Abstract
Although Chlamydia pneumoniae is an important human pathogen, the antigens eliciting a specific humoral immune response remain elusive. We scrutinized several recombinant chlamydial surface proteins for species-specific recognition by a panel of human sera previously tested for the presence of anti-C. pneumoniae and anti-C. trachomatis antibodies by microimmunofluorescence and enzyme-linked immunosorbent assay. The 15-kDa cysteine-rich protein (CrpA), porin-b (PorB), 9-kDa outer membrane protein (OMP3), 60-kDa outer membrane protein (OMP2), and four fragments of the major outer membrane protein (MOMP) representing each variable domain (VD) were overexpressed in Escherichia coli, affinity purified, and employed for Western blot analysis. None of the sera tested contained antibodies recognizing PorB and OMP3 of C. pneumoniae. Sera from C. pneumoniae-immune patients cross-reacted with OMP2 of C. trachomatis, and sera from C. trachomatis-immune patients cross-reacted with CrpA of C. pneumoniae, indicating that some of chlamydial surface molecules share antigenic epitopes. In contrast, the VD2, as well as the VD3, regions of the MOMP of C. pneumoniae were only recognized by C. pneumoniae-positive sera, suggesting the existence of species-specific epitopes. The identification of such epitopes of cell surface molecules provides new insights into C. pneumoniae-specific immune responses and may be of value for the improvement of C. pneumoniae-specific diagnostic assay systems based on defined recombinant antigens.
The obligate intracellular bacterium Chlamydia pneumoniae has been recognized as an important cause of several respiratory diseases in humans that gives rise to an average of 10% of cases of community-acquired pneumonia (17). Infection with C. pneumoniae has been implicated in the pathogenesis of severe cardiovascular diseases, especially atherosclerosis (32); however, the role of this pathogen as etiologic agent has not yet been proven and is still controversial (4, 13, 23, 31). Likewise, the possible involvement of C. pneumoniae in the pathogenesis of neurodegenerative diseases and lung cancer is intensely debated (18, 43). Antibodies to this organism are spread worldwide in the adult population (17). Analyses of various independent C. pneumoniae isolates revealed a high level of sequence homology (14, 29, 33). In contrast to C. trachomatis, for which 19 different serovars exist (20), only a few differences between different C. pneumoniae strains on a genetic, pathogenic, or serologic level have been revealed (9, 12, 17, 36). Although infections with C. pneumoniae can be monitored by direct isolation of the agent or by PCR-based detection of species-specific nucleotide sequences, serology represents the current routine method for a fast and convenient diagnosis. The microimmunofluorescence (MIF) assay, originally developed for C. trachomatis (38), is being considered as a sensitive and specific serologic method for the detection of C. pneumoniae infections (37). However, the diagnostic value of the results of the MIF test strongly depends on the antigen preparation and the experience of the individual investigator (7, 25). Partially automated enzyme-linked immunosorbent assays (ELISAs) have also been developed for routine chlamydial diagnostics (10, 27). Both MIF and ELISAs are based on preparations of entire elementary bodies, which explains their inherent problems concerning cross-reactivities between Chlamydia species and even unrelated microorganisms (10, 15, 24, 34).
Parts of the chlamydial outer membrane complex and especially the major outer membrane protein (MOMP) have long been considered as primary targets of the humoral immune response in humans (5, 17). Despite extensive studies on the antigenic composition of C. pneumoniae, the existence of species-specific and/or immunodominant surface antigens is still puzzling. The MOMP of C. pneumoniae has been described as an immunodominant antigen (11, 12, 28). Most of the immunogenic epitopes of MOMP appear to be conformation dependent because monoclonal antibodies raised against denatured MOMP display low binding affinity (8, 40). MOMP is considered a porin, spanning the outer chlamydial membrane (30, 41). Sequence comparisons of the MOMPs from several Chlamydia species revealed the existence of four variable domains (VD1 to VD4), presumably surface exposed, flanked by five strictly conserved regions (26, 42). The murine antibody response to peptides representing the VDs of C. pneumoniae MOMP has been previously characterized (28, 40). However, the individual humoral immune response to each of these VDs in humans has not yet been resolved.
Besides MOMP, other members of the outer membrane complex have been described in Chlamydia as being immunogenic, e.g., porin-b or the cysteine-rich outer membrane proteins CrpA, OMP2, and OMP3 (16, 19, 22, 39, 44). However, only little information is available about species-specific antibodies reacting with the corresponding surface antigens of C. pneumoniae.
In the present study we performed a comparative serologic analysis of several prominent surface antigens of C. pneumoniae and C. trachomatis in immunoblot assays with particular emphasis on their MOMPs. Since these proteins possess an obviously polyantigenic character, fragments of MOMPs were expressed as recombinant fusion proteins. The immunoreactivity of these fragments was analyzed by using a panel of human sera that were previously tested for anti-C. pneumoniae and anti-C. trachomatis antibodies.
MATERIALS AND METHODS
Chlamydial strains and genomic DNA.
C. pneumoniae strain CWL-029 and C. pneumoniae isolates were obtained from M. Maass, University of Lübeck, Lübeck, Germany. Genomic DNA from C. trachomatis serovar D was provided by E. Straube, University of Jena, Jena, Germany.
Serum samples.
Human sera were collected from independent clinic patients seropositive for C. pneumoniae and/or C. trachomatis, as well as from apparently healthy donors.
MIF and ELISAs.
The levels of immunoglobulins (immunoglobulin G [IgG] and IgA) against C. pneumoniae and C. trachomatis in serum were determined semiquantitatively with SeroCP/CT ELISA kits (Hain Diagnostika, Nehren, Germany) according to the manufacturer's instructions, respectively. Optical densities at 450 nm of ≥1.4 for C. pneumoniae and ≥1.2 for C. trachomatis were considered positive. Additionally, detection of specific IgG was carried out by MIF assay with a C. pneumoniae MIF kit (Virion, Munich, Germany). Serum titers of ≥1:20 were considered positive.
Molecular cloning of C. pneumoniae and C. trachomatis genes.
C. pneumoniae was propagated in HEp-2 cells (35) and elementary bodies were isolated (21). Bacterial DNA was purified by using a DNeasy DNA extraction kit (Qiagen). Genes and gene fragments coding for CrpA, PorB, OMP2, OMP3, and the separate variable MOMP domains VD1 to VD4 were amplified by PCR by using Pfu polymerase (Stratagene) and oligonucleotide primers flanked with specific restriction sites (Table 1 and Fig. 1). The primer design was based on chlamydial sequences obtained from published data banks (14). After restriction of the PCR products with the indicated restriction enzymes, DNA fragments were cloned into the protein expression vectors pET-14b (Novagen) and pGEX-2T (Pharmacia), respectively (Table 1). The identity of clones was confirmed by nucleotide sequencing in both directions over the entire DNA insert.
TABLE 1.
Oligonucleotides used to generate recombinant chlamydial fusion proteins
| Organism and protein | Oligonucleotide pairs (5′→3′)a | Expression vector, cloning site(s) |
|---|---|---|
| C. pneumoniae | ||
| CrpA | GAGctcgagATGTCATCAAATCTACATCC | pET-14b, XhoI |
| GATctcgagCTAAACGCGAGCTATTTTACT | ||
| PorB | TAggatccCTTTGACATGAATAGCAAGATG | pET-14b, BamHI |
| TTggatccTCTAAAACTGCAGACCTGACGT | ||
| OMP2 | AGctcgagATGTCCAAACTCATCAGACGAG | pET-14b, XhoI |
| CTTctcgagTTAATACACGTGGGTATTTTC | ||
| OMP3 | AAActcgagAAGAAAGCTGTTTTAATTGCT | pET-14b, XhoI |
| CCActcgagTTACTGTTTGCATCTGCCATC | ||
| MOMP-VD1 | GTGCGACGgaattcGCTTACGTGCTGG | pGEX-2T, EcoRI |
| AGCTCCTgaattcCAGAAAACATCAAAGCG | ||
| MOMP-VD2 | CTTTGATggatccTGTACTTTAGGAGC | pGEX-2T, BamHI |
| AACTTTAGGTTTggatccTGCATATTGG | ||
| MOMP-VD3 | CTTATGggatccGGTTGTGCAACTTTGG | pGEX-2T, BamHI |
| GGATGTTATCAggatccAAAGTTGCTCG | ||
| MOMP-VD4 | CCATACATTGgaattcAATGGTCTCGA | pGEX-2T, EcoRI |
| GAATCTgaattcACCAGATACGTGAGC | ||
| C. trachomatis | ||
| CrpA | AGGAATGTAActcgagAGCACTGTACCCGT | pET-14b, XhoI |
| TAGAGCAATctcgagTCATTGGGTCTGATCC | ||
| OMP2 | AAAATAActcgagCGAATAGGAGATCCTATG | pET-14b, XhoI |
| CATCGATAAAActcgagATTAATAGATGTGTG | ||
| MOMP-VD2 | ATCGTTTTGATggatccTGTACATTAGGAG | pGEX-2T, BamHI/EcoRI |
| TAGGTTTgaattcAGCATATTGGAATGAAGC | ||
| MOMP-VD3 | CTTTGTGGggatccGGATGTGCAACTTTAGG | pGEX-2T, BamHI/EcoRI |
| TCGTATCgaattcAAAGCTTGCTCGAGACC | ||
| MOMP-VD4 | AGTTAAAggatccCGAGCAAGCTTTGATGC | pGEX-2T, BamHI/EcoRI |
| TTAGAAGCGgaattcTGCATTTACGTGAGC |
Inserted restriction sites are indicated in lowercase letters. Chlamydial gene sequences (in boldface) are derived from C. pneumoniae CWL-029 and C. trachomatis serovar D, respectively.
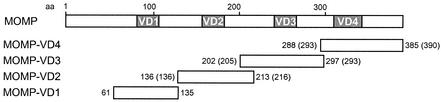
FIG. 1.
Molecular cloning of separate VDs of the C. pneumoniae and C. trachomatis MOMPs. The positions of the VDs are depicted schematically within the chlamydial full-length MOMP. The coding regions of the indicated MOMP fragments VD1 to VD4, along with flanking sequences, were individually cloned into a pGEX-2T expression vector, respectively (see Table 1). Amino acid (aa) positions are indicated for C. pneumoniae and C. trachomatis (in parentheses).
Expression and purification of recombinant fusion proteins.
As indicated in Table 1, chlamydial genes were overexpressed in Escherichia coli BL21(DE3) as proteins fused to either glutathione S-transferase (GST) or an N-terminal hexahistidine tag (His6). Bacteria were grown at 37°C in 2×YT medium (Difco) containing 200 μg of ampicillin/ml and 1% glucose with constant shaking at 225 min−1. Overexpression of fusion proteins was induced at an optical density at 600 nm of 0.5 to 1.0 by the addition of 0.4 mM (for His6 proteins) or 0.25 mM (for GST proteins) IPTG (isopropyl-β-d-thiogalactopyranoside). After a 2-h incubation at 30°C, cells were harvested and lysed by three consecutive freeze-thaw cycles in the presence of lysis buffer (for His6 proteins, 20 mM Tris-HCl [pH 8.0], 0.5 M NaCl, 0.2% Triton X-100, 2 mM 2-mercaptoethanol, 8 M urea; for GST proteins, 20 mM HEPES-KOH [5], 140 mM NaCl, 1% Triton X-100, 2 mM 2-mercaptoethanol) and protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 1 μg of pepstatin A/ml, 1 μg of leupeptin/ml), followed by short ultrasonication. After centrifugation (20,000 × g, 30 min), supernatants were subjected to affinity chromatography by using glutathione-Sepharose 4B or Chelating Sepharose Fast Flow (both from Pharmacia) according to the manufacturer's protocols, respectively. GST fusion proteins were eluted by adding 10 mM reduced glutathione and His-fused proteins by successively increasing the imidazole concentration to 0.5 M. The quality of the recovered protein fractions was judged after sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and silver staining (1). Protein concentrations were determined with a colorimetric BCA kit (Pierce). Finally, in-frame expression of the proteins was confirmed by mass spectrometry as previously described (3).
Immunoblot analysis.
The reactions of recombinant fusion proteins with human serum immunoglobulins were analyzed by Western blot assays according to standard protocols (1). In brief, protein preparations were subjected to a SDS-13% PAGE. The gels were then electroblotted onto a Hybond C nitrocellulose membrane (Amersham). Proper expression and size of the fusion proteins was confirmed by using a monoclonal anti-His6 antibody (dilution, 1:3,000; Qiagen) or goat anti-GST antiserum (dilution, 1:1,000; Pharmacia), followed by the horseradish peroxidase-conjugated secondary antibodies goat anti-mouse IgG and rabbit anti-goat IgG (both form Sigma at dilutions of 1:3,000 each), respectively.
The detection of interactions between antibodies of each MIF- or ELISA-tested serum sample and the recombinant chlamydial fusion proteins was performed with ca. 0.1 μg of purified protein per lane and serum dilutions of 1:200. Human immunoglobulins (IgG and IgA) were detected with horseradish peroxidase-conjugated secondary antibodies from Dako (Glostrup, Denmark) used at a 1:1,000 dilution. Signals were detected by enhanced chemiluminescence (Amersham).
RESULTS AND DISCUSSION
Selection of human sera by ELISA and MIF.
IgG and IgA titers to both C. pneumoniae and C. trachomatis were detected by ELISA. In addition, the sera were subjected to a IgG MIF test. When the results of both ELISA and MIF were consistent, the sera were chosen for further experiments. Four serum groups were distinguished: group I (Cp+/Ct+) represents sera positive for both C. pneumoniae and C. trachomatis (n = 8); group II (Cp+/Ct−) represents sera only positive for C. pneumoniae (n = 20); group III (Cp−/Ct+) represents sera only positive for C. trachomatis (n = 6); group IV (Cp−/Ct−) represents sera that tested negative for chlamydial antibodies (n = 7).
Western blot analysis of recombinant chlamydial proteins.
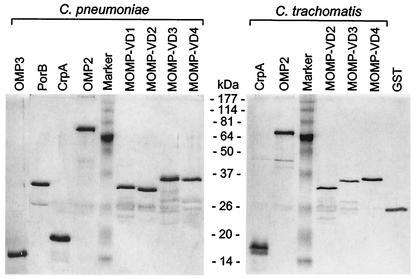
All chlamydial fusion proteins were obtained with a correct molecular weight and high purity (Fig. 2). Additional faint bands of lower molecular size were also visible in some of our protein preparations. They were identified as cleavage products of the fusion proteins in Western blots with anti-GST antiserum (data not shown).
FIG. 2.
Purified recombinant fusion proteins of C. pneumoniae and C. trachomatis. The indicated protein preparations were separated by SDS-PAGE, followed by silver staining.
In order to evaluate the immunoreactivity of the purified chlamydial surface antigens, each of the 41 sera belonging to one of the four different serum groups I to IV was analyzed by Western blotting. A reaction pattern of representative sera of each serogroup is shown in Fig. 3. The results obtained by Western blot analysis of all sera are summarized in Fig. 4. Porin-b (PorB) and the 9-kDa outer-membrane protein (OMP3) of C. pneumoniae were not recognized by IgG or IgA antibodies in any of the human sera tested (Fig. 4). Whether PorB and OMP3 are poorly immunogenic or whether putative linear or conformation-dependent epitopes were lost due to denaturing Western blot conditions remains unclear.
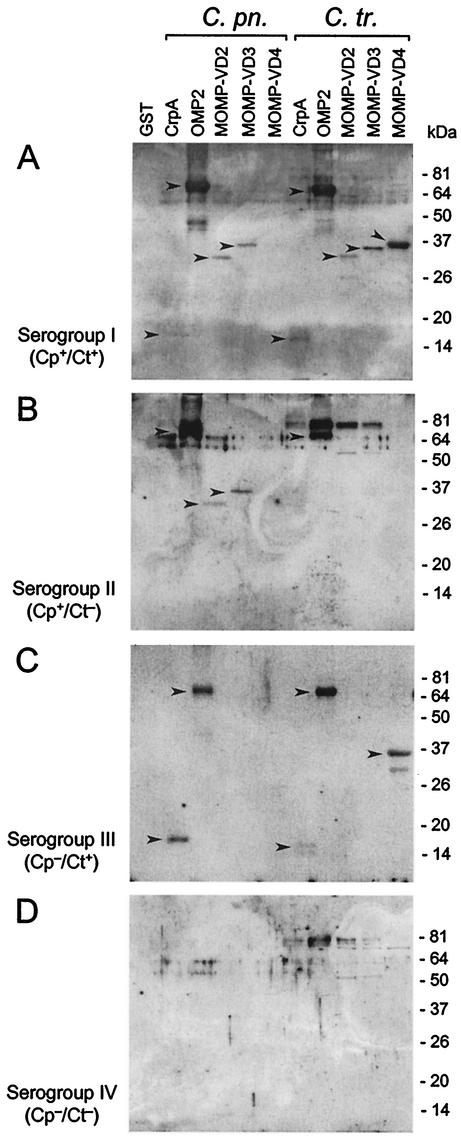
FIG. 3.
Western blot analysis of recombinant surface proteins of C. pneumoniae (C. pn.) and C. trachomatis (C. tr.) with human sera previously tested for the presence of Chlamydia-specific antibodies. IgG reactivity is shown for representative serum samples corresponding to four groups. (A) Group I, seropositive for both C. pneumoniae and C. trachomatis. (B) Group II, seropositive for C. pneumoniae only. (C) Group III, seropositive for C. trachomatis only. (D) Group IV, seronegative for both C. pneumoniae and C. trachomatis. Chlamydia-specific reactions are highlighted by arrowheads.
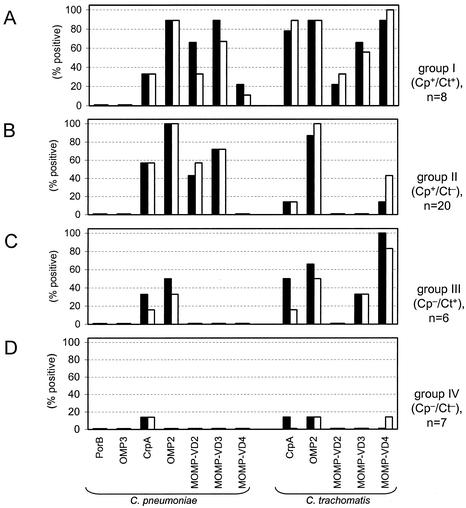
FIG. 4.
Immunoreactivity of serum antibodies with recombinant chlamydial surface proteins. Immunoglobulins (IgA and IgG) recognizing the indicated fusion proteins of C. pneumoniae and C. trachomatis were detected by Western blotting in a cohort of 41 human sera belonging to serogroups I to IV (panels A to D, respectively). The frequencies of positive reactions are presented for each serogroup and each individual chlamydial antigen. Secondary antibodies were directed against IgG (solid bars) and IgA (open bars).
As shown in Fig. 3A to C and 4A to C, the strongest and most frequent reactions were observed toward the recombinant OMP2 proteins of both C. pneumoniae and C. trachomatis. Notably, all sera that tested positive only for C. pneumoniae reacted with OMP2 from both C. pneumoniae and C. trachomatis (Fig. 4B). Likewise, C. trachomatis immune sera displayed OMP2 reactivity at considerably less frequency but with a significant degree of cross-reactivity to OMP2 of C. pneumoniae (Fig. 3C and 4C). This indicates that the OMP2 proteins from C. pneumoniae and C. trachomatis share immunodominant epitope(s). Indeed, the primary structure of both polypeptides is highly conserved (14, 22). The question arose as to why this strong cross-reaction of C. pneumoniae immune sera with OMP2 of C. trachomatis does not yield positive results in ELISA or MIF assay for C. trachomatis. It has been shown that the chlamydial cysteine-rich 60-kDa proteins are scarcely accessible, if at all, for antibody binding at the surface of intact bacteria or when whole elementary bodies were presented (22, 39). However, denatured and truncated chlamydial OMP2 fusion proteins have been reported to be major immunogens in chlamydial infections in a genus-specific manner (6, 22, 39). As a result of host defense reactions during the course of infection, determinants of previously inaccessible bacterial proteins may be released from disintegrated cell walls and become exposed to B-cell antigen receptors. Furthermore, upon protein unfolding after limited proteolysis, previously inaccessible linear motifs can be exposed to serve as B-cell epitopes.
Antibody binding to the recombinant 15-kDa cystein-rich proteins (CrpA) of both Chlamydia species was mainly observed with sera belonging to groups I, II, and III (Fig. 3 and 4). The apparent cross-reactivities can be explained by a strong sequence homology (38%) between CrpA of C. pneumoniae and C. trachomatis (14). However, sera from the uninfected group IV also contained antibodies to CrpA of both Chlamydia species (Fig. 4D). These observations are consistent with previous reports that showed weak or nonspecific reactivities with chlamydial 15-kDa outer membrane proteins (5, 8). Seroreactivity with the proteins CrpA, OMP2, and MOMP-VD4 was also shown for one of the seven sera, which were previously classified into group IV (Cp−/Ct−) (Fig. 4D). However, since the reliability and specificity of commercial MIF assay and ELISA is limited (10, 24, 34), this serum may have tested false negative for the presence of chlamydial antibodies.
The immunoreactivity against the chlamydial MOMP was investigated by using defined protein fragments representing VDs 1 to 4 (MOMP-VD1 to MOMP-VD4) (Fig. 1). Since the immunoblot analysis of the different MOMPs was performed with proteins fused to GST, GST was included as a control. Nonspecific reactions with the GST polypeptide were not observed with any of the sera under investigation, indicating that the immunoreactivities detected can be sensed to be Chlamydia specific (Fig. 3). Although fragment MOMP-VD1 from C. pneumoniae contains a predicted extracellular VD, it showed no reactivity with IgG or IgA from human sera in Western blots (data not shown). As shown in Fig. 3A and B and 4A and B, the MOMP-VD2 and MOMP-VD3 fragments of C. pneumoniae were only recognized by sera that tested positive in a MIF assay and an ELISA specific for C. pneumoniae. No significant immunoreactivity to this domain was observed when C. pneumoniae-negative sera from group III (Cp−/Ct+, Fig. 3C and 4C) or group IV (Cp−/Ct−, Fig. 3C and 4C) were used. Correspondingly, the MOMP VD3 region of C. trachomatis was only recognized by C. trachomatis positive sera (Fig. 4C). Notably, IgA immunoreactivity patterns were similar to those of IgG with regard to species-specific recognition of VD2 and VD3. Since at least 19 serovars of C. trachomatis exist, the possibility remains that antibodies to the VD2/VD3 regions of MOMP of a specific C. trachomatis serovar not represented in the patient group III might cross-react with the corresponding regions of C. pneumoniae MOMP.
Reactions with the MOMP VD4 region of C. pneumoniae were scarce (Fig. 4). In contrast, MOMP-VD4 of C. trachomatis yielded strong signals when C. trachomatis-positive sera were tested (Fig. 3A and C and 4A and C). This finding is consistent with a previous report wherein species- and serovar-specific epitopes were mapped to the VD4 region of the C. trachomatis MOMP by using murine monoclonal antibodies and different synthetic peptides (2). Some additional bands were visible in a size range between 50 and 75 kDa, in particular when C. trachomatis protein preparations were used (Fig. 3). These were probably due to interactions of human antibodies with E. coli proteins contaminating our preparations of recombinant chlamydial proteins.
We have confirmed and extended previous reports describing the immunogenicity of surface-exposed chlamydial proteins. Up to the present, it has been difficult to show species-specific immunoreactivity of surface antigens of C. pneumoniae. This held especially true for the MOMP of C. pneumoniae, which is an immunodominant and functionally important protein within the chlamydial infection cycle (11, 12, 36). We show here that it is possible to avoid frequently reported cross-reactivities of antibodies when the MOMP protein is dissected into fragments according to its extracellular VDs. Although VD4 may allow for the detection of C. trachomatis-specific antibodies, the identification of VD2 and VD3 regions of C. pneumoniae as species-specific domains within the MOMP may prove valuable for the improvement of species-specific assay systems for C. pneumoniae. Furthermore, the identification of species-specific recognition of well-defined proteins should allow new insights into humoral immune responses against C. pneumoniae and C. trachomatis.
Acknowledgments
We are grateful to T. Hoppe and F. Zahedi-Homayoun for supplying patient sera. We also thank M. Maass, University of Luebeck, Luebeck, Germany, and E. Straube, University of Jena, Jena, Germany, for providing chlamydial strains or DNA.
This work was partly supported by a grant from the Maria-Pesch-Stiftung.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1997. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 2.Batteiger, B. E., P. M. Lin, R. B. Jones, and B. J. Van Der Pol. 1996. Species-, serogroup-, and serovar-specific epitopes are juxtaposed in variable sequence region 4 of the major outer membrane proteins of some Chlamydia trachomatis serovars. Infect. Immun. 64:2839-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernardo, K., S. Fleer, N. Pakulat, O. Krut, F. Hünger, and M. Krönke. 2002. Identification of Staphylococcus aureus exotoxins by combined sodium dodecyl sulfate gel electrophoresis and matrix-assisted laser desorption/ionization-time of flight mass spectrometry. Proteomics 2:740-746. [DOI] [PubMed] [Google Scholar]
- 4.Boman, J., and M. R. Hammerschlag. 2002. Chlamydia pneumoniae and atherosclerosis: critical assessment of diagnostic methods and relevance to treatment studies. Clin. Microbiol. Rev. 15:1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell, L. A., C. C. Kuo, and J. T. Grayston. 1990. Structural and antigenic analysis of Chlamydia pneumoniae. Infect. Immun. 58:93-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ciervo, A., P. Visca, A. Petrucca, L. M. Biasucci, A. Maseri, and A. Cassone. 2002. Antibodies to 60-kilodalton heat shock protein and outer membrane protein 2 of Chlamydia pneumoniae in patients with coronary heart disease. Clin. Diagn. Lab. Immunol. 9:66-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dowell, S. F., R. W. Peeling, J. Boman, G. M. Carlone, B. S. Fields, J. Guarner, M. R. Hammerschlag, L. A. Jackson, C. C. Kuo, M. Maass, T. O. Messmer, D. F. Talkington, M. L. Tondella, and S. R. Zaki. 2001. Standardizing Chlamydia pneumoniae assays: recommendations from the Centers for Disease Control and Prevention (USA) and the Laboratory Centre for Disease Control (Canada). Clin. Infect. Dis. 33:492-503. [DOI] [PubMed] [Google Scholar]
- 8.Essig, A., U. Simnacher, M. Susa, and R. Marre. 1999. Analysis of the humoral immune response to Chlamydia pneumoniae by immunoblotting and immunoprecipitation. Clin. Diagn. Lab. Immunol. 6:819-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gieffers, J., R. J. Belland, W. Whitmire, S. Ouellette, D. Crane, M. Maass, G. I. Byrne, and H. D. Caldwell. 2002. Isolation of Chlamydia pneumoniae clonal variants by a focus-forming assay. Infect. Immun. 70:5827-5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hermann, C., K. Graf, A. Groh, E. Straube, and T. Hartung. 2002. Comparison of eleven commercial tests for Chlamydia pneumoniae-specific immunoglobulin G in asymptomatic healthy individuals. J. Clin. Microbiol. 40:1603-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iijima, Y., N. Miyashita, T. Kishimoto, Y. Kanamoto, R. Soejima, and A. Matsumoto. 1994. Characterization of Chlamydia pneumoniae species-specific proteins immunodominant in humans. J. Clin. Microbiol. 32:583-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jantos, C. A., S. Heck, R. Roggendorf, M. Sen-Gupta, and J. H. Hegemann. 1997. Antigenic and molecular analyses of different Chlamydia pneumoniae strains. J. Clin. Microbiol. 35:620-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jantos, C. A., A. Nesseler, W. Waas, W. Baumgartner, H. Tillmanns, and W. Haberbosch. 1999. Low prevalence of Chlamydia pneumoniae in atherectomy specimens from patients with coronary heart disease. Clin. Infect. Dis. 28:988-992. [DOI] [PubMed] [Google Scholar]
- 14.Kalman, S., W. Mitchell, R. Marathe, C. Lammel, J. Fan, R. W. Hyman, L. Olinger, J. Grimwood, R. W. Davis, and R. S. Stephens. 1999. Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nat. Genet. 21:385-389. [DOI] [PubMed] [Google Scholar]
- 15.Kern, D. G., M. A. Neill, and J. Schachter. 1993. A seroepidemiologic study of Chlamydia pneumoniae in Rhode Island: evidence of serologic cross-reactivity. Chest 104:208-213. [DOI] [PubMed] [Google Scholar]
- 16.Kubo, A., and R. S. Stephens. 2000. Characterization and functional analysis of PorB, a Chlamydia porin and neutralizing target. Mol. Microbiol. 38:772-780. [DOI] [PubMed] [Google Scholar]
- 17.Kuo, C. C., L. A. Jackson, L. A. Campbell, and J. T. Grayston. 1995. Chlamydia pneumoniae (TWAR). Clin. Microbiol. Rev. 8:451-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laurila, A. L., T. Anttila, E. Laara, A. Bloigu, J. Virtamo, D. Albanes, M. Leinonen, and P. Saikku. 1997. Serological evidence of an association between Chlamydia pneumoniae infection and lung cancer. Int. J. Cancer 74:31-34. [DOI] [PubMed] [Google Scholar]
- 19.Melgosa, M. P., C. C. Kuo, and L. A. Campbell. 1993. Outer membrane complex proteins of Chlamydia pneumoniae. FEMS Microbiol. Lett. 112:199-204. [DOI] [PubMed] [Google Scholar]
- 20.Millman, K. L., S. Tavare, and D. Dean. 2001. Recombination in the ompA gene but not the omcB gene of Chlamydia contributes to serovar-specific differences in tissue tropism, immune surveillance, and persistence of the organism. J. Bacteriol. 183:5997-6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murdin, A. D., P. Dunn, R. Sodoyer, J. Wang, J. Caterini, R. C. Brunham, L. Aujame, and R. Oomen. 2000. Use of a mouse lung challenge model to identify antigens protective against Chlamydia pneumoniae lung infection. J. Infect. Dis. 181(Suppl. 3):S544-S551. [DOI] [PubMed] [Google Scholar]
- 22.Mygind, P., G. Christiansen, K. Persson, and S. Birkelund. 1998. Analysis of the humoral immune response to Chlamydia outer membrane protein 2. Clin. Diagn. Lab. Immunol. 5:313-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ngeh, J., V. Anand, and S. Gupta. 2002. Chlamydia pneumoniae and atherosclerosis: what we know and what we don't. Clin. Microbiol. Infect. 8:2-13. [DOI] [PubMed] [Google Scholar]
- 24.Ozanne, G., and J. Lefebvre. 1992. Specificity of the microimmunofluorescence assay for the serodiagnosis of Chlamydia pneumoniae infections. Can. J. Microbiol. 38:1185-1189. [DOI] [PubMed] [Google Scholar]
- 25.Peeling, R. W., S. P. Wang, J. T. Grayston, F. Blasi, J. Boman, A. Clad, H. Freidank, C. A. Gaydos, J. Gnarpe, T. Hagiwara, R. B. Jones, J. Orfila, K. Persson, M. Puolakkainen, P. Saikku, and J. Schachter. 2000. Chlamydia pneumoniae serology: interlaboratory variation in microimmunofluorescence assay results. J. Infect. Dis. 181(Suppl. 3):S426-S429. [DOI] [PubMed] [Google Scholar]
- 26.Perez Melgosa, M., C. C. Kuo, and L. A. Campbell. 1991. Sequence analysis of the major outer membrane protein gene of Chlamydia pneumoniae. Infect. Immun. 59:2195-2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Persson, K., and J. Boman. 2000. Comparison of five serologic tests for diagnosis of acute infections by Chlamydia pneumoniae. Clin. Diagn. Lab. Immunol. 7:739-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peterson, E. M., X. Cheng, Z. Qu, and L. M. de La Maza. 1996. Characterization of the murine antibody response to peptides representing the variable domains of the major outer membrane protein of Chlamydia pneumoniae. Infect. Immun. 64:3354-3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Read, T. D., R. C. Brunham, C. Shen, S. R. Gill, J. F. Heidelberg, O. White, E. K. Hickey, J. Peterson, T. Utterback, K. Berry, S. Bass, K. Linher, J. Weidman, H. Khouri, B. Craven, C. Bowman, R. Dodson, M. Gwinn, W. Nelson, R. DeBoy, J. Kolonay, G. McClarty, S. L. Salzberg, J. Eisen, and C. M. Fraser. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 28:1397-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodriguez-Maranon, M. J., R. M. Bush, E. M. Peterson, T. Schirmer, and L. M. de la Maza. 2002. Prediction of the membrane-spanning beta-strands of the major outer membrane protein of Chlamydia. Protein Sci. 11:1854-1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rose, A. 2002. Failure to detect Chlamydia pneumoniae in senile calcific aortic stenosis or calcified congenital bicuspid aortic valve by immunofluorescence, polymerase chain reaction, and electron microscopy. Cardiovasc. Pathol. 11:300-304. [DOI] [PubMed]
- 32.Saikku, P., M. Leinonen, K. Mattila, M. R. Ekman, M. S. Nieminen, P. H. Makela, J. K. Huttunen, and V. Valtonen. 1988. Serological evidence of an association of a novel Chlamydia, TWAR, with chronic coronary heart disease and acute myocardial infarction. Lancet ii:983-986. [DOI] [PubMed]
- 33.Shirai, M., H. Hirakawa, M. Kimoto, M. Tabuchi, F. Kishi, K. Ouchi, T. Shiba, K. Ishii, M. Hattori, S. Kuhara, and T. Nakazawa. 2000. Comparison of whole genome sequences of Chlamydia pneumoniae J138 from Japan and CWL029 from USA. Nucleic Acids Res. 28:2311-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stralin, K., H. Fredlund, and P. Olcen. 2001. Labsystems enzyme immunoassay for Chlamydia pneumoniae also detects Chlamydia psittaci infections. J. Clin. Microbiol. 39:3425-3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tjhie, J. H., R. Roosendaal, D. M. MacLaren, and C. M. Vandenbroucke-Grauls. 1997. Improvement of growth of Chlamydia pneumoniae on HEp-2 cells by pretreatment with polyethylene glycol in combination with additional centrifugation and extension of culture time. J. Clin. Microbiol. 35:1883-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wagels, G., S. Rasmussen, and P. Timms. 1994. Comparison of Chlamydia pneumoniae isolates by Western blot (immunoblot) analysis and DNA sequencing of the omp2 gene. J. Clin. Microbiol. 32:2820-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang, S. 2000. The microimmunofluorescence test for Chlamydia pneumoniae infection: technique and interpretation. J. Infect. Dis. 181(Suppl. 3):S421-S425. [DOI] [PubMed] [Google Scholar]
- 38.Wang, S. P., and J. T. Grayston. 1970. Immunologic relationship between genital TRIC, lymphogranuloma venereum, and related organisms in a new microtiter indirect immunofluorescence test. Am. J. Ophthalmol. 70:367-374. [DOI] [PubMed] [Google Scholar]
- 39.Watson, M. W., P. R. Lambden, J. S. Everson, and I. N. Clarke. 1994. Immunoreactivity of the 60-kDa cysteine-rich proteins of Chlamydia trachomatis, Chlamydia psittaci, and Chlamydia pneumoniae expressed in Escherichia coli. Microbiology 140:2003-2011. [DOI] [PubMed] [Google Scholar]
- 40.Wolf, K., E. Fischer, D. Mead, G. Zhong, R. Peeling, B. Whitmire, and H. D. Caldwell. 2001. Chlamydia pneumoniae major outer membrane protein is a surface-exposed antigen that elicits antibodies primarily directed against conformation-dependent determinants. Infect. Immun. 69:3082-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wyllie, S., D. Longbottom, A. J. Herring, and R. H. Ashley. 1999. Single channel analysis of recombinant major outer membrane protein porins from Chlamydia psittaci and Chlamydia pneumoniae. FEBS Lett. 445:192-196. [DOI] [PubMed] [Google Scholar]
- 42.Yuan, Y., Y. X. Zhang, N. G. Watkins, and H. D. Caldwell. 1989. Nucleotide and deduced amino acid sequences for the four variable domains of the major outer membrane proteins of the 15 Chlamydia trachomatis serovars. Infect. Immun. 57:1040-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yucesan, C., and S. Sriram. 2001. Chlamydia pneumoniae infection of the central nervous system. Curr. Opin. Neurol. 14:355-359. [DOI] [PubMed] [Google Scholar]
- 44.Zhang, Y. X., N. G. Watkins, S. Stewart, and H. D. Caldwell. 1987. The low-molecular-mass, cysteine-rich outer membrane protein of Chlamydia trachomatis possesses both biovar- and species-specific epitopes. Infect. Immun. 55:2570-2573. [DOI] [PMC free article] [PubMed] [Google Scholar]