathogenicities and replication kinetics of the wholly human virus and a triple-reassortant virus in 7-week-old pigs that were infected intranasally with 2 × 103 to 2 × 106 50% tissue culture infective doses of virus. Our results demonstrate that the wholly human virus replicated to significantly lower titers and that the onset of virus shedding was delayed compared to the replication titers and the time of onset of virus shedding in triple-reassortant viruses. In addition, infection with the triple-reassortant virus was associated with moderate to severe gross pathological and histological pulmonary lesions, while infection with the wholly human virus induced only mild pulmonary changes.
Infection of pigs with influenza A viruses is of substantial importance to the swine industry and to the epidemiology of human influenza (5, 19). At present, three main subtypes of influenza viruses are circulating in the swine population throughout the world: subtypes H1N1, H3N2, and H1N2 (18). In North America, influenza virus outbreaks among pigs have historically been due almost exclusively to infection with H1N1 viruses (3, 8, 20). Since 1997-1998, however, H3N2 viruses have emerged and spread widely within the swine population (13, 18, 37, 40). Triple-reassortant H3N2 viruses containing hemagglutinin (HA), neuraminidase (NA), and PB1 polymerase genes of human influenza virus origin, the matrix (M), nucleoprotein (NP), and nonstructural (NS) genes of classical swine influenza virus origin, and the PA and PB2 polymerase genes of avian influenza virus origin have been isolated widely throughout the United States. These viruses have been associated with outbreaks of respiratory disease in pigs of all ages and abortions in pregnant sows (13, 37, 40). In addition, reassortment between these viruses and classical H1N1 viruses has led to the subsequent development of H1N2 viruses, which have also spread throughout the swine population of the United States (11, 12). In contrast, an H3N2 virus in which all eight RNA segments were of human influenza virus origin was isolated from a single baby pig in 1997 on a farm in Ontario, Canada. This virus did not spread within the farm of origin and has not been recovered from pigs subsequent to its initial isolation (13).
Influenza A viruses have been isolated from various species, including humans, pigs, horses, birds, sea mammals, and mink (39). However, wild waterfowl serve as the reservoir from which all influenza viruses are thought to have emerged (39). Despite their common origin, influenza A viruses are generally restricted in host range. In particular, avian influenza viruses replicate poorly in humans and nonhuman primates and human influenza viruses do not replicate well in birds (1, 7, 8, 14, 17, 19, 38, 39). In contrast, swine influenza viruses have been shown repeatedly to infect humans as zoonotic infections, and conversely, human influenza viruses have also been isolated from pigs (19).
The host range restriction of influenza viruses is a polygenic trait (16, 27, 32, 33), but the HA gene is considered to be a particularly important determinant since HA is responsible for attachment of the virus to sialic acid receptors on the host cell surface (9, 34, 39). While human influenza viruses preferentially bind to sialic acid bound to galactose by α2,6 linkages, avian viruses preferentially recognize sialic acid bound by α2,3 linkages (9, 15, 25, 36). This is consistent with the fact that human tracheal epithelial cells predominantly express α2,6-linked receptors, whereas avian intestinal cells predominantly express α2,3-linked receptors (4, 10). However, avian and human influenza viruses can both infect pigs because porcine respiratory epithelial cells express both N-acetylneuraminic acid-α2,3-galactose and N-acetylneuraminic acid-α2,6-galactose (10). Pigs can therefore function as intermediate “mixing vessel” hosts in establishing new influenza virus lineages by supporting coinfection, replication, and reassortment among human, avian, and swine influenza viruses (19, 26, 27, 39).
Even though influenza viruses can cross species barriers and infect pigs, it is not known what properties are necessary to allow a virus to form a stable lineage and to spread efficiently within the pig population. In particular, a wholly human H3N2 virus, A/Swine/Ontario/00130/97 (H3N2) (Sw/ONT), initially crossed the species barrier to infect a pig in Canada in 1997, but it did not infect other pigs in the herd (13). Its disappearance may have been due to characteristics of the virus (i.e., its replication efficiency in pigs) or epidemiological factors (i.e., the availability of susceptible animals). In contrast, triple-reassortant H3N2 viruses containing genes from human, classical swine, and avian influenza viruses have spread throughout the swine population of the United States since 1998 (13, 37, 40). The critical factors that affect replication ability in pigs may include the overall constellation of genes present in the triple-reassortant viruses. However, we have also identified specific differences in the sequences of the HA genes of triple-reassortant viruses that may represent swine adaptation mutations (13). As an initial step in understanding the pathogenesis of these viruses in pigs, the present study was designed to define the specific characteristics of these viruses, including their infectivities, replication kinetics, and ability to induce pathological lesions under experimental infection conditions.
MATERIALS AND METHODS
Animals and influenza viruses.
Thirty-six domestic pigs were purchased from a commercial herd and shown to be serologically negative for prior exposure to porcine reproductive and respiratory syndrome virus, Mycoplasma hyopneumoniae, and H1 and H3 influenza viruses. The pigs were 6 weeks old at the start of the experiment and were maintained in accordance with the guidelines of the U.S. Department of Agriculture and the University of Wisconsin Research Animal Resources Center. Field isolates of A/Swine/Minnesota/593/99 (Sw/MN) (H3N2; generously provided by G. Anderson, ImmTech Biologics, LLC, Bucyrus, Kans.) and Sw/ONT (H3N2; generously provided by S. Carman, University of Guelph, Guelph, Ontario, Canada) were cultivated in Madin-Darby canine kidney (MDCK) cells. The viruses were grown in Eagle minimal essential medium (GIBCO/BRL) supplemented with 0.5% bovine serum albumin (GIBCO/BRL), penicillin-streptomycin (GIBCO/BRL), amphotericin B (Fungizone; GIBCO/BRL) and tolylsulfonyl phenylalanyl chloromethyl ketone-treated trypsin (1 μg/ml; Worthington Biochemical Corporation, Lakewood, N.J.). Sw/MN virus has previously been shown to contain the HA, NA, and PB1 genes of the human influenza virus lineage, the NP, M, and NS genes of the classical swine influenza virus lineage, and the PA and PB2 polymerase genes of the avian influenza virus lineage, whereas all eight RNA segments of Sw/ONT are of human influenza virus origin (13).
Experimental design.
The pigs were randomly assigned to nine cohorts of four pigs each and acclimated to biosafety level 2 housing for 1 week. On day 0 of the experiment, prior to inoculation, a short physical examination was performed to assess the general appearance of the animals (attitude, sneezing, and coughing were quantified on a scale of 0 to 3, with 0 indicating a clinically normal animal and 3 indicating a severely abnormal condition), their food intakes (with 0 indicating that the animal is eating and 1 indicating that the animal is off feed), respiratory rates (number of breaths per minute), and rectal temperatures. Thereafter, the pigs were weighed and preinoculation nasal swab (Dacron polyester; Hardwood Products Company, Guilford, Maine) samples for virus isolation and blood samples for serology and complete blood counts (CBCs) were collected. The pigs were then sedated by intramuscular injection of 0.5 mg of xylazine (Phoenix Pharmaceutical) per kg of body weight and 2 mg of tiletamine plus zolazepam (Telazol; Fort Dodge) per kg and inoculated intranasally with either Sw/MN or Sw/ONT at 2 × 103, 2 × 104, 2 × 105, or 2 × 106 50% tissue culture infective doses (TCID50s). Four pigs were inoculated with each dose of virus, and four pigs in a ninth cohort were mock inoculated to serve as uninfected controls. At daily intervals after inoculation, the animals were assessed clinically and nasal swab samples were obtained from each animal. The swabs were placed in 1 ml of viral transport medium containing phosphate-buffered saline (PBS), 0.5% bovine serum albumin, and the antimicrobials penicillin G, streptomycin, nystatin, and gentamicin and stored at −80°C until further analysis. Blood samples for serology were collected from all animals on day 7, and repeat CBCs were performed on days 3 and 7 for animals infected with 2 × 106 TCID50s of either virus and the uninfected control animals.
Evaluation of pathological lesions.
On day 7 the animals were euthanized by exsanguination following anesthesia by intramuscular injection of 4 mg of xylazine per kg and 4 mg of tiletamine-zolazepam per kg. The lungs were removed for postmortem analysis. Macroscopic pulmonary lesions were graded on the basis of the percentage of the surface area of each lung lobe exhibiting gross pathological evidence of tissue consolidation. The values for the individual lobes were averaged to obtain the overall percentage of lung tissue consolidation for each pig. Sections of lung tissue were obtained from a standardized location (left cranial lung lobe) and fixed in 4% buffered formalin, embedded in paraffin, cut in 6-μm-thick sections, and stained with hematoxylin-eosin. Each histological section was evaluated by a board-certified pathologist, without knowledge of the animal's infection status, for a variety of parameters of pulmonary pathology (atelectasis, epithelial necrosis, hemorrhage, airway plugging, epithelial hyperplasia, interstitial changes, leukocyte infiltration). Each individual parameter was given a score ranging from 0 to 3, with 0 indicating no abnormalities and 3 indicating severe abnormalities. These scores were added to obtain an overall score for each section.
Evaluation of virus shedding.
To determine the extent of virus shedding in nasal secretions, 10-fold serial dilutions of the viral transport medium containing the nasal swabs were inoculated (in duplicate) onto MDCK cells grown in 48-well tissue culture plates, and the plates were incubated for 72 h at 37°C with 5% CO2. The presence of virus replication in each well was confirmed by immunocytochemical staining with a mouse anti-NP monoclonal antibody (monoclonal antibody 68D2; kindly provided by M. McGregor and Y. Kawaoka, University of Wisconsin—Madison). All incubation steps were carried out at room temperature. Prior to staining, the cells were fixed for 30 min in 10% phosphate-buffered formalin and washed with PBS. Subsequently, the cells were incubated for 30 min with 0.3% H2O2-methanol and washed in PBS. Following fixation, the cells were incubated with serum blocking solution (nonimmune horse serum) for 30 min and subsequently incubated with a 1:1,000 dilution of anti-NP monoclonal antibody-containing ascitic fluid for 1 h. After the cells were washed in PBS, they were incubated with biotinylated secondary antibody for 30 min and washed in PBS; this was followed by 45 min of incubation with the avidin-biotin conjugate (Vectastain Elite kit; Vector Laboratories Inc., Burlingame, Calif.). Finally, the color was developed with the chromogen aminoethyl carbazole (Zymed Laboratories Inc., San Francisco, Calif.). Each staining procedure contained an isotype-matched negative control antibody and mock-infected negative control cells. The titer of virus in each specimen, expressed as the number of TCID50s per milliliter, was calculated by the method of Reed and Muench (24).
HA inhibition assay.
The levels of hemagglutination (HA)-inhibiting (HI) antibodies in serum samples were determined as described previously (22). Briefly, sera were incubated overnight at 37°C with 4 volumes of receptor-destroying enzyme (Denka Seiken Co., Tokyo, Japan) prepared from Vibrio cholerae. After inactivation of the receptor-destroying enzyme by incubation of the samples at 56°C for 60 min, twofold serial dilutions of sera were mixed with 4 HA units of Sw/MN or Sw/ONT. The assays were developed by adding 0.5% (vol/vol) chicken red blood cells, and the HI antibody titers were defined as the reciprocal of the highest dilution causing complete inhibition of agglutination.
Statistical analysis.
All values are expressed as the means ± standard errors of the means (SEMs). Data between treatment groups were analyzed by the t test or one-way analysis of variance. When F was significant (P ≤ 0.05), further differences between means were determined by Tukey's omega procedure. Means were considered significantly different at P values ≤0.05.
RESULTS AND DISCUSSION
Clinical responses to infection and CBC data.
Only mild changes in the clinical appearances of the animals were observed in this study. However, this is not unusual in our experience (unpublished results) when pigs are infected intranasally with influenza virus in a clean isolation environment. The general condition of the animals and food intake were unchanged in all treatment groups during the present experiment. In addition, no significant changes in the amount of sneezing and coughing between animals in the virus challenge groups and the control animals were observed (data not shown). Animals infected with Sw/MN showed a mild increase in respiratory rates compared to those of animals infected with Sw/ONT on days 6 and 7 of infection, but these changes were significant only on day 7 for the groups receiving 2 × 104 TCID50s of virus. On day 2 after infection, animals challenged with Sw/MN showed a mild increase in rectal temperature compared to the rectal temperatures of pigs infected with Sw/ONT, but these differences were significantly different only between pigs infected with the different viruses at 2 × 103 and 2 × 106 TCID50s. The changes in CBCs observed between pigs infected with the different viruses were minimal. There were no significant differences in the number of total white blood cells, neutrophils, lymphocytes, or monocytes/macrophages between and within the virus challenge groups and the controls on days 0, 3, and 7. Finally, during the course of the experiment, animals infected with Sw/MN had overall decreased rates of weight gains compared to those for the control animals and the Sw/ONT-infected pigs. These changes were significantly different for pigs challenged with 2 × 106 TCID50s of Sw/MN when the results were compared to those for the pigs challenged with the corresponding dose of Sw/ONT.
Macroscopic and histological pulmonary lesions.
The lungs of all control animals had no grossly visible signs of pulmonary disease at the time of necropsy. With the exception of one animal that received the highest challenge dose, pigs inoculated with Sw/ONT developed only very mild macroscopic pulmonary lesions. However, all pigs infected with Sw/MN at any dose exhibited obvious signs of pulmonary pathology. The percentages of overall lung tissue consolidation among the Sw/MN-inoculated pigs were higher compared to those among the Sw/ONT-inoculated animals, and these differences were statistically significant at 2 × 104 and 2 × 105 TCID50s (Table 1).
TABLE 1.
Responses of pigs to intranasal infection with Sw/ONT or Sw/MN
| Dose (TCID50) and virus | % Lung tissue consolidationa | Overall histopathology scorea,b | HI antibody titer in seruma |
|---|---|---|---|
| Controls | 0 | 2.25 ± 0.95 | 0 |
| 2 × 103 | |||
| Sw/MN | 4.66c ± 1.75 | 6.75cd ± 0.45 | 245 ± 135.74 |
| Sw/ONT | 0.095 ± 0.05 | 2.0d ± 0.85 | 0 |
| 2 × 104 | |||
| Sw/MN | 7.2c,d ± 1.2 | 5.0d ± 0.9 | 90c,d ± 10 |
| Sw/ONT | 0.29d ± 0.2 | 2.0d ± 0.7 | 0d |
| 2 × 105 | |||
| Sw/MN | 6.5c,d ± 0.7 | 7.25c,d ± 1.1 | 260c,d ± 20 |
| Sw/ONT | 0.27d ± 0.15 | 1.25d ± 0.25 | 7.5d ± 4.70 |
| 2 × 106 | |||
| Sw/MN | 14.54c ± 7.05 | 7.5c,d ± 1.1 | 280c,d ± 40 |
| Sw/ONT | 0.982 ± 0.7 | 3.5d ± 0.85 | 20d ± 20 |
Data are means ± SEMs; n = 4 animals per treatment group.
Cumulative mean scores for overall histological changes per treatment group, reflecting the sum of scores for atelectasis, epithelial necrosis, hemorrhage, airway plugging, epithelial hyperplasia, interstitial changes, and leukocyte infiltration. Scores for each criterion ranged from 0 to 3, with 0 indicating no abnormalities and 3 indicating severe abnormalities.
P ≤ 0.05 versus controls.
P ≤ 0.05 between viruses.
The degrees of macroscopic lung pathology were mirrored by the changes found histologically. Characteristic swine influenza lesions consisting of epithelial cell damage, airway plugging, and peribronchial and perivascular infiltration by inflammatory cells (35) were present in all animals infected with the Sw/MN triple reassortant. Pigs infected with the wholly human Sw/ONT virus generally showed only mild signs of pneumonia, and the overall histological scores were significantly lower at all challenge doses compared to those for the pigs challenged with the triple-reassortant virus (Table 1).
Virus shedding.
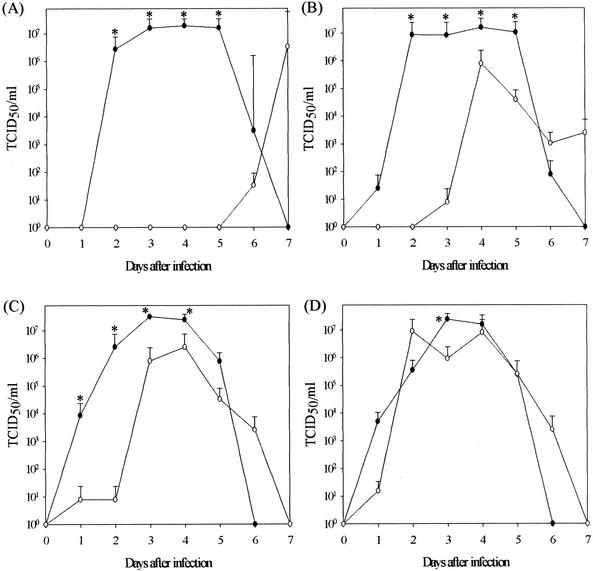
The kinetics and absolute levels of virus shedding were remarkably different for the two viruses (Fig. 1). All pigs inoculated with the Sw/MN triple reassortant shed detectable levels of virus for at least 3 days over the course of the experiment. The mean duration of shedding was 4.3 days. Animals infected with 2 × 105 and 2 × 106 TCID50s of Sw/MN generally began shedding virus on day 1, and the highest titers (2 × 107.5 TCID50s/ml) were reached by day 3, while pigs infected with the lower doses generally began shedding on day 2 or 3 and the highest titers were reached on day 4. In contrast, among animals inoculated with Sw/ONT, three pigs (one each receiving a challenge dose of 2 × 103 TCID50s, 2 × 105 TCID50s, and 2 × 106 TCID50s) never shed influenza virus at any detectable levels. In addition, the overall mean shedding time was only 2.7 days for the Sw/ONT-inoculated pigs. While three pigs infected with 2 × 106 and 2 × 105 TCID50s shed virus beginning on day 1, animals inoculated with the lower doses did not begin shedding until much later, day 4 to day 6.
FIG. 1.
Group means ± SEMs of nasal virus shedding from pigs infected with either Sw/MN (•) or Sw/ONT (○) from the day of inoculation (day 0) to day 7 after inoculation. (A) Pigs infected with 2 × 103 TCID50s of virus; (B) pigs infected with 2 × 104 TCID50s of virus; (C) pigs infected with 2 × 105 TCID50s of virus; (D) pigs infected with 2 × 106 TCID50s of virus. Control animals did not shed virus at any point during the experiment and are not represented in the graphs. *, P ≤ 0.05 between viruses.
The nasal virus titers were also higher in triple-reassortant Sw/MN-infected animals than in the pigs infected with wholly human Sw/ONT influenza virus. At the lowest challenge dose, nasal virus titers in the Sw/MN-infected pigs were significantly higher on days 2, 3, and 4. In pigs inoculated with 2 × 104 or 2 × 105 TCID50s of virus, nasal virus titers in the Sw/MN-infected pigs were significantly higher on days 2 to 5 and days 1 to 4, respectively. Finally, the pigs infected with Sw/MN at 2 × 106 TCID50s shed significantly more virus on day 3.
While the triple-reassortant H3N2 influenza virus was able to infect all pigs at each challenge dose, the wholly human virus did not. These findings are consistent with the observation made at the time of the initial isolation of Sw/ONT that this virus did not spread to other pigs in the population. This observation and our experimental results suggest that the wholly human Sw/ONT influenza virus is not as replication competent in pigs as Sw/MN is. Specifically, Sw/ONT may not enter host cells as readily as Sw/MN, and/or Sw/ONT may not be able to replicate efficiently enough and may not be shed in sufficient quantities to promote pig-to-pig transmission. Preliminary results of an in vitro study comparing the infectivities and replication kinetics of these two viruses (data not shown) suggest that there are substantial differences between the Sw/Ont and Sw/MN viruses during the initial steps of host cell infection and the initiation of replication. Such differences in virus entry and/or replication initiation at the cellular level could explain the differences in infectivities and virus shedding kinetics observed in our pig experiment. While the pigs in the cohorts infected with 2 × 103 TCID50s of either virus showed substantial differences in nasal virus titers and shedding kinetics, these differences became less obvious at higher initial infectious doses. These findings suggest that with an inoculum of 2 × 103 TCID50s of Sw/MN, most of the available respiratory cells became infected and an increase in the infectious dose did not substantially change nasal virus titers or shedding kinetics. In comparison, the number of respiratory cells that were infected with 2 × 103 TCID50s of Sw/ONT was likely much lower, and by increasing the infectious dose, the number of infected cells also increased proportionally until it reached levels comparable to those of Sw/MN.
HI antibody responses.
All pigs were serologically negative for swine influenza virus on day 0. With the exception of one pig in the group receiving the lowest challenge dose, virus-specific antibody titers, as measured by the HI assay, were present in the sera of all Sw/MN-infected pigs by day 7 (Table 1). In contrast, with the exception of three pigs (one in the group receiving 2 × 106 TCID50s, two in the group receiving 2 × 105 TCID50s), none of the pigs inoculated with Sw/ONT developed detectable antibody titers by day 7, and the titers in these three pigs were low (1:60 for the pig in the group receiving 2 × 106 TCID50s and 1:10 and 1:20 for the pigs in the group receiving 2 × 105 TCID50s). The Sw/ONT-inoculated pig that had the highest antibody titer (1:60) by day 7 also showed the most severe macroscopic and histological pulmonary lesions compared to the severities of the lesions for the rest of the animals infected with Sw/ONT. The mean antibody titers in the Sw/ONT- and Sw/MN-inoculated pigs were significantly different in the groups receiving 2 × 104, 2 × 105, and 2 × 106 TCID50s.
The pattern of antibody response generally mimics the differences in the extent of replication of Sw/MN and Sw/ONT. We suspect that the poor antibody responses in the pigs inoculated with Sw/ONT reflects limited antigen presentation due to delayed virus replication. It is possible, however, that the pigs infected with Sw/ONT would have seroconverted at a later time point if the time course of the experiment had been extended.
The aim of this study was to compare the pathogenicities, replication efficiencies, and virus shedding kinetics of two genetically different H3N2 influenza A viruses in pigs. The H3N2 influenza viruses used in this study were both initially isolated from naturally infected pigs. However, while the H3N2 influenza viruses with a triple-reassortant genotype were able to establish a stable lineage within the pig population of North America, the wholly human H3N2 influenza virus did not spread and has never been reisolated from pigs subsequent to the initial isolation in 1997. Human influenza viruses have been shown to readily infect pigs in experimental settings, and in some cases human viruses have established themselves in swine populations in nature (2, 21, 23, 29, 30). However, it is not known what properties are necessary to allow an influenza virus to establish a stable lineage within a new host population. Influenza viruses do not establish latent infections (39), so the successful survival of a particular virus strain within a population is likely dependent on the efficient infection of new hosts (6). This implies that in order for a virus to establish a stable lineage within a host species, the virus must infect and replicate efficiently in these hosts.
The results of our study indicate that triple-reassortant H3N2 virus Sw/MN infected and replicated efficiently in pigs, while the levels of replication and shedding of wholly human H3N2 virus Sw/ONT were much lower. The first step of influenza virus entry is dependent on the interaction of the viral HA with cellular sialic acid residues (16), and as such, the viral HA is also thought to be a major contributor to the host range of the virus (34). Previous work has shown that many of the triple-reassortant H3N2 viruses (including Sw/MN) contain 12 amino acid differences in their HA proteins that are unique to the swine viruses and that are not present in the most closely related human H3 virus HA proteins (including the Sw/ONT HA protein) (13, 40). It is possible that these amino acid differences in the HA protein could account for at least part of the observed differences in infectivity and replication efficiency. In addition, the triple-reassortant H3N2 swine viruses contain gene segments of the classical swine influenza virus lineage (NP, M, and NS genes) and the avian influenza virus lineage (PA and PB2 polymerase genes), while they maintain the human influenza virus HA, NA, and PB1 genes (13, 40), and the NP of influenza virus is considered to be a strong determinant of the influenza virus host range (27, 28, 31). Therefore, it is also possible that this overall constellation of genes is important for efficient replication of these viruses in the porcine host. Studies being conducted in our laboratory with reverse genetically engineered viruses are addressing which gene segment or combination of gene segments is responsible for the differences in replication efficiencies between Sw/ONT and Sw/MN observed in this study.
Acknowledgments
This work was supported by funding from the U.S. Department of Agriculture National Research Initiative Competitive Grants Program.
We thank Martha McGregor for technical assistance and Christian Hofer, Katie Grawe, Mike Bonds, Mary Krempaski, Jerry Brokish, and Claudia Hirsch for assistance during the animal experiments. We also thank Yoshihiro Kawaoka for critically reviewing the manuscript.
REFERENCES
- 1.Beare, A. S., and R. G. Webster. 1991. Replication of avian influenza viruses in humans. Arch. Virol. 119:37-42. [DOI] [PubMed] [Google Scholar]
- 2.Bikour, M. H., E. H. Frost, S. Deslandes, B. Talbot, J. M. Weber, and Y. Elazhary. 1995. Recent H3N2 swine influenza virus with haemagglutinin and nucleoprotein genes similar to 1975 human strains. J. Gen. Virol. 76:697-703. [DOI] [PubMed] [Google Scholar]
- 3.Chambers, T. M., V. S. Hinshaw, Y. Kawaoka, B. C. Easterday, and R. G. Webster. 1991. Influenza viral infection of swine in the United States 1988-1989. Arch. Virol. 116:261-265. [DOI] [PubMed] [Google Scholar]
- 4.Couceiro, J. N. S. S., J. C. Paulson, and L. G. Baum. 1993. Influenza virus strains selectively recognize sialyloligosaccharides on human respiratory epithelium: the role of the host cell in selection of hemagglutinin receptor specificity. Virus Res. 29:155-165. [DOI] [PubMed] [Google Scholar]
- 5.Easterday, B. C., and V. S. Hinshaw. 1992. Swine influenza, p. 349-357. In A. D. Leman, B. E. Straw, W. L. Mengeling, S. D. D'Allaire, and D. J. Taylor (ed.), Diseases of swine. Iowa State University Press, Ames.
- 6.Flint, S. J., L. W. Enquist, R. M. Krug, V. R. Racaniello, and A. M. Skalka. 2000. Patterns of infection: a delicate balance, p. 518-552. In Principles of virology, molecular biology, pathogenesis, and control. ASM Press, Washington, D.C.
- 7.Hinshaw, V. S., R. G. Webster, C. W. Naeve, and B. R. Murphy. 1983. Altered tissue tropism of human-avian reassortant influenza viruses. Virology 128:260-263. [DOI] [PubMed] [Google Scholar]
- 8.Hinshaw, V. S., W. J. Bean, R. G. Webster, and B. C. Easterday. 1978. The prevalence of influenza viruses in swine and the antigenic and genetic relatedness of influenza viruses from man and swine. Virology 84:51-62. [DOI] [PubMed] [Google Scholar]
- 9.Ito, T. 2000. Interspecies transmission and receptor recognition of influenza A viruses. Microbiol. Immunol. 44:423-430. [DOI] [PubMed] [Google Scholar]
- 10.Ito, T., J. N. S. S. Couceiro, S. Kelm, L. G. Baum, S. Krauss, M. R. Castrucci, I. Donatelli, H. Kida, J. C. Paulson, R. G. Webster, and Y. Kawaoka. 1998. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J. Virol. 72:7367-7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karasin, A. I., G. Anderson, and C. W. Olsen. 2000. Genetic characterization of an H1N2 influenza virus isolated from a pig in Indiana. J. Clin. Microbiol. 38:2453-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karasin, A. I., J. Landgraf, S. Swenson, G. Erickson, S. Goyal, M. Woodruff, G. Scherba, G. Anderson, and C. W. Olsen. 2002. Genetic characterization of H1N2 influenza viruses isolated from pigs throughout the United States. J. Clin. Microbiol. 40:1073-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karasin, A. I., M. M. Schutten, L. A. Cooper, C. B. Smith, K. Subbarao, G. A. Anderson, S. Carman, and C. W. Olsen. 2000. Genetic characterization of H3N2 influenza viruses isolated from pigs in North America, 1977-1999: evidence for wholly human and reassortant genotypes. Virus Res. 68:71-85. [DOI] [PubMed] [Google Scholar]
- 14.Kida, H., T. Ito, J. Yasuda, Y. Shimizu, C. Itakura, K. F. Shortridge, Y. Kawaoka, and R. G. Webster. 1994. Potential for transmission of avian influenza viruses to pigs. J. Gen. Virol. 75:2183-2188. [DOI] [PubMed] [Google Scholar]
- 15.Matrosovich, M. N., A. S. Gambaryan, S. Teneberg, V. E. Piskarev, S. S. Yamnikova, D. K. Lvov, J. S. Robertson, and K. A. Karisson. 1997. Avian influenza A viruses differ from human viruses by recognition of sialyloligosaccharides and gangliosides and by a higher conservation of the HA receptor-binding site. Virology 233:224-234. [DOI] [PubMed] [Google Scholar]
- 16.Murphy, B. R., and R. G. Webster. 1996. Orthomyxoviruses, p. 1397-1445. In N. Fields, D. M. Knipe, P. M. Howley, et al. (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 17.Murphy, B. R., V. S. Hinshaw, D. L. Sly, W. T. London, N. T. Hosier, F. T. Wood, R. G. Webster, and R. M. Chanock. 1982. Virulence of avian influenza A viruses for squirrel monkeys. Infect. Immun. 37:103-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olsen, C. W. 2002. The emergence of novel swine influenza viruses in North America. Virus Res. 85:199-210. [DOI] [PubMed] [Google Scholar]
- 19.Olsen, C. W. Influenza in pigs and their role as the intermediate host. In K. G. Nicholoson, R. G. Webster, A. J. Hay, and N. C. Cox (ed.), Textbook of influenza, 2nd ed., in press. Blackwell Science, Oxford, United Kingdom.
- 20.Olsen, C. W., S. Carey, L. Hinshaw, and A. I. Karasin. 2000. Virologic and serologic surveillance for human, swine and avian influenza virus infections among pigs in the north-central United States. Arch. Virol. 145:1399-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ottis, K., L. Sidoli, P. A. Bachman, R. G. Webster, and M. M. Kaplan. 1982. Human influenza viruses in pigs: isolation of a H3N2 strain antigenically related to A/England/42/72 and evidence for continuous circulation of human viruses in the pig population. Arch. Virol. 73:103-108. [DOI] [PubMed] [Google Scholar]
- 22.Palmer, D. F., W. R. Dowdle, M. T. Coleman, and G. C. Schild. 1975. Advanced laboratory techniques for influenza diagnosis. U.S. Department of Health, Education and Welfare Immunology Series. U.S. Department of Health, Education and Welfare, Washington, D.C.
- 23.Peiris, J. S. M., Y. Guan, D. Markwell, P. Ghose, R. G. Webster, and K. F. Shortridge. 2001. Cocirculation of avian H9N2 and contemporary “human” H3N2 influenza A viruses in pigs in southeastern China: potential for genetic reassortment? J. Virol. 75:9679-9686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty per cent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 25.Rogers, G. N., and J. C. Paulson. 1983. Receptor determinants of human and animal influenza virus isolates: differences in receptor specificity of the H3 hemagglutinin based on species of origin. Virology 127:361-373. [DOI] [PubMed] [Google Scholar]
- 26.Scholtissek, C., and E. Naylor. 1988. Fish farming and influenza pandemics. Nature 331:215.. [DOI] [PubMed] [Google Scholar]
- 27.Scholtissek, C., H. Burger, O. Kistner, and K. F. Shortridge. 1985. The nucleoprotein as a possible major factor in determining host specificity of influenza H3N2 viruses. Virology 147:287-294. [DOI] [PubMed] [Google Scholar]
- 28.Schultz, U., W. M. Fitch, S. Ludwig, J. Mandler, and C. Scholtissek. 1991. Evolution of pig influenza virus. Virology 183:61-73. [DOI] [PubMed] [Google Scholar]
- 29.Shortridge, K. F., A. Cherry, and A. P. Kendal. 1979. Further studies on the antigenic properties of H3N2 strains of influenza A viruses isolated from swine in Southeast Asia. J. Gen. Virol. 44:251-254. [DOI] [PubMed] [Google Scholar]
- 30.Shortridge, K. F., R. G. Webster, W. K. Butterfield, and C. H. Campbell. 1977. Persistence of Hong Kong influenza virus variants in pigs. Science 196:1454-1455. [DOI] [PubMed] [Google Scholar]
- 31.Shu, L. L., W. J. Bean, and R. G. Webster. 1993. Analysis of the evolution and variation of the human influenza A virus nucleoprotein gene from 1933 to 1990. J. Virol. 67:2723-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Snyder, M. H., M. L. Clements, D. De Borde, H. F. Maassab, and B. R. Murphy. 1987. Attenuation of wild-type human influenza A virus by acquisition of the PA polymerase and matrix protein genes of influenza A/Ann Arbor/6/60 cold-adapted donor virus. J. Clin. Microbiol. 22:719-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Subbarao, E. K., W. London, and B. R. Murphy. 1993. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J. Virol. 67:1761-1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suzuki, Y., I. Toshihiro, T. Suzuki, R. E. Holland, T. M. Chambers, M. Kiso, H. Ishida, and Y. Kawaoka. 2000. Sialic acid species as a determinant of the host range of influenza A viruses. J. Virol. 74:11825-11831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thacker, E. L., B. J. Thacker, and B. H. Janke. 2001. Interaction between Mycoplasma hyopneumoniae and swine influenza virus. J. Clin. Microbiol. 39:2525-2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vines, A., K. Wells, M. Matrosovich, M. R. Castrucci, T. Ito, and Y. Kawaoka. 1998. The role of influenza A virus hemagglutinin residues 226 and 228 in receptor specificity and host range restriction. J. Virol. 72:7626-7631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Webby, R. J., S. L. Swenson, S. L. Krauss, P. J. Gerrish, S. M. Goyal, and R. G. Webster. 2000. Evolution of swine H3N2 influenza viruses in the United States. J. Virol. 74:8243-8251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Webster, R. G., M. Yakhno, V. S. Hinshaw, W. J. Bean, and K. G. Murti. 1978. Intestinal influenza: replication and characterization of influenza viruses in ducks. Virology 842:268-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Webster, R. G., W. J. Bean, O. T. Gorman, T. M. Chambers, and Y. Kawaoka. 1992. Evolution and ecology of influenza A viruses. Microbiol. Rev. 56:152-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou, N. N., D. A. Senne, J. S. Landgraf, S. L. Swenson, G. Erickson, K. Rossow, L. Liu, K.-J. Yoon, S. Krauss, and R. G. Webster. 1999. Genetic reassortment of avian, swine, and human influenza A viruses in American pigs. J. Virol. 73:8851-8856. [DOI] [PMC free article] [PubMed] [Google Scholar]