Abstract
An electrophoretically purified allophycocyanin–linker complex, AP⋅LC7.8, from phycobilisomes of Mastigocladus laminosus has been crystallized in the orthorhombic space group P212121. Cryocrystallographic x-ray measurements enabled the structural analysis of the complex at a resolution of 2.2 Å. The asymmetric unit contains two side-to-side associated “trimeric” (αβ)3 allophycocyanin complexes comprising the linker polypeptide in a defined orientation inside the trimer. The linker representing a protein fold related to the prosegment of procarboxypeptidase A is in contact with only two of the three β-subunits and directly interacts with the corresponding chromophores of these proteins. In addition to a modulation of the chromophores’ spectral properties, the linker polypeptide attracts the αβ-subcomplexes, thereby bringing the β-chromophores closer together. These results will enable interpretations of energy-transfer mechanisms within phycobiliproteins.
Keywords: cyanobacteria, linker polypeptides, crystallization, photosynthesis, energy transfer
Phycobilisomes are supramolecular light-harvesting protein–pigment complexes of cyanobacteria and Rhodophyceae with Mrs ranging from 5 million to 20 million (1). The complexes are anchored to the cytoplasmic surface and localized above the rows of dimeric or tetrameric photosystem II particles of the thylakoid membrane (2). The phycobilisomes have been characterized in numerous electron microscope (2), spectroscopic (3), biochemical (4), genetic (5), and physiological (6) studies. Two structural and functional domains, a centrally situated allophycocyanin core, and a species-specific number of rods radiating from this core are typical for all phycobilisomes. The rods are composed of phycocyanin alone or, in addition, phycoerythrin and, in some cases, phycoerythrocyanin (7). The core consists of a bi- or tricylindrical central high-molecular-weight protein complex (APCM) comprising allophycocyanin (AP) and two copies of the anchor polypeptide (linker protein LCM) and serves as backbone of the phycobilisome and as structural bridge to photosystem II (8, 9). At both faces of APCM, trimeric complexes of allophycocyanin B and allophycocyanin AP⋅LC7.8, containing the low-molecular-weight linker polypeptide, LC7.8 (formerly LC10, LC8.9), are associated (9–11). The three-dimensional structure, molecular composition, spectral properties, and adaptability of trimeric allophycocyanins have been characterized (11–15).
In their native environment in phycobilisomes, all phycobiliprotein complexes are associated with mostly uncolored linker polypeptides that are intrinsic parts of the subcomplexes. The strict sequential assembly of trimeric phycobiliprotein complexes to high-molecular-weight structures is only achieved by the participation of these proteins (7, 16). The linker molecules are strongly basic, and although they contain a significant amount of hydrophilic amino acids, they have a strong tendency to aggregate in aqueous solutions. Distinct interactions between linkers and phycobiliproteins are proposed to be involved in the folding of the tertiary structure of the linker polypeptides as well as in the modulation of the spectral and molecular properties of the phycobiliproteins (1, 4, 7, 8, 17).
The previously determined crystal structures of the main phycobiliprotein classes allophycocyanin (12), phycocyanin (18–21), phycoerythrocyanin (22), and phycoerythrin (23–25) either do not contain linker molecules or show only their general location because of disorder. The phycobiliproteins with and without linker polypeptides crystallized mostly in trigonal or hexagonal space groups occupying threefold axis of symmetry, thus leading to statistical disorder of the linker moiety (24). Therefore, neither structural analyses of the linkers nor the elucidation of the linker-induced modulation of the polypeptides and chromophores were possible. Here we describe the crystallization and structure determination of a phycobiliprotein–linker complex in an orthorhombic space group where both phycobiliprotein and linker moieties are ordered, allowing a detailed structural analysis and providing insight into the complex molecular events within the naturally assembled phycobilisomes.
MATERIALS AND METHODS
Strain, Growth Conditions and Preparation of AP⋅LC7.8.
Cells of Mastigocladus laminosus (Fischerella sp. PCC6703) were cultivated under conditions where only traces of phycoerythrocyanin but the maximum amount of AP⋅LC7.8 are incorporated into the phycobilisomes (15). The purification and analysis of AP⋅LC7.8 has been described in detail (9, 11, 15). For storage of the sample and the following crystallization experiments, the isolation buffer has been changed to ammonium/boric acid (100 mM:100 mM, pH 7.9). At this buffer condition and 4°C, the concentrated (15 mg/ml) biliprotein–linker complex was stable for at least 3 months.
Crystallization Conditions and Cryocrystallography.
Crystallization was performed in hanging drops of 5–20 μl by vapor diffusion in Linbro plates (Hampton Research, Riverside, CA) at different temperatures. The growth of homogenous crystals in the orthorhombic space group has been reproducibly observed only at 27°C. Two crystallization conditions were successful, and the resulting crystals have been analyzed. Protein solution (7.5 μl; 15 mg/ml) was mixed with 7.5 μl of precipitation solution containing 640 mM Li2SO4 or 360 mM MgSO4, 8% or 6% (wt/vol) polyethyleneglycol 6000 and 24 mM or 18 mM Tris/boric acid (pH 7.9). The samples were concentrated against 0.3 ml of the precipitation solution at 27°C for 72 hr. For cryomeasurements, the crystals were stored at 4°C before transferring stepwise into the cryoprotectant [300 mM Li2SO4 and 40% (wt/vol) polyethyleneglycol 1000]. After the last step, storage at room temperature was possible.
Data Collection and Crystallographic Evaluation.
The best data set of the AP⋅LC7.8 complex (diffraction limit 2.2 Å) was collected on the wiggler beamline BW6 at DORIS (Deutsches Elektronensynchrotron, Hamburg, Germany) with one flash-frozen crystal at 100 K using a Cryostream cryosystem (Oxford Cryosystems, Oxford) and a 345-mm image plate detector (Mar Research, Hamburg, Germany). The data were collected in two parts: high-resolution data were collected to 2.2 Å in frames of 0.5° through a continuous angular range of 90° followed by the collection of low-resolution data to 3.5 Å in frames of 1.2°. The data were processed with mosflm version 5.51 (26) and scaled with scala version 2.43 (27).
Patterson searches were computed by using amore (28) and a trimeric linker-free allophycocyanin model from Spirulina platensis (12). The search model was generated from the αβ-monomer structure of S. platensis by applying the corresponding crystallographic symmetry operators. For each of the two molecules in the asymmetric unit, three distinct solutions with correlation coefficients 13.7% above the highest noise peak were found, each of which differed by a 120° rotation around the trimer axis. The correlation coefficient containing both asymmetric molecules was 56.0% above the noise. These results were inspected and the complete model including the linker molecules built by using main (29). In a first step, residues that differed between the search model and the published M. laminosus sequence (30) were exchanged. At this stage the crystallographic R factor was 37.33 (Rfree = 36.12). The linker molecule was initially introduced as poly-alanine model by using both the Fobs − Fcalc difference electron density and a 2Fobs − Fcalc-based density resulting from cyclic twofold averaging about the local twofold in the asymmetric unit and solvent flattening by using main (29). After addition of the side chains (31), water molecules were introduced by using arp (32). The entire process involved several cycles of model inspection followed by positional and B factor refinement. In some instances, simulated annealing was introduced with xplor version 3.851 (33). The model inspection concentrated exclusively on the linker model in the beginning and included the entire molecular model of the complex as well as the water molecules in the end. Initially the refinement was carried out with noncrystallographic symmetry restraints between either all six αβ-monomers or the two molecules of the asymmetric unit, but inspection of the relative change in R and Rfree with and without noncrystallographic symmetry restraints led us to abandon these restraints. During the final refinement cycles, bulk solvent correction was applied. A direct space search for alternate orientations of the linker within the trimer using xplor proved that only one unique orientation of the linker molecule within each trimer occurs.
The model quality was checked by using procheck (34) and sfcheck (27), and the asymmetric unit was defined as the two molecules that show the highest number of contacts by using contact (27). Except for Thr-74 of the β-subunits, which is very well defined in the electron density and makes contacts to the β-chromophore, all residues are in the most favored (95.6%) or additionally allowed (4.1%) regions in the Ramachandran Plot (35). Data collection and refinement are summarized in Table 1. The rotation of the monomers in the trimer on binding of the linker were initially observed in Cα plots comparing the symmetric allophycocyanin model (12) with the two AP⋅LC7.8 molecules in the asymmetric unit.
Table 1.
Summary of crystallographic analysis
| Measurement | Value |
|---|---|
| Data collection | |
| Resolution limit, Å | 2.2 |
| Total reflections | 637,968 |
| Unique reflections | 180,426 |
| Completeness, % | 96.6 (96.6) |
| Rsymm,*† % | 6.9 (31.8) |
| Refinement | |
| Resolution, Å | 25–2.2 |
| Number of non-H protein Atoms | 16,184 |
| Number of water molecules‡ | 1,364 |
| Rcryst,§¶ % | 21.11 (30.74) |
| Rfree,§ % | 25.68 (32.46) |
| Average B, Å2 | 34.6 |
| rms deviation bond length, Å | 0.009 |
| rms deviation bond angles | 1.806 |
| rms deviation bonded B factors, Å2 | 2.586 |
Values in parentheses correspond to the last resolution shell from 2.28 to 2.20 Å.
Rsymm = Σ|Ihi − 〈Ih〉|/Σ〈Ih〉
This count includes the 20 non-hydrogen atoms of the four B(OH)4− molecules.
Values in parentheses correspond to the last resolution shell from 2.24–2.20 Å.
Rcryst = Σ|Foh − Fch|/ΣFoh, where Foh and Fch are the observed and calculated structure factor amplitudes for reflection h.
Value of Rcryst for 5% of reflections selected in 20 thin shells and excluded from refinement. The Rfree selection was based on thin shells rather than a random selection to reduce the correlation between the Rfree and refinement reflections caused by NCS.
RESULTS
Crystallization, Data Collection, and Assembly of the Unit Cell.
Crystallization of AP⋅LC7.8 in an orthorhombic space group preferentially succeeds at higher temperatures (25°–27°C) in the presence of boric acid, high salt concentrations, and polyethylene glycol. The best crystals grew when the solutions of polyethylene glycol and salt separated into two phases. At similar conditions, regular crystallization of linker-free allophycocyanin complexes has never been observed, thus demonstrating the influence of the linker on the physical properties of the phycobiliproteins. The crystallized complexes contain the linkers in their natural amount and functional activity, as shown by spectral analyses and electrophoreses. Neither the starting sample nor the crystals show linker-free allophycocyanin (results not shown).
The P212121 crystals have unit-cell dimensions of a = 179.28 Å, b = 156.51 Å, c = 140.45 Å and α = β = γ = 90°. The problems of an initial diffraction limit of 3.5–3.2 Å and crystal degradation during measurement at 18°C could be solved by cryocrystallography. A soaking procedure at 4°C, stepwise transferal of the crystals from their mother liquor (harvesting buffer) to the cryobuffer, data collection at 100 K, and the use of more intense synchrotron radiation significantly improved the resolution to 2.2 Å. During the soaking procedure and flash freezing, the cell constants decreased to a = 176.12 Å, b = 151.90 Å, c = 137.85 Å and α = β = γ = 90°.
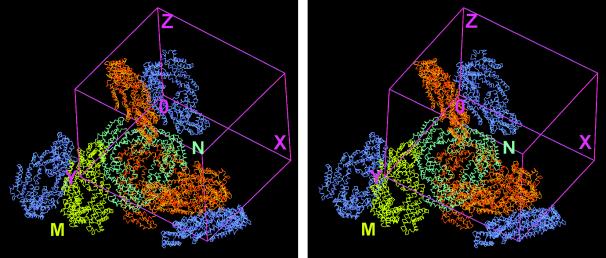
The solvent content is 70% in the frozen crystal (Matthews parameter 4.07). The assembly of the unit cell of the crystal including all eight trimers is shown in Fig. 1. The asymmetric unit consists of two trimeric complexes, referred to in the text as molecule M and N. The different interactions between the two molecules and their symmetry-mates within the unit cell can be described as three almost orthogonal side-to-side contacts as well as a relatively loose, slightly staggered back-to-back contact. The closest association—one of the side-to-side contacts—involves both α- and β-subunits. This contact is located on a noncrystallographic diad, where each trimer is tilted to opposite directions of the plane perpendicular to this twofold axis, resulting in an approximate angle of 90° between the two complexes. In principle, the complex composition of the crystal cell can be attributed to the lateral association of distinct α- and β-subunits of the allophycocyanin complexes. A tetrahedrally shaped electron density interpreted as B(OH)4− was observed at the positively charged side chain of Arg-24 of the two α-subunits in each trimer not involved in the contacts between the two molecules in the asymmetric unit.
Figure 1.
Stereo view of the arrangement of AP⋅LC7.8 within the unit cell of the orthorhombic P212121 crystals. The two trimeric complexes building the asymmetric unit are shown in yellow and cyan, respectively. Their crystallographic symmetry-mates creating the entire unit cell are in orange and blue. The picture demonstrates that there are two types of contact between the two sets of molecules that can form the asymmetric unit, either side to side or back to back. The figure was prepared in main (29).
Structure of the Linker Polypeptide and Its Interaction with the Trimer.
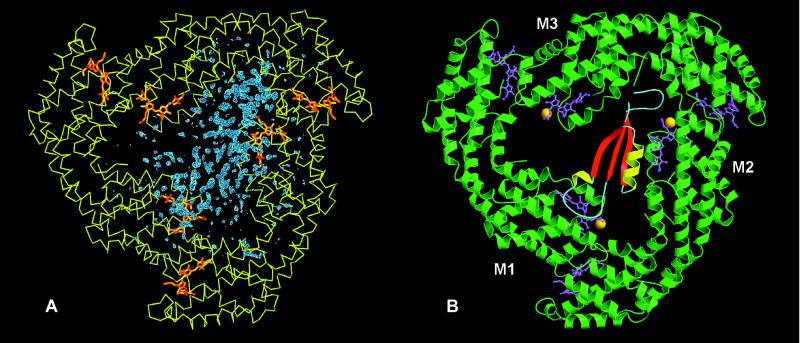
The ordered orientation of the AP⋅LC7.8 complexes within the crystal cell, as confirmed by direct space searches for alternative linker orientations was a prerequisite for a successful structural analysis of the intrinsic linker polypeptide (24). Fig. 2A shows the Fobs − Fcalc electron-density map contoured at 2.5σ for molecule M resulting from the Patterson search. This density map clearly shows the molecular boundaries of the linker polypeptide in contact with two of the three β-subunits.
Figure 2.
Representation of the entire AP⋅LC7.8 complex. (A) Fobs − Fcalc Fourier map phased with the Patterson solution after exchange of those residues that differ between allophycocyanin from M. laminosus and the replacement model from S. platensis (12). Blue, Fobs − Fcalc difference electron density; yellow, Cα plot of the α and β subunits; orange, chromophores. (B) Ribbon plot. The α/β-subunits are represented in green and the chromophores in pink. The methylated Asn-19 of the β-subunit is present in all three monomers and is shown as an orange ball. The secondary structural elements of the linker polypeptide are represented in red, yellow, and blue. The linker polypeptide interacts directly with the chromophores of only two of the three β-chromophores. Fig. 2A was prepared in main (29) and Fig. 2B was made with molscript (48) and rendered with raster3d (49).
The linker polypeptide has an elongated shape and consists of a three-stranded β-sheet (β1, Leu-3–Leu-9; β2, Tyr-26–Pro-32; β3, Lys-49–Leu-55), two α-helices (α1, Leu-22–Thr-25; α2, Tyr-33–Met-46), one of which has only about one turn, and the connecting random coil segments, as determined by dssp (36). A search in the database for similar secondary structural elements by using dali (37) revealed several hits with a good alignment to single domains of larger structures, e.g., topoisomerase II (38) or the prosegment of procarboxypeptidase A, which has two additional helices (39).
The linker polypeptide is predominantly located between two distinct β-subunits and directly interacts with the corresponding chromophores of these subunits (Fig. 2). Despite the generally acidic character of the trimer and the basic character of the linker polypeptide, no clear clustering of charged, polar, or hydrophilic residues is found at the protein–protein interface. The surface area of the linker buried in the complex is 45.3%.
Linker-Induced Structural Changes of the Complex.
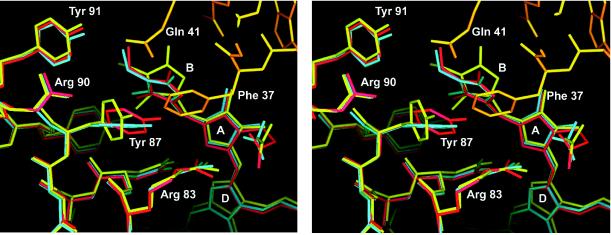
The specific interaction of the linker polypeptide with the β chromophores can be described as a tight contact including polar and hydrophobic interactions between the N-terminal residues of the long linker helix and monomer 2 of the trimer (M2, N2). Insertion of the linker Phe-37 between Tyr-87 and chromophore pyrrole ring B of monomer 2 results in a displacement of both Tyr-87 and pyrrole ring B to opposite directions, which leads to a change in conformation of this chromophore as well as a break of the stacking interaction between Tyr-87 and pyrrole ring B observed for the other two monomers in the complex structure (Fig. 3).
Figure 3.
Stereo representation of the interaction between the linker polypeptide and monomer 2. The Cα-aligned structures are shown for the symmetric, linker-free allophycocyanin (blue), monomer 3, which has no interaction with the linker polypeptide (red), and monomer 2 (yellow), which interacts with the N terminus of the long linker α-helix (orange). Water molecules are omitted for clarity. Phe-37 of the linker inserts between Tyr-87 and the pyrrole ring B of the corresponding β-subunit displacing both to opposite directions and disrupting the stacking interaction between the two aromatic ring systems, which is seen at the other two β-chromophores of the linker containing trimer. This figure was prepared in main (29).
The second linker–chromophore interaction is characterized almost exclusively by charged and polar residues in the loop between β strand 1 and the short α-helix of the linker polypeptide, which is located on top of the otherwise solvent exposed side of the chromophore of monomer 1 (M1, N1). No distinct conformational change of this chromophore is observed. However, the presence of this loop certainly changes the surrounding of this chromophore with respect to either the linker-free allophycocyanin or the third chromophore (monomer 3), which has no linker contacts. These two clearly different interactions between the linker polypeptide and two of the three β-chromophores, along with the fact that the third β-chromophore remains solvent-exposed, break the threefold symmetry between the three β-chromophores as observed in the crystal structure of linker-free allophycocyanin (12).
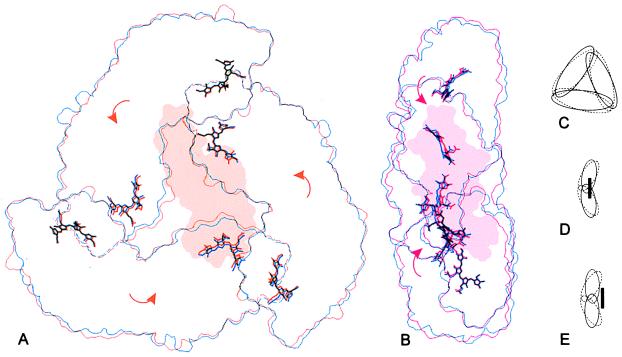
In more general terms, the trimer is contracted toward the pseudo-threefold axis and its hourglass-like shape becomes more flattened on binding of the linker polypeptide (Fig. 4). This change is achieved by a slight rotation of the three monomers bringing mainly the β-chromophores closer to each other. The distance between the β- and their nearest α-chromophores remains unchanged within the error of the crystallographic structural analysis. Interestingly, the two trimers within the asymmetric unit show distinctly different positions of the linker polypeptide with respect to their corresponding α- and β-subunits. The structure of the two linker molecules, i.e., the positions of the Cα atoms, is very similar (Fig. 5). In molecule M, the linker is inserted deeper in the interior of the trimer, resulting in a stronger contraction of the chromophores. Molecule N is located about 3 Å closer to the edge of the trimer, forcing a stronger displacement of the chromophores in that direction and less contraction of the chromophores toward the pseudo-threefold axis (Figs. 4 and 5). These two different positions of the linker polypeptides, together with the relatively high B factors of the linkers in comparison to the α- and β-subunits and water molecules (average B factors: LC_M, 56.1 Å2; LC_N, 74.4 Å2; trimer M, 34.1 Å2; trimer N, 30.0 Å2; chromophores, 27.8 Å2; waters, 40.6 Å2), indicate some variability in the positioning of the linker polypeptides with respect to their associated α- and β-subunits within the complex.
Figure 4.
Comparison of the molecular outlines of the symmetric, linker-free allophycocyanin (12) and the linker-containing complex (A and B) as well as an exaggerated schematic representation (C–E): The symmetric allophycocyanin (blue or dashed lines) is compared with molecule M (orange) in front view (A) and molecule N (red) in side view (B). On binding of the linker, the entire complex collapses toward the pseudo-threefold axis (C) and the α/β-ring becomes flattened (D and E) because of a rotation of the monomers as indicated by the arrows. In molecule M the linker inserts further into the trimer (D), resulting in a larger movement of the chromophores toward the pseudo-threefold axis, but with a slighter displacement of the chromophores to the right as compared with molecule N (E). These outlines were prepared by using an edge-detection filter and grasp surface representations of each molecule (50).
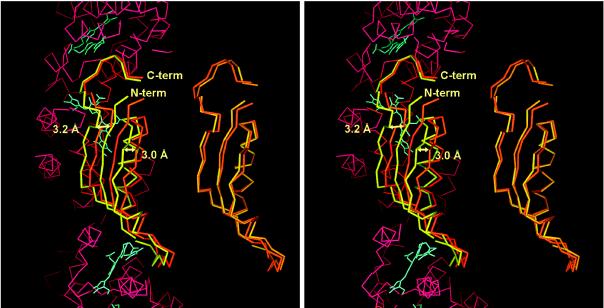
Figure 5.
Comparison of the structure of the linker polypeptides of the two molecules in the asymmetric unit. On the right the linker Cα-atoms are aligned directly by an rms procedure. On the left, the linkers are shown after rms alignment of the Cα-atoms of the two trimers in the asymmetric unit. The two linker molecules are very similar. However, the linker in molecule M (yellow) is shifted by about 3 Å to the left with respect to the molecule N linker (orange). Part of the Cα-trace, as well as the chromophores of molecule N, is shown as reference in red and blue, respectively. The figure was prepared in main (29).
DISCUSSION
Arrangement of the Unit Cell.
The occurrence of a hitherto uncharacterized microheterogeneity of the α- and β-subunits within nearly all allophycocyanin core complexes of various cyanobacteria and even rhodophytes reflects the multiple functions and recognition mechanisms that arise from the face-to-face, side-to-side, and side-to-face interactions of the core constituents (4, 9, 11, 15, 40, 41). Because of the heterogeneous subunit composition, especially in the α-subunits, the naturally occurring side-to-side association, and the excellent stability of AP⋅LC7.8 in a wide range of experimental conditions, this molecule has been selected for crystallization of a biliprotein–linker complex in a space group deviating from the threefold symmetry.
The close lateral contacts involving distinct α- and β-subunits of AP⋅LC7.8 are primarily responsible for the architecture of the unit cell. The ordered location of the linker within the trimer may be a consequence of the asymmetric crystal packing. The linker is, however, not directly involved in crystal contacts. A biological significance of the side-to-side contacts cannot be deduced, because the angle of approximately 90° between the side-to-side-associated AP⋅LC7.8 trimers within the crystals is incompatible with results from electron microscopy studies of the phycobilisome core (2, 10, 42). The weak back-to-back contacts seen in the AP⋅LC7.8 crystals, however, may be comparable to the native interactions of neighboring cores stabilizing the phycobilisome arrangement within rows (2, 15, 43).
Interactions of the Linker Within the Allophycocyanin Trimer.
The linker polypeptide is preferentially located between only two monomers of the trimeric complex and binds via multiple charged, polar, and hydrophobic contacts to the protein chains of these monomers and especially their β-chromophores. This result is not consistent with a previously postulated threefold symmetric shape of the LC7.8 coordinating the three αβ-monomers by hydrophobic interactions (13). The linker fits into the trimeric allophycocyanin readily and causes only very small deformations of its structure (Figs. 2 and 4). Other studies of the reconstitution behavior of allophycocyanin with and without linker revealed that the trimers assemble without participation of the linker, whereas the folding of the linker depends on the presence of at least monomeric subcomplexes (44, 45).
Concerning the chromophore modulation, it is clearly shown that the linker differentiates the energetically coupled αβ-chromophore pairs into three species. Interestingly, the location and orientation of the α chromophores in relation to the β chromophores is not significantly affected by the association of the linker polypeptide. This fact gives evidence for the continued strong coupling of the αβ-chromophores, which is confirmed by energy-transfer studies (3, 14). The influence of the linker polypeptide on the spectroscopic properties of allophycocyanin can be exemplified by the comparison of its absorbance with and without linker. The enhanced near-UV absorbance ratio of the linker-free allophycocyanin originates from a statistical proportion of weakly stabilized chromophores (46). In addition to the general coordination and stabilization effect, the influence of the linker is expressed by the bathochromic absorbance shift to 653 nm and the decreased shoulder near 620 nm of AP⋅LC7.8. Resonance coherent anti-Stokes raman scattering (CARS) spectroscopy demonstrated the effect of the aggregation states (monomer and trimer) on the geometry of the chromophores of both subunits and in addition, geometrical variations of chromophores depending on the linker association (47). The results of the present study prove that the bathochromic shifts are exclusively caused by the charged and polar contacts of the linker with the β-chromophore of monomer 1 as well as the conformational change of the β-chromophore of monomer 2. The entirety of the linker-induced effects, modulation and stabilization as well as attraction of the chromophores, optimizes the directed energy transfer within the molecule and between the constituents of the core.
The Protein Fold of the Linker.
The flat and elongated shape of the linker polypeptide as well as the small core formed by its secondary structural elements raises the question whether this peptide adopts a stable fold in solution. A definite answer must await structural studies of isolated AP⋅LC7.8 molecules, but several experiments indicate that the shown structure is stabilized in complex with allophycocyanin: the presence of at least monomeric subcomplexes is required for successful refolding of the linker polypeptide (44, 45). The linker shows a general high tendency toward aggregation even though its surface properties are typical for soluble proteins. The extended coil segments at both termini and the large loop between the first β-strand and the short α-helix have no tertiary contacts to the core of the structure.
Functional Aspects of the Linker–Allophycocyanin Association in the Core Environment.
One noteworthy result of this work is the different penetration depth of the LC7.8 into the trimers, resulting in a varied structure of the allophycocyanin complexes. The high B factors of the linkers and the different structures of chemically identical trimers may be indicative of a dynamic fluctuation between distinct energetic states, which also are realized in the phycobilisome core. The adjustment of the energetic state will probably be influenced by the anchor polypeptide, LCM, coordinating the assembly of the complexes and the energy distribution within the core. The complete integration of the linkers into the trimers makes the previously postulated purpose of the LC7.8 as a capping protein terminating the association of trimeric allophycocyanins to the core unlikely (13). Instead, specific interactions between LCM and LC7.8 are suggested to enable the correct positioning of AP⋅LC7.8 and allophycocyanin B at both sides of the integral allophycocyanin complex, APCM (9, 15, 40).
Acknowledgments
This study was supported by the Deutsche Forschungsgemeinschaft with a Habilitationsstipendium to W.R. and by the Sonderforschungsbereich 533 Lichtinduzierte Dynamik von Biopolymeren.
ABBREVIATIONS
- AP
allophycocyanin
- L
linker
- C
core
- CM
core-membrane
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, Brookhaven National Laboratory, Upton, NY 11973 (PDB ID code 1B33).
References
- 1.Glazer A N. Annu Rev Biophys Biophys Chem. 1985;14:47–77. doi: 10.1146/annurev.bb.14.060185.000403. [DOI] [PubMed] [Google Scholar]
- 2.Mörschel E, Riehl E. In: Membranous Structures. Harris J R, Horne R W, editors. London: Academic; 1987. pp. 210–254. [Google Scholar]
- 3.Holzwarth A R. Physiol Plant. 1991;83:518–528. [Google Scholar]
- 4.Sidler W. In: The Molecular Biology of Cyanobacteria. Bryant D A, editor. Dordrecht, The Netherlands: Kluwer; 1994. pp. 139–216. [Google Scholar]
- 5.Bryant D A. In: Cell Culture and Somatic Cell Genetics of Plants. Bogorad L, Vasil I K, editors. San Diego: Academic; 1991. pp. 257–300. [Google Scholar]
- 6.Reuter W, Müller C. J Photochem Photobiol. 1993;B 21:3–27. [Google Scholar]
- 7.Glazer A N. J Biol Chem. 1989;264:1–4. [PubMed] [Google Scholar]
- 8.Capuano V, Thomas J-C, Tandeau de Marsac N, Houmard J. J Biol Chem. 1993;268:8277–8283. [PubMed] [Google Scholar]
- 9.Reuter W, Wehrmeyer W. Arch Microbiol. 1990;153:111–117. [Google Scholar]
- 10.Anderson K A, Eiserling F A. J Mol Biol. 1986;191:441–451. doi: 10.1016/0022-2836(86)90139-7. [DOI] [PubMed] [Google Scholar]
- 11.Reuter W, Wehrmeyer W. Arch Microbiol. 1988;150:534–540. [Google Scholar]
- 12.Brejc K, Ficner R, Huber R, Steinbacher S. J Mol Biol. 1995;249:424–440. doi: 10.1006/jmbi.1995.0307. [DOI] [PubMed] [Google Scholar]
- 13.Füglistaller P, Mimuro M, Zuber H. Biol Chem Hoppe-Seyler. 1987;368:353–367. doi: 10.1515/bchm3.1987.368.1.353. [DOI] [PubMed] [Google Scholar]
- 14.Holzwarth A R, Bittersmann E, Reuter W, Wehrmeyer W. Biophys J. 1990;51:1–12. doi: 10.1016/S0006-3495(90)82514-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reuter W, Nickel-Reuter C. J Photochem Photobiol Biol. 1993;B 18:51–66. [Google Scholar]
- 16.Tandeau de Marsac N, Cohen-Bazire G. Proc Natl Acad Sci USA. 1977;74:1635–1639. doi: 10.1073/pnas.74.4.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu M, Glazer A N. J Biol Chem. 1982;257:3429–3433. [PubMed] [Google Scholar]
- 18.Schirmer T, Bode W, Huber R, Sidler W, Zuber H. J Mol Biol. 1985;184:257–277. doi: 10.1016/0022-2836(85)90379-1. [DOI] [PubMed] [Google Scholar]
- 19.Schirmer T, Huber R, Schneider M, Bode W, Miller M, Hackert M L. J Mol Biol. 1986;188:651–676. doi: 10.1016/s0022-2836(86)80013-4. [DOI] [PubMed] [Google Scholar]
- 20.Schirmer T, Bode W, Huber R. J Mol Biol. 1987;196:677–695. doi: 10.1016/0022-2836(87)90040-4. [DOI] [PubMed] [Google Scholar]
- 21.Duerring M, Schmidt G B, Huber R. J Mol Biol. 1991;217:577–592. doi: 10.1016/0022-2836(91)90759-y. [DOI] [PubMed] [Google Scholar]
- 22.Duerring M, Huber R, Bode W, Rümbeli R, Zuber H. J Mol Biol. 1990;211:633–644. doi: 10.1016/0022-2836(90)90270-v. [DOI] [PubMed] [Google Scholar]
- 23.Ficner R, Lobeck K, Schmidt G, Huber R. J Mol Biol. 1992;228:935–950. doi: 10.1016/0022-2836(92)90876-l. [DOI] [PubMed] [Google Scholar]
- 24.Ficner R, Huber R. Eur J Biochem. 1993;218:103–106. doi: 10.1111/j.1432-1033.1993.tb18356.x. [DOI] [PubMed] [Google Scholar]
- 25.Chang W, Jiang T, Wan Z, Zhang J, Yang Z, Liang D. J Mol Biol. 1996;262:721–731. doi: 10.1006/jmbi.1996.0547. [DOI] [PubMed] [Google Scholar]
- 26.Leslie A G W. mosflm. Darebury, Warrington, U.K.: SERC Laboratory; 1991. , Version 5.51. [Google Scholar]
- 27.Collaborative Computational Project, Number 4. Acta Crystallogr. 1994;D 50:760–763. [Google Scholar]
- 28.Navaza J. Acta Crystallogr. 1994;D 50:157–163. [Google Scholar]
- 29.Turk D. Ph.D. thesis. München, Germany: Technische Universität; 1992. [Google Scholar]
- 30.Sidler W, Gysi J, Isker E, Zuber H. Hoppe-Seyler’s Z Physiol Chem. 1981;362:611–628. doi: 10.1515/bchm2.1981.362.1.611. [DOI] [PubMed] [Google Scholar]
- 31.Füglistaller P, Ruembeli R, Suter F, Zuber H. Hoppe-Seyler’s Z Physiol Chem. 1984;365:1085–1096. [Google Scholar]
- 32.Lanzin V S, Wilson K S. Acta Crystallogr. 1993;D 49:129–147. doi: 10.1107/S0907444992008886. [DOI] [PubMed] [Google Scholar]
- 33.Brünger A. xplor. New Haven, CT: Yale Univ. Press; 1992. , Version 3.851. [Google Scholar]
- 34.Laskowski R A, McArthur M W, Moss D S, Thornton J M. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 35.Ramachandran G N, Sasisekharan V. Adv Protein Chem. 1968;23:283–437. doi: 10.1016/s0065-3233(08)60402-7. [DOI] [PubMed] [Google Scholar]
- 36.Kabsch W, Sander C. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 37.Holm L, Sander C. Nucleic Acids Res. 1994;22:3600–3609. [PMC free article] [PubMed] [Google Scholar]
- 38.Berger J M, Gamblin S J, Harrison S C, Wang J C. Nature (London) 1996;379:225. doi: 10.1038/379225a0. [DOI] [PubMed] [Google Scholar]
- 39.Gomis-Ruth F X, Gomez M, Bode W, Huber R, Aviles F X. EMBO J. 1995;14:4387–4394. doi: 10.1002/j.1460-2075.1995.tb00117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Redecker D, Wehrmeyer W, Reuter W. Eur J Cell Biol. 1993;62:442–450. [PubMed] [Google Scholar]
- 41.Reuter W, Nickel C, Wehrmeyer W. FEBS Lett. 1990;273:155–158. doi: 10.1016/0014-5793(90)81073-w. [DOI] [PubMed] [Google Scholar]
- 42.Isono T, Katoh T. Plant Cell Physiol. 1983;24:257–368. [Google Scholar]
- 43.Bald D, Kruip J, Rögner M. Photosynth Res. 1996;49:103–118. doi: 10.1007/BF00117661. [DOI] [PubMed] [Google Scholar]
- 44.Betz M, Rüesegger U, Esteban A M, Sidler W, Zuber H. Biol Chem Hoppe-Seyler. 1993;374:435–443. doi: 10.1515/bchm3.1993.374.7-12.435. [DOI] [PubMed] [Google Scholar]
- 45.Gottschalk L, Lottspeich F, Scheer H. Photochem Photobiol. 1994;58:761–767. [Google Scholar]
- 46.Sharkov A V, Kryukov I V, Khoroshilov E V, Kryukov P G, Fischer R, Scheer H, Gillbro T. Chem Phys Lett. 1992;191:633–638. [Google Scholar]
- 47.Schneider S, Prenzel C J, Brehm G, Gottschalk L, Zhao K H, Scheer H. Photochem Photobiol. 1995;62(5):847–854. [Google Scholar]
- 48.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 49.Merritt E A, Murphy M E P. Acta Crystallogr. 1994;D 50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- 50.Nicholls A, Bharadwaj R, Honig B. Biophys J. 1993;A 64:166. [Google Scholar]