Abstract
The aim of this study was to develop a national model and analyze the value of a molecular epidemiological Mycobacterium tuberculosis DNA fingerprint-outbreak database. Incidents were investigated by the United Kingdom PHLS Mycobacterium Reference Unit (MRU) from June 1997 to December 2001, inclusive. A total of 124 incidents involving 972 tuberculosis cases, including 520 patient cultures from referred incidents and 452 patient cultures related to two population studies, were examined by using restriction fragment length polymorphism IS6110 fingerprinting and rapid epidemiological typing. Investigations were divided into the following three categories, reflecting different operational strategies: retrospective passive analysis, retrospective active analysis, and retrospective prospective analysis. The majority of incidents were in the retrospective passive analysis category, i.e., the individual submitting isolates has a suspicion they may be linked. Outbreaks were examined in schools, hospitals, farms, prisons, and public houses, and laboratory cross-contamination events and unusual clinical presentations were investigated. Retrospective active analysis involved a major outbreak centered on a high school. Contact tracing of a teenager with smear-positive pulmonary tuberculosis matched 14 individuals, including members of his class, and another 60 cases were identified in schools clinically and radiologically and by skin testing. Retrospective prospective analysis involved an outbreak of 94 isoniazid-resistant tuberculosis cases in London, United Kingdom, that began after cases were identified at one hospital in January 2000. Contact tracing and comparison with MRU databases indicated that the earliest matched case had occurred in 1995. Subsequently, the MRU changed to an active prospective analysis targeting linked isoniazid-monoresistant isolates for follow up. The patients were multiethnic, born mainly in the United Kingdom, and included professionals, individuals from the music industry, intravenous drug abusers, and prisoners.
Mycobacterium tuberculosis is the causative organism of tuberculosis (TB) and produces 8 million new cases of TB and leads to 2 million deaths annually (14, 16, 49).
The development of novel molecular DNA fingerprinting techniques over the last decade has improved our understanding of TB transmission, supplementing information from conventional epidemiological methods such as contact tracing (47). Molecular fingerprinting can be used at a local level to identify or exclude laboratory cross-contamination or establish or refute the existence of an outbreak. Outbreaks of drug-sensitive and -resistant TB in human immunodeficiency virus (HIV)-positive or -negative individuals have been established in hospitals, schools, bars, prisons, nursing homes, and homeless shelters in the United States, Europe, and elsewhere (8, 9, 10, 13, 17, 20, 21, 22, 23, 29, 36, 39). These techniques have also been used in population studies to detect outbreaks missed by conventional methods and to estimate transmission levels by comparing the proportion of strains with the same or very similar fingerprint to those of unique isolates (1, 2, 27, 33, 40).
In the last few years several new techniques have been developed with different advantages and disadvantages. Arguably, the “gold standard” method is that of restriction fragment length polymorphism (RFLP) analysis based on the insertion sequence IS6110 (32, 44, 47). In general, RFLP methods are the most discriminating (32) but are slow, as they require viable organisms and, as there is no amplification stage, sufficient mycobacterial growth to extract enough DNA for analysis. More-rapid techniques employ a nucleic acid amplification step first (which is usually PCR) and so can be performed on cultures with little bacterial growth, on nonviable organisms, or with limited success, on heavily smear-positive sputum (30, 47). The principal methods are divided into those such as spacer-oligonucleotide typing (spoligotyping) (31) and variable number tandem repeat (VNTR) (24), which can be codified and stored electronically for comparative purposes, and rapid and/or arbitrary primed methods. Alternatively, rapid PCR-based epidemiological typing (RAPET) (50) and comparable methods can analyze small numbers of strains, which can be compared visually, but the results are usually not amenable for storage in electronic databases.
All the above methods are employed at the United Kingdom Public Health Laboratory Service (PHLS) Mycobacterium Reference Unit (MRU), but one of these techniques, RFLP IS6110, has been used to type all TB isolates of suspected outbreaks or hospital or laboratory cross-contamination submitted to the MRU since 1995. Since January 2000, our strategy has been to type all TB isolates by the RAPET method immediately (50) followed by RFLP IS6110 analysis. The strategy is to rapidly fingerprint TB cultures on arrival to distinguish laboratory or hospital cross-contamination quickly for hospital infection-control doctors and those responsible for controlling community outbreaks. These results are then confirmed by the definitive RFLP IS6110 method once the cultures have grown further.
There is no definitive fingerprinting method for Mycobacterium bovis, a cause of approximately 1% of the bacteriologically proven TB cases in the United Kingdom. The strategy at the MRU is to employ a combination of IS6110, spoligotyping, VNTR, and RAPET in combination with supportive epidemiology to reach a conclusion. This strategy has been developed in partnership with the United Kingdom Veterinary Laboratory Service so that transmission across all mammalian groups, including humans, may be determined.
This study analyzes suspected outbreaks and cross-contamination events from TB cultures submitted to the United Kingdom PHLS MRU by hospitals and laboratories in the United Kingdom and Ireland between June 1997 and December 2001. It attempts to address the question of what the value is of a national molecular database and describes framework investigative models or strategies for national TB reference centers.
MATERIALS AND METHODS
Mycobacterial isolates.
All isolates submitted were cultured onto two Lowenstein-Jensen solid medium slopes and, where necessary, into MB BacT liquid culture medium (Biomerieux, Basingstoke, United Kingdom). Isolates were subsequently stored at −70°C in Middlebrook 7H9 medium for at least 2 years and, wherever possible, for a longer period of time.
DNA extraction.
Extraction of DNA for RAPET was carried out as described by Yates et al. (50). Briefly, a 1-μl loop of culture was removed from a slope and placed in 100 μl of sterile distilled water; 100 μl of chloroform (Sigma-Aldrich, Poole, England) was added to the mixture, which was then vortexed for approximately 10 s. The mixture was heated at 80°C for 20 min and then held at −20°C. When needed, the mixture was allowed to thaw. While still chilled, the mixture was centrifuged at 12,000 × g for 5 min in a minifuge; 10 μl of supernatant was used for PCR.
Extraction of DNA for RFLP analysis was performed by using the standardized protocol (47) once there was a sufficient growth on the slopes.
RAPET.
Since January 2000, our strategy has been to type all TB isolates by using the RAPET method immediately (50) followed by RFLP IS6110 analysis. RAPET was performed as described previously by Yates et al. (50). Briefly, a PCR with a single primer (5′-GAGTCTCCGGACTCACCGG-3′) was used which targets an inverted repeat sequence of the IS6110 element. The PCR was performed in a reaction volume of 40 μl. The reaction conditions were as follows: an initial denaturation at 95°C for 120s; 1 cycle of 95°C for 20 s, 45°C for 360 s, and 72°C for 120 s; 30 cycles of 95°C for 20 s, 62°C for 30 s, and 72°C for 180 s; and a final extension at 72°C for 10 min. The PCR products were analyzed on a 0.8% agarose gel and visualized by using 0.5 mg of ethidium bromide per ml. The gel was run at 100 V for 30 min initially and then for a further 20 min, if further separation was required. PCR products giving similar fingerprints were subjected to restriction enzyme analysis with HaeIII (Promega, Southhampton, England). The digested products were analyzed on a 2% (wt/vol) agarose gel containing ethidium bromide at a concentration of 0.5 mg/ml.
RFLP.
RFLP typing targeting the insertion sequence IS6110 was performed accordingly to the internationally standardized protocol described by Van Embden et al. (44).
Epidemiological information.
All those submitting an isolate were asked to complete a standard molecular epidemiological form (available from the PHLS website and in reference 15), which requests information about the patient (name, gender, date of birth or age, hospital name, address, and location, clinical details, and the names of the treating physician and laboratory director). Further information may be provided on the form, by accompanying letter, or in practice by prior telephone consultation. All data were collated and stored electronically with the IS6110 fingerprint.
RESULTS
Over the period of 55 months from June 1997 to December 2001, inclusive, a total of 124 incidents occurred. This involved a total of 972 TB cases. These included a total number of 520 patients whose samples were examined by RFLP IS6110 from small referred incidents and 452 patients related to two population incidents involving isoniazid-resistant TB in London, England, and an outbreak of drug-sensitive TB in Leicester, England. Three analytical strategies are examined separately and subdivided by incident categories illustrating how these strategies are implemented operationally within the MRU.
Incident strategy 1: retrospective passive analysis.
The majority of incidents (n = 122) fell (and continue to fall) into this category. In this group, an initial suspicion usually exists on the part of the individual submitting the isolates that they may be part of a linked event, e.g., a laboratory cross-contamination event, multiple positive results in a short time period, or clinical doubt over TB diagnosis. In these incidents, the numbers of cases involved are usually small and there is a high probability that some or most of the cases are linked. Epidemiological evidence supporting linkage of cases is usually available.
Within this group, there were 122 incidents (121 were M. tuberculosis and 1 was M. bovis), with 520 patient isolates examined by using RFLP IS6110; 205 isolates were also examined by using RAPET. Incidents were characterized as (i) outbreaks, (ii) clinical confirmation, (iii) cross-contamination of specimens, or (iv) reinfection or reactivation of a past infection.
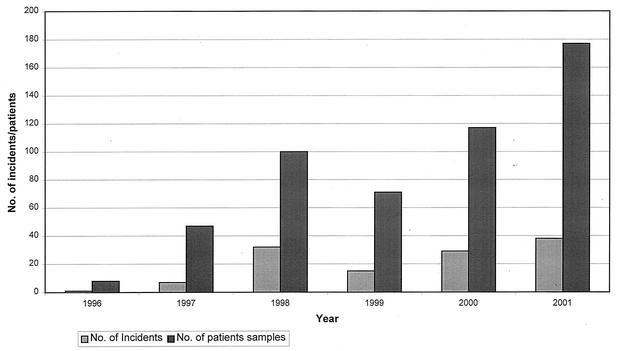
The average number of incidents investigated annually by the MRU was 28.5, but there was an increasing trend from 1996 to 2001 (Fig. 1). The majority of incidents (65 of 122, or 53.3%) submitted for molecular investigation were from possible outbreaks of TB, which were all confirmed by RFLP. One outbreak involved M. bovis (see below).
FIG. 1.
Number of incidents investigated by the MRU.
The RAPET method, newly developed and introduced as a rapid screening method, gave results concordant with the RFLP results in all incidents investigated (58 of 124, or 46.8%, where both techniques were performed).
The average number of cases per outbreak was 4.8 (range, 2 to 25). The following incidents describe the range and nature of the work of the MRU. Incidents or outbreaks involving schools, hospitals, public houses, nurseries, and other congregate settings dominate this category of work.
Category 1: outbreaks. (i) Category 1a: schools.
The largest outbreak in the retrospective passive analysis category was recorded in 2001 (one of two major incidents centered on schools in the whole database). The outbreak centered on a secondary school in Wales and involved 25 patients. These included school children, relatives, and friends at other schools. The index case, that of a 17-year-old boy, was identified. After RAPET and RFLP analysis, two clusters emerged. One cluster of 10 cases, included the index case, and a second but different cluster of 2 cases were also identified. The remaining 13 isolates were unique. Outbreaks commonly occur in schools and have been well documented. In total, six outbreaks occurred in schools during the study period. This incident and a larger one described in the next category reflect on the vulnerability of school children. Child-to-child transmission is believed to be rare in young children, but we should be cautious in extrapolating this to older children. School children, in any case, are exposed to potentially infectious adults, including teachers, parents, and other support staff who should not be overlooked.
(ii) Category 1b: public houses.
Twenty TB cases in 4 incidents involving bars or public houses were investigated. One incident occurred in 2001 in a public house in Newton Abbot, England, and involved 9 individuals (6 males and 3 females), 8 of whom drank in the pub. The RFLP IS6110 analysis (supported by RAPET analysis) indicated that the TB isolates from the 8 individuals had indistinguishable fingerprints. The ninth TB case was found to have a unique fingerprint. The patient lived in close proximity to the public house but did not actually drink there.
(iii) Category 1c: hospital.
The majority of potential outbreaks investigated occurred in hospitals (20%) (Table 1). An outbreak of drug-sensitive TB at a hospital in Cheltenham, England, involved a medical student, two nurses, and a patient who were all HIV negative. The index case was that of the male patient, and a further three cases were identified. The investigation is continuing.
TABLE 1.
Place of contact for each retrospective passive outbreak investigated
| Place or form of contact | No. of outbreaks | % of total no. of outbreaks |
|---|---|---|
| School | 5 | 7.7 |
| Hospital | 13 | 20.0 |
| Nursing home | 3 | 4.6 |
| Public house | 4 | 6.2 |
| Work | 5 | 7.7 |
| Shared house or flat | 4 | 6.2 |
| Nursery | 1 | 1.5 |
| Farm | 1 | 1.5 |
| Family and/or friends | 9 | 13.8 |
| Community and/or prisona | 12 | 18.5 |
| Other | 8 | 12.3 |
| Total | 65 | 100 |
Five of 20 cases in one community outbreak had occurred in prison. Similarly, in another incident of isoniazid-resistant TB in London, England, several cases were associated with a prison. The prison outbreaks have been counted as part of community outbreaks rather than as a specific prison incident.
DNA fingerprinting has also been used to exclude the existence of outbreaks. For example, multiple TB cases in a renal unit at a London teaching hospital in 1995 were thought to be linked epidemiologically but were subsequently shown to be multiple separate events (7). In this study, a suspected outbreak involving a doctor and four patients with TB occurred at a Birmingham, England, hospital in 1998. As the doctor had been in contact with all the other patients, he was presumed to be the source, but his isolate was unique. Two of the patients had indistinguishable isolates which differed from that of the doctor, and the remaining two had unique isolates.
(iv) Category 1d: nursery.
At a nursery in Leicester, England, TB was identified in a nursery nurse, the school cook, and two children, one of whom subsequently died (of TB meningitis). Conventional epidemiological follow-up suggested that the school nurse had had the source case, but subsequent analysis indicated that the DNA fingerprint of the isolate was unique and that, in fact, the isolate from the school cook was indistinguishable from that seen in the children. This incident had ramifications for a subsequent incident in the same town (see below).
(v) Category 1e: farm.
This represents the only incident in our database involving M. bovis in humans. Two teenagers, a brother and sister working on their family farm, were both diagnosed with TB. There had been cattle herd breakdowns and badgers on the farm, from which M. bovis had been isolated in previous years. Although there is no definitive molecular typing system, working in partnership with the United Kingdom Veterinary Laboratory Agency, the MRU applied spoligotyping, VNTR, RFLP IS6110, and RAPET to the human and animal isolates. All the systems applied suggested that the isolates were indistinguishable. The male teenager had helped on the farm and had been covered with cattle nasal secretions as he restrained the animals. He became symptomatic, and several months later, his sister became unwell with classic symptoms of TB. His sister was diabetic and pregnant, and considering the relative time intervals, it appears most likely that, while the brother was probably infected from the cattle, she was infected by exposure to her brother. In both cases, standard chemotherapy was successful.
Category 2: clinical case confirmation.
In this incident, DNA fingerprinting techniques were used to confirm clinical transmission. Most analyses have focused on pulmonary cases, as these are the most common and are likely to be infectious and transmissible; extrapulmonary TB can be infectious, and outbreaks have been associated with aerosol-generating procedures, e.g., irrigation of wound abscesses. DNA fingerprinting was used to establish an unusual case of sexually transmitted TB in a married couple (3). Fully drug-sensitive M. tuberculosis was cultured from a painless ulcer at the tip of the penis of a 50-year-old Indian man. Three early morning urine samples were negative for TB. Histopathological analysis showed the presence of caseating granulomas in a punch biopsy taken from the lesion. He was placed on appropriate TB chemotherapy for 6 months and was cured. A year later, his 49-year-old wife presented to the gynecologist with a 3-month history of menorrhagia, fever, night sweats, and weight loss. Endometrial biopsies were taken, caseating granulomas were seen, and drug-sensitive M. tuberculosis was isolated. RAPET and RFLP IS6110 analysis indicated that the isolates were indistinguishable. This was the first instance of the use of molecular fingerprinting to prove that sexual transmission of TB can occur.
Category 3: cross-contamination.
The 39 incidents investigated were divided into three main types: laboratory cross-contamination (37 incidents) and bronchoscopic-related (2 incidents) and ward-based (1 incident) contamination. In the ward incident, the fingerprint results confirmed that specimens from two patients had been mislabeled and mixed up. Mislabeling, contamination of reagents, or poor laboratory techniques in the generation of aerosols accounted for the majority of the laboratory incidents.
Category 4: reinfection and/or reactivation.
Prior to the development of molecular fingerprinting techniques, it was impossible to determine whether a second episode of TB was a relapsed case or a new infection. In eight episodes which were investigated, DNA fingerprinting established which of the above two events had occurred. In one case, a male patient was diagnosed and apparently treated successfully for three episodes of TB in 1997, 1998, and 2000. RFLP IS6110 analysis gave indistinguishable fingerprints, indicating that his subsequent illness was due to relapse of the original disease. In two cases, disease which was initially due to drug sensitive isolates was followed by disease caused by drug-resistant strains, including a multiple-drug-resistant isolate. Fingerprinting established that the strains were the same, i.e., they were cases of acquired drug resistance rather than reinfection.
Pseudoincidents.
In the final 9 (9 of 122, or 7.4%) episodes investigated in which either an outbreak or laboratory contamination was suspected, RFLP IS6110 and RAPET analysis independently confirmed that the isolates were unique and that there had been no incident.
Incident strategy 2: retrospective active analysis.
This usually develops from retrospective passive analysis. The most significant example relates to an outbreak centered on a high school in Leicester, England.
Contact tracing of a teenager with smear-positive pulmonary TB indicated that several other members of the same class had signs and symptoms of TB or radiological and/or bacteriological evidence of TB or were tuberculin skin test positive, suggesting infection. The source case patient had been symptomatic for 10 months and was highly infectious.
DNA fingerprint matches of 90% similarity or greater were obtained from 14 individuals in the main incident investigated, which included the source case patient's father and fellow students. Subsequently linked cases were seen at other educational establishments, and over 60 TB cases were subsequently identified in the town's schools clinically and radiologically and by skin testing. M. tuberculosis was cultured in only a small proportion of cases, which is not unusual in pediatric cases. Nevertheless, without bacteria for molecular fingerprinting it is impossible to distinguish disease due to different multiple exposures.
Concern was expressed that the schools might be acting as a sentinel for a wider-ranging unnoticed epidemic in the town. Six months previously, a young child in the same city had died of TB (see above), and there was concern that the TB strain might be the same. Using rapid PCR-based RAPET and spoligotyping, the MRU established by the first meeting of the outbreak committee that the incident was not linked to the earlier nursery one and that the strains of the source case patient and his or her parent were the same.
Overall, it seemed likely that the long duration of exposure to a highly infectious case could explain the high attack rate, although the strain is under continuing investigation by the incident team for intrinsic properties that might have made it either more infectious or virulent.
All TB isolates from patients within the town isolated during the preceding year were DNA fingerprinted. One hundred seventy-four isolates were fingerprinted by RFLP IS6110 analysis. Strains clustering together or those with 5 or fewer IS6110-containing bands were reanalyzed by spoligotyping. Isolates within the original school or with the school fingerprint were also analyzed by VNTR analysis. In addition to the recognized cluster, 17 other groups of indistinguishable isolates were noticed with two clusters acting as internal controls in that they were subsequently shown to be repeat isolates from the same individuals. In most of the remaining 15 groups, case numbers were small (2 to 6 cases per incident), with the majority represented by pairs of incident cases in household contact. A detailed report of the incident is in preparation.
Incident strategy 3: retrospective prospective analysis.
This group is represented by an outbreak of isoniazid-monoresistant TB in London, United Kingdom. Until 1 May 2002, 94 cases had been identified as part of the outbreak (84 in London and 10 cases outside London). The outbreak began as a retrospective passive analysis: astute medical microbiologists realized that they might have an outbreak after three cases of isoniazid-monoresistant TB were identified by the PHLS MRU at their hospital in January 2000. DNA fingerprinting (RAPET and RFLP IS6110) of all isoniazid-resistant TB isolates from the hospital from the beginning of 1999 identified a further 11 cases.
Contract tracing and analysis of the outbreak fingerprint with MRU databases indicated that the earliest matched case had occurred in a Nigerian student in 1995, and two further cases were identified from 1998. All isoniazid-monoresistant isolates from neighboring hospitals from 1999 onwards were DNA fingerprinted retrospectively, and 15 further cases were identified, nearly all of them in London.
Subsequently, the analysis changed from a passive to active retrospective one and then to an active prospective analysis with all London isoniazid-monoresistant isolates fingerprinted and linked cases identified. These cases were targeted for detailed follow up, and other unlinked isoniazid-resistant cases were targeted for routine contact tracing. The patients were multiethnic and mainly United Kingdom born, and some were professional or connected to the music industry, secondhand car sales, and intravenous drug abuse.
In the initial investigation, a link was identified with prisoners with TB. This has been the first documented case of TB transmission within a United Kingdom prison. Subsequently, 16 cases were found to be associated with the outbreak, and 9 transmissions probably occurred in the prison.
Overall, the local TB notification rate increased in London from 29.1 per 100,000 in 1995 to 40.3 per 100,000 in 2000 with the proportion of isoniazid-resistant TB remaining approximately the same at 6 to 7% nationally (but higher [7.6%] in London) (12). Detailed accounts of this outbreak are in preparation.
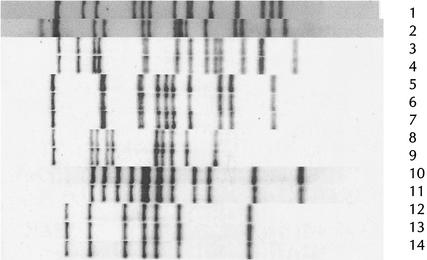
IS6110 fingerprints from outbreaks in each of the three categories are given in Fig. 2.
FIG. 2.
IS6110 fingerprints from outbreaks in London and Leicester and from some of the smaller proven outbreaks. Lanes: 1, Leicester outbreak strain; 2, London InhR strain; 3 to 14, strains from five different outbreaks, each of which contained between two and three cases.
DISCUSSION
The development of robust molecular fingerprinting systems for M. tuberculosis in the last 5 to 10 years has revolutionized our understanding of TB transmission. Comparison can be made between isolates of M. tuberculosis complementing conventional contact-tracing techniques. These techniques are of value clinically, for surveillance and to address public health issues. Methods must be based on genetic elements which can generate sufficient variability but which are sufficiently stable for longitudinal comparisons in population analyses.
Definitive RFLP analyses such as IS6110 (32, 47) are highly discriminating but require viable bacterial growth; alternatively, molecular amplification techniques are more rapid, do not require viable growth, and for some systems, such as spoligotyping and VNTR, can be digitalized and stored in databases (19, 32). Combinations of these methods may prove as discriminating as RFLP IS6110. Some methods such as spoligotyping have added value in that they are able to identify to subspecies level M. bovis BCG and M. tuberculosis and can be used to analyze dead cultures, smear-positive smears, and formalin-fixed specimens (30, 32). They aid in our understanding of evolution at the molecular level. For example, specific strains may be dominant and have an impact on TB transmission and the spread of drug-resistant TB, such as the Beijing genotype. This genotype has had a high impact on the TB epidemics in Asia, Vietnam, the former Union of Soviet Socialist Republics, and increasingly, in other geographic regions (11, 35, 45). The occurrence of this strain in Vietnam has been correlated with young age, suggesting recent transmission (4).
Other PCR methods including mixed-linker PCR (32) and RAPET (50) may be as discriminatory but are not amenable to storage on computerized databases. Spoligotyping requires a commercial membrane product, which makes it more expensive than RAPET, but spoligotyping analysis is easier, can be captured digitally, and provides diagnostic information as it can differentiate between members of the M. tuberculosis complete. Spoligotyping and RAPET are rapid methods offering time advantages over the IS6110 method.
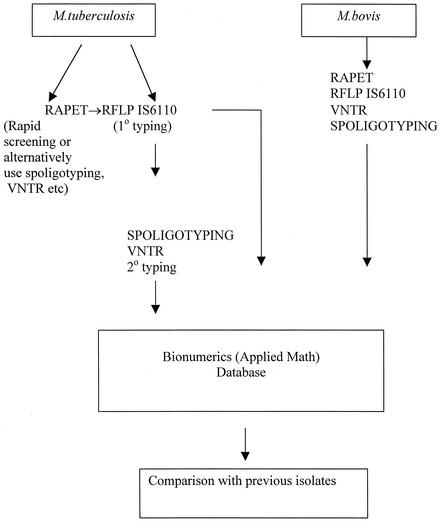
For any center maintaining a comparative longitudinal database, RFLP IS6110 fingerprinting is the current de facto gold standard, as it is highly discriminating, with relatively stable fingerprints, well-developed database analytical systems, and an international comparative methodology. The MRU maintains a fingerprint database of RFLP IS6110 isolates from a combined multicenter study from 1995 to 1997 (33) and a database of RFLP IS6110 fingerprints from TB isolates from England in 1998. In addition the MRU maintains a molecular outbreak database as described in this analysis. In total, the MRU database contains approximately 5,000 IS6110 fingerprints as well as substantial databases of spoligotyping profiles. In considering all the above factors the overall strategy adopted by the MRU is illustrated in Fig. 3.
FIG. 3.
Molecular epidemiological outbreak and incident investigation strategy. 1°, primary typing; 2°, secondary typing.
The value of a single center maintaining a longitudinal database was illustrated by the isoniazid-monoresistant TB outbreak in London, where cases were identified from earlier years by comparative examination of the database.
Undiagnosed outbreaks and/or incidents can be identified through widespread retrospective and prospective analyses. This can be confined to screening of laboratory cultures, for example, to detect cross-contamination. These techniques have established novel chains of transmission of smear-negative pulmonary cases (6) and of extrapulmonary cases (such as open abscesses), which have informed public health intervention strategies. Proof of transmission within the United Kingdom prison service was only obtained recently through molecular analysis.
Two major outbreaks of multidrug-resistant TB at two London teaching hospitals among mainly HIV-positive individuals were investigated by the MRU in 1995 and 1996 prior to the introduction of routine analysis. Isolates were found to be indistinguishable (8). TB is no more infectious to HIV-positive individuals than to HIV-negative individuals, but HIV infection increases susceptibility to clinical TB following infection. These events are not described in this study but are important, as they illustrated the high fatality rates associated with this combination of diseases and the importance of rapid identification and intervention within the institutions to prevent further nosocomial spread.
TB carries a stigma, and there may be a personal stigma for healthcare staff if they or others believe that they have transmitted the disease to their patients. The potential for transfer to sick or immunocompromised patients with consequent media and political attention complicates medical management, as the case in Birmingham, England, indicated. Conventional epidemiological analysis suggested that the likely source was the attending doctor, but molecular analysis demonstrated that this could not have been the case. Similarly, in the incident involving a child who died after apparently being infected by the nursery nurse, molecular analysis established that she could not have been the source of the infection.
Where these techniques can influence health policy most is when they are used in population studies, although the interpretation of studies is frequently controversial. In countries where the rates of endemic TB are low, the proportion of new cases due to recent infection is estimated to be 38 to 41% in New York, N. Y. (2, 22), 40% in San Francisco, Calif. (40), 28% in Berne, Switzerland (25), 46% in Amsterdam, The Netherlands (43), 28% in France (42), 38% in Seville, Spain (38), and 27% in London, England (27).
Fewer molecular epidemiological studies of TB transmission have been undertaken in countries or regions where HIV and TB are endemic. Estimates range from only 13% transmission of 200 isolates from a drug susceptibility study in Botswana (S. Lockman, J. W. Tappero, C. R. Braden, et al., Int. Union Tuberc. Lung Dis. UICTMR, abstr. no. 298-PA 02, 1997.), to 32% of 239 TB patients in San Paulo, Brazil, of whom approximately half were HIV infected (18), 25% of 84 patients in Honduras (37), 33% of 51 patients in Guadeloupe (41), 45% of 38 patients in Havana, Cuba (34), 41% in a study of 41 HIV-positive TB patients from Nairobi, Kenya (26), 45% of 246 isolates clustered in South Africa (48), 62% in Tunisia (28), and 53.6% in a small study of 28 TB patients in Harare, Zimbabwe (30). In a recent study, using a combination of spoligotyping and VNTR on isolates from Harare, Zimbabwe, 78.6% of 187 patients were identified as being part of a cluster (P. J. Easterbrook, A. L. Gibson, and F. A. Drobniewski, unpublished results); this high rate of clustering suggested a high rate of recent transmission.
Tuberculosis is more likely to develop in the first 12 months following acquisition of infection in HIV-positive individuals (5). Among HIV-infected patients living in New York and San Francisco, more than 60% of new TB cases were the result of recent transmission (2, 40). Other studies have also found the frequency of cluster pattern strains is significantly higher among seropositive patients (2, 18) and AIDS patients (40), although this has not been a consistent finding (46, 48).
New techniques such as single nucleotide polymorphism analysis are being investigated at the MRU and at other centers to develop fingerprint systems which are method independent, such as DNA sequencing, where analysis of the DNA sequence is independent of the system used to produce it, and do not require the comparison of optical data. Further attempts to automate the IS6110 methodology combine digital type methods, such as spoligotyping and VNTR, and develop method-independent techniques based ideally on sequence analysis need to be developed. They should be tested against epidemiologically well-defined culture panels to which the investigators are blinded. The development of new methods which are used to investigate the epidemiology of TB transmission in a population such as a town while using the epidemiology itself to simultaneously validate the new method are inappropriate.
Cross-contamination events and small outbreaks can be investigated locally by using a variety of techniques. The investigation by a single center by using standardized discriminating techniques such as RFLP IS6110 or RFLP with polymorphic GC-rich repeat sequences (which remain difficult to standardize at multiple centers) produces added value in that longitudinal and population analyses can be performed. The latter is essential where overall transmission in relatively low-incidence regions is to be analyzed and has been introduced with great success in diverse countries such as The Netherlands, Germany, Denmark, South Africa, Switzerland, and Estonia. The United Kingdom Veterinary Laboratory Service maintains a single database, which permits tracking of TB in mobile animal populations. Humans are at least as mobile, and multiple small collections will be insufficient to identify and track large incidents such as are occurring in London. Such a center can operate by using all three modes as described above, depending on the scope of work. A center performing analyses on a small number of outbreak strains can prove or disprove the event by using rapid PCR techniques, but the results cannot be placed in their wider context unless definitively typed by using a method whose results can be stored electronically.
Acknowledgments
We thank all the clinical microbiologists, consultants in communicable disease control, chest physicians, TB nurses, and public health doctors who so kindly provided information and who so ably managed the incidents discussed above.
REFERENCES
- 1.Agerton, T. B., S. E. Valway, R. J. Blinkhorn, K. L. Shilkret, R. Reves, W. W. Schluter, B. Gore, C. J. Pozsik, B. B. Plikaytis, C. Woodley, and I. M. Onorato. 1999. Spread of strain W, a highly drug-resistant strain of Mycobacterium tuberculosis, across the United States. Clin. Infect. Dis. 29:85-92. [DOI] [PubMed] [Google Scholar]
- 2.Alland, D., G. E. Kalkut, A. R. Moss, R. A. McAdam, J. A. Hahn, W. Bosworth, E. Drucker, and B. Bloom. 1994. Transmission of tuberculosis in New York City-An analysis by DNA fingerprinting and conventional epidemiological methods. N. Engl. J. Med. 330:1710-1716. [DOI] [PubMed] [Google Scholar]
- 3.Angus, B. J., M. Yates, C. Conlon, and I. Byren. 2001. Cutaneous tuberculosis of the penis and sexual transmission of tuberculosis confirmed by molecular typing. Clin. Infect. Dis. 33:132-134. [DOI] [PubMed] [Google Scholar]
- 4.Anh, D. D., M. W. Van Borydorff, L. N. Van, N. T. Lan, T. van Gorkom, K. Kremer, and D. van Soolingen. 2000. Mycobacterium tuberculosis Beijing genotype emerging in Vietnam. Emerg. Infect. Dis. 6:302-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barnes, P. F., A. B. Bloch, and P. T. Davidson. 1991. Tuberculosis in patients with human immunodeficiency virus infection. N. Engl. J. Med. 324:1644-1650. [DOI] [PubMed] [Google Scholar]
- 6.Behr, M. A., S. A. Warren, H. Salamon, P. C. Hopewell, A. Ponce de Leon, C. L. Daley, and P. M. Small. 1999. Transmission of Mycobacterium tuberculosis from AFB smear-negative patients. Lancet 353:444-449. [DOI] [PubMed] [Google Scholar]
- 7.Bendall, R. P., F. A. Drobniewski, D. Jayasena, P. M. Nye, A. H. C. Uttley, and G. M. Scott. 1995. Restriction fragment length polymorphism analysis rules out cross-infection among renal patients with tuberculosis. J. Hosp. Infect. 30:51-56. [DOI] [PubMed] [Google Scholar]
- 8.Breathnach, A. S., A. De Ruiter, G. M. C. Holdsworth, N. T. Bateman, D. G. M. O'Sullivan, P. J. Rees, H. Snashall, H. J. Milburn, B. S. Peters, J. Watson, F. A. Drobniewski, and G. L. French. 1998. An outbreak of multi-drug resistant tuberculosis in a London teaching hospital. J. Hosp. Infect. 39:111-117. [DOI] [PubMed] [Google Scholar]
- 9.Coronado, V. G., C. M. Beck-Sague, M. D. Hutton, B. J. Davis, P. Nicholas, C. Villareal, C. L. Woodley, J. O. Kilburn, J. T. Crawford, T. R. Frieden, R. L. Sinkowitz, and W. R. Jarvis. 1993. Transmission of multidrug-resistant Mycobacterium tuberculosis among persons with human immunodeficiency virus infection in an urban hospital: epidemiologic and restriction fragment length polymorphism analysis. J. Infect. Dis. 168:1052-1055. [DOI] [PubMed] [Google Scholar]
- 10.Daley, C. L., P. Small, G. Schector, G. Schoolnik, R. McAdam, W. Jacobs, and P. Hopewell. 1992. An outbreak of tuberculosis with accelerated progression among persons infected with human immunodeficiency virus. N. Engl. J. Med. 326:231-235. [DOI] [PubMed] [Google Scholar]
- 11.Diaz, R., K. Kremer, P. E. de Haas, R. I. Gomez, A. Marrero, J. A. Valdivia, J. D. van Embden, and D. van Soolingen. 1998. Molecular epidemiology of tuberculosis in Cuba, outside of Havana, July 1994-June 1995: utility of spoligotyping versus IS6110 restriction fragment length polymorphism. Int. J. Tuberc Lung Dis. 2:743-750. [PubMed] [Google Scholar]
- 12.Djuretic, T., J. Herbert, F. Drobniewski, M. Yates, E. G. Smith, J. G. Magee, R. Williams, P. Flanagan, B. Watt, A. Rayner, M. Crowe, M. V. Chadwick, A. M. Middleton, and J. M. Watson. 2002. Antibiotic resistant tuberculosis in the United Kingdom: 1993-1999. Thorax 57:477-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dooley, S. W., M. E. Villarino, M. Lawrence, L. Salinas, S. Amil, J. V. Rullan, W. R. Jarvis, A. B. Bloch, and G. M. Cauthen. 1992. Nosocomial transmission of tuberculosis in a hospital unit for HIV-infected patients. JAMA 267:2632-2634. [PubMed] [Google Scholar]
- 14.Drobniewski, F., A. Pablos-Mendez, and M. C. Raviglione. 1997. Epidemiology of tuberculosis in the world. Sem. Resp. Crit. Care Med. 18:419-429 [Google Scholar]
- 15.Drobniewski, F. A., J. G. Magee, E. G. Smith, and R. Williams. 1997. PHLS mycobacteriology reference services in England and Wales. Commun. Dis. Rep. CDR Rev. 7:R106-R109. [PubMed] [Google Scholar]
- 16.Dye, C., S. Scheele, P. Dolin, V. Pathania, and M. C. Raviglione. 1999. Consensus statement. Global burden of tuberculosis: estimated incidence, prevalence, and mortality by country. W. H. O. Global Surveillance and Monitoring Project. JAMA 282:677-686. [DOI] [PubMed] [Google Scholar]
- 17.Edlin, B. R., J. I. Tokars, M. H. Grieco, J. T. Crawford, J. Williams, E. M. Sordillo, K. R. Ong, J. O. Kilburn, S. W. Dooley, and K. G. Castro. 1992. An outbreak of multidrug-resistant tuberculosis among hospitalized patients with the acquired immunodeficiency syndrome. N. Engl. J. Med. 326:1514-1521. [DOI] [PubMed] [Google Scholar]
- 18.Ferrazoli, L., M. Palaci, L. R. Marques, L. F. Jamal, J. B. Afiune, E. Chimara, M. C. Martins, M. A. Silva Telles, C. A. Oliveira, M. C. Palhares, D. T. Spada, and L. W. Riley. 2000. Transmission of tuberculosis in an endemic urban setting in Brazil. Int. J. Tuberc. Lung Dis. 4:18-25. [PubMed] [Google Scholar]
- 19.Filliol, I., S. Ferdinand, L. Negroni, C. Sola, and N. Rastogi. 2000. Molecular typing of Mycobacterium tuberculosis based on variable number of tandem DNA repeats used alone and in association with spoligotyping. J. Clin. Microbiol. 38:2520-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fischl, M. A., R. B. Uttamchandani, G. L. Daikos, R. Poblete, J. Moreno, R. Reyes, A. M. Boota, L. M. Thompson, T. J. Cleary, and S. Lai. 1992. An outbreak of TB caused by multi-drug resistant tubercle bacilli among patients with HIV infection. Ann. Intern. Med. 117:177-183. [DOI] [PubMed] [Google Scholar]
- 21.Frieden, T. R., T. Sterling, A. Pablos-Mendez, J. O. Kilburn, G. M. Cauthen, and S. W. Dooley. 1993. The emergence of drug-resistant tuberculosis in New York City. N. Engl. J. Med. 328:521-526. [DOI] [PubMed] [Google Scholar]
- 22.Frieden, T. R., P. I. Fujiwara, R. M. Washko, and M. A. Hamburg. 1995. Tuberculosis in New York City-turning the tide. N. Engl. J. Med. 333:229-233. [DOI] [PubMed] [Google Scholar]
- 23.Frieden, T. R., L. F. Sherman, K. L. Maw, P. I. Fujiwara, J. T. Crawford, B. Nivin, V. Sharp, D. Hewlett, K. Brudney, Jr., D. Alland, and B. N. Kreisworth. 1996. A multi-institutional outbreak of highly drug-resistant tuberculosis: epidemiology and clinical outcomes. JAMA 276:1229-1235. [PubMed] [Google Scholar]
- 24.Frothingham, F., and W. A. Meeker-O'Connell. 1998. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem repeats. Microbiology 44:1189-1196. [DOI] [PubMed] [Google Scholar]
- 25.Genewein, A., A. Telenti, C. Bernasconi, C. Mordasini, S. Weiss, A. M. Maurer, H. L. Rieder, K. Schopfer, and T. Bodmer. 1993. Molecular approach to identifying route of transmission of tuberculosis in the community. Lancet 342:841-844. [DOI] [PubMed] [Google Scholar]
- 26.Gilks, C. F., P. Godfrey-Faussett, B. I. Batchelor, J. C. Ojoo, S. J. Ojoo, R. J. Brindle, J. Paul, J. Kimari, M. C. Bruce, J. Bwayo, F. A. Plummer, and D. A. Warrell. 1997. Recent transmission of tuberculosis in a cohort of HIV-1 infected female sex workers in Nairobi, Kenya. AIDS 11:911-918. [DOI] [PubMed] [Google Scholar]
- 27.Hayward, A. C., S. Goss, F. Drobniewski, N. Saunders, R. J. Shaw, M. Goyal, A. Swan, A. Uttley, A. Pozniak, J. Grace-Parker, and J. M. Watson. 2002. The molecular epidemiology of tuberculosis in inner London. Epidemiol. Infect. 128:175-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hermans, P. W., F. Messadi, H. Guebrexabher, D. van Soolingen, P. E. de Haas, H. Heersma, H. de Neeling, A. Ayoub, F. Portaels, D. Frommel, et al. 1995. Analysis of the population structure of Mycobacterium tuberculosis in Ethiopia, Tunisia, and The Netherlands: usefulness of DNA typing for global tuberculosis epidemiology. J. Infect. Dis. 171:1504-1513. [DOI] [PubMed] [Google Scholar]
- 29.Herrera, D., R. Cano, and P. Godoy. 1996. Multidrug-resistant tuberculosis outbreak on an HIV ward-Madrid, Spain 1991-1995. Morb. Mortal. Wkly. Rep. 45:330-333. [PubMed] [Google Scholar]
- 30.Heyderman, R. S., M. Goyal, P. Roberts, S. Ushewokunze, S. Zizhou, B. G. Marshall, R. Makombe, J. D. Van Embden, P. R. Mason, and R. J. Shaw. 1998. Pulmonary tuberculosis in Harare, Zimbabwe: analysis by spoligotyping. Thorax 53:346-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kremer, K., D. van Soolingen, R. Frothingham, W. H. Haas, P. W. Hermans, C. Martin, P. Palittapongarnpim, B. B. Plikaytis, L. W. Riley, M. A. Yakrus, J. M. Musser, and J. D. van Embden. 1999. Comparison of methods based on different molecular epidemiological markers for typing of Mycobacterium tuberculosis complex strains: interlaboratory study of discriminatory power and reproducibility. J. Clin. Microbiol. 37:2607-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maguire, H., J. W. Dale, T. D. McHugh, P. D. Butcher, S. H. Gillespie, A. Costetsos, H. Al-Ghusein, R. Holland, A. Dickens, L. Marston, P. Wilson, R. Pitman, D. Strachan, F. A. Drobniewski, and D. K. Banerjee. 2002. Molecular epidemiology of tuberculosis in London 1995-7 showing low rate of active transmission. Thorax 57:617-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Montoro, E., J. Valdivia, and S. Cardoso Leao. 1998. Molecular fingerprinting of Mycobacterium tuberculosis isolates obtained in Havana, Cuba, by IS6110 restriction fragment length polymorphism analysis and by the double-repetitive element PCR method. J. Clin. Microbiol. 36:3099-3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Niemann, S., G. S. Rusch, and E. Richter.1997. IS6110 fingerprinting of drug-resistant Mycobacterium strains isolated in Germany during 1995. J. Clin. Microbiol. 35:3015-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pearson, M. L., J. A. Jereb, T. R. Frieden, J. T. Crawford, B. J. Davis, S. W. Dooley, and W. R. Jarvis. 1992. Nosocomial transmission of multi-drug resistant Mycobacterium tuberculosis: a risk to patients and health care workers. Ann. Intern. Med. 117:191-196. [DOI] [PubMed] [Google Scholar]
- 37.Pineda-Garcia, L. A., A. Ferreira, and S. E. Hoffner. 1997. DNA fingerprinting of Mycobacterium tuberculosis strains from patients with pulmonary tuberculosis in Honduras. J. Clin. Microbiol. 35:2393-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Safi, H., J. Aznar, and J. C. Palomares. 1997. Molecular epidemiology of Mycobacterium tuberculosis strains isolated during a 3-year period (1993 to 1995) in Seville, Spain. J. Clin. Microbiol. 35:2472-2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Small, P. M., R. W. Shafer, P. C. Hopewell, S. P. Singh, M. J. Murphy, E. Desmond, M. F. Sierra, and G. K. Schoolnik. 1993. Exogenous reinfection with multi-drug resistant Mycobacterium tuberculosis in patients with advanced HIV infection. N. Engl. J. Med. 328:1137-1144. [DOI] [PubMed] [Google Scholar]
- 40.Small, P. M., P. C. Hopewell, S. P. Singh, A. Paz, J. Parsonnet, D. C. Ruston, G. F. Schecter, C. L. Daley, and G. K Schoolnik. 1994. The epidemiology of tuberculosis in San Francisco—a population-based study using conventional and molecular methods. N. Engl. J. Med. 330:1703-1709. [DOI] [PubMed] [Google Scholar]
- 41.Sola, C. L., K. S. Horgen, S. Goh, and N. Rastogi. 1997. Molecular fingerprinting of Mycobacterium tuberculosis on a Caribbean island with IS6110 and DRr probes. J. Clin. Microbiol. 35:843-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Torrea, G., C. Offredo, M. Simonet, B. Gicquel, P. Berche, and C. Pierre-Audigier. 1996. Evaluation of tuberculosis transmission in a community by 1 year of systematic typing of Mycobacterium tuberculosis clinical isolates. J. Clin. Micrbiol. 34:1043-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Deutekom, H., J. J. Gerritsen, D. van Soolingen, E. J. van Ameijden, J. D. van Embden, and R. A. Coutinho. 1997. A molecular epidemiological approach to studying the transmission of tuberculosis in Amsterdam. Clin. Infect Dis. 25:1071-1077. [DOI] [PubMed] [Google Scholar]
- 44.Van Embden, J. D. A., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, T. M. Shinnick, and P. M. Small. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van Soolingen, D., L. Qian, P. E. de Haas, J. T. Douglas, H. Traore, F. Portaels, H. Z. Qing, D. Enkhsaikan, P. Nymadawa, and J. D. van Embden. 1995. Predominance of a single genotype of Mycobacterium tuberculosis in countries of east Asia. J. Clin. Microbiol. 33:3234-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Soolingen, D., M. W. Borgdorff, P. E. de Haas, M. M. Sebek, J. Veen, M. Dessens, K. Kremer, and J. D. van Embden. 1999. Molecular epidemiology of tuberculosis in the Netherlands: a nationwide study from 1993 through 1997. J Infect. Dis. 180:726-736. [DOI] [PubMed] [Google Scholar]
- 47.Van Soolingen, D. 2000. Molecular epidemiology of tuberculosis and other mycobacterial infections: main methodologies and achievements. J. Intern. Med. 249:1-26. [DOI] [PubMed] [Google Scholar]
- 48.Wilkinson, D., M. Pillay, J. Crump, C. Lombard, G. R. Davies, and A. W. Sturm. 1997. Molecular epidemiology and transmission dynamics of Mycobacterium tuberculosis in rural Africa. Trop. Med. Int. Health 2:747-753. [DOI] [PubMed] [Google Scholar]
- 49.World Health Organization and International Union Against Tuberculosis and Lung Disease. 1997. Anti-tuberculosis drug resistance in the world. World Health Organization, Geneva, Switzerland.
- 50.Yates, M. D., F. A. Drobniewski, and S. M. Wilson. 2002. Evaluation of a rapid PCR-based epidemiological typing method for routine studies of Mycobacterium tuberculosis. J. Clin. Microbiol. 40:712-714. [DOI] [PMC free article] [PubMed] [Google Scholar]