Abstract
Group B streptococci (GBS) are the most frequent pathogens in neonates with sepsis. A rapid screening method is required to identify carriage of GBS in pregnant women at the time of delivery. In order to detect GBS in vaginal specimens, the efficiency of the standard culture versus fluorescent in situ hybridization (FISH) was investigated. In 258 examined vaginal specimens, FISH identified 58 of the 59 GBS-positive samples (98.3%), whereas by means of standard culture only 38 specimens were positive (64.4%). We recommend FISH as a rapid, specific, highly sensitive screening technique for the detection of GBS in pregnant women at delivery.
Group B streptococci (GBS) are the most frequent pathogens isolated from neonates with invasive bacterial disease. The intrapartum fetal transmission may lead to invasive disease in 1 to 2 infants per 1,000 live births (5). The early onset form of the disease accounts for approximately 80% of GBS infection in infants. The mortality rate ranges from 10 to 20%. Infants suffering from GBS infection require prolonged hospitalization, and those who survive may have neurological sequelae (2, 3).
Approximately 20% of pregnant women are asymptomatically colonized with GBS. The diagnostic standard is the culture of anal and genital specimens obtained at 35 to 37 weeks of gestation or at delivery when at least one risk factor associated with neonatal infection is present (7).
Intrapartum chemoprophylaxis decreases the incidence of early onset from 1.7 per 1,000 live births to 0.6 per 1,000 live births (11). The recommended agents are intravenous penicillin G or ampicillin (12).
While the standard culture requires at least 36 h and may be a false negative (1, 2), fluorescent in situ hybridization (FISH) is a rapid and reliable method for detection and identification of microorganisms (8).
The aim of our work was to compare the FISH technique with standard culture to identify GBS in pregnant women at delivery and to determine which method is able to detect GBS earlier and more reliably.
We studied 258 pregnant women selected independent of diagnosis or hospitalization. The vaginal specimens obtained at prenatal visits or at admission for delivery were collected with a commercially prepared collection and transport system for aerobes and anaerobes, BBL CultureSwab Plus (Becton Dickinson, Heidelberg, Germany). All specimens were transported at room temperature and were cultured and tested by FISH.
For identification of GBS, the specimens were incubated on standard agar medium (β-streptococcus elective agar base, used according to the recommendations of Liebermeister and Braveny, Darmstadt, Germany), and colonies of streptococci suspected of being beta-hemolytic were subcultured for serological grouping by the method of Lancefield by using a commercially available system (Slidex Strepto-Kit A, B, C, D, E, and F; bioMérieux sa, Marcy l'Etoile, France). Finally, a minimum of 36 h was required for GBS identification. Positive cultures were graded as negative (no growth of GBS), + (slight colonization, less than 100 CFU/agar plate), ++ (moderate colonization, between 100 and 300 CFU/agar plate), or +++ (heavy colonization, more than 300 CFU/agar plate).
For FISH, all nucleotide probes used in this study have been previously described and evaluated (8). Probe Saga 67 a/b 5′-Cy3-GTAAACACCCMTCAGCG-3′ targeted to a 16S rRNA position was used to identify Streptococcus agalactiae in vaginal specimens (Fig. 1). For hybridization, vaginal swabs were smeared onto glass slides. The hybridization was performed at 46°C for 90 min. In short, streptococci were incubated with lysozyme (1 mg/ml for 20 min at 30°C) dissolved in 10 mM Tris (pH 8.0). Thereafter, the slides were washed, and 5 ng of each oligonucleotide was added to 10 μl of hybridization buffer containing 20% formamide. The probes were applied simultaneously with probe EUB 338-fluorescein isothiocyanate (FITC) and stained with DAPI (4′,6′-diamidino-2-phenylindol) (8). The slides were analyzed with a DMRE HC microscope (Leica Microsystem, Bensheim, Germany) by two independent investigators.
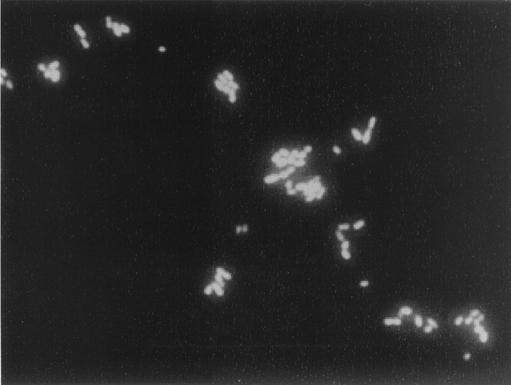
FIG. 1.
Detection of S. agalactiae within vaginal specimens of pregnant women by whole-cell hybridization (FISH) with the fluorescence (Cy3)-labeled oligonucleotide Saga 67 a/b. S. agalactiae was identified due to (i) its typical morphology (cocci in chains) and (ii) the specific red fluorescence. Magnification, ×630.
Microscopic sensitivity testing with serially diluted bacterial suspensions revealed a limit of detection by FISH between 103 and 104 CFU/ml. Positive FISH results were quantitatively processed (data not shown) and graded as negative (less than 103 CFU/ml), + (103 to 104 CFU/ml), ++ (105 to 106/ml), or +++ (107 to 108 CFU/ml).
Statistical analysis was performed. The Pearson χ2 test method was used to determine trends in discrepancies of isolation of GBS obtained by standard culture and those obtained by FISH. A P value of <0.05 defined statistically significant association. The statistical analysis was performed with Excel 2000 software (SR2b; Microsoft Corporartion, Redmond, Wash.) and JMP software (version 4.0; SAS Institute, Inc., Cary, N.C.).
Among 258 pregnant women, a total of 59 (22.8%) were identified as carriers of GBS by both culture and FISH. By using the standard culture, 38 (14.7%) vaginal specimens were positive and 220 (85.3%) were negative for GBS (Table 1). In order to semiquantitatively characterize the colonization status, 26 (68.4%) of the 38 positive cultures had heavy colonization (+++), 7 (18.4%) showed a moderate colonization (++), and only 5 (13.2%) had a low level of colonization (+) (Table 2).
TABLE 1.
Examination of 258 vaginal specimens by standard culture and FISH for detection of S. agalactiae
| Group no.a | Result by:
|
No. of specimensd | |
|---|---|---|---|
| Cultureb | FISHc | ||
| I | Negative | Negative | 199 |
| II | Positive | Positive | 37 |
| III | Positive | Negative | 1 |
| IV | Negative | Positive | 21 |
Group I specimens were negative by culture and FISH; group II specimens were positive by culture and FISH; group III specimens were positive by culture and negative by FISH; group IV specimens were negative by culture and positive by FISH.
A total of 258 specimens were examined by standard culture; 38 had a positive result and 220 had a negative result.
A total of 258 specimens were examined by FISH; 58 had a positive result and 200 had a negative result.
A total of 258 specimens were examined by both standard culture and FISH; 59 had a positive result and 199 had a negative result.
TABLE 2.
Quantitative evaluation of GBS-positive vaginal specimens identified by both standard culture and FISH
| Culture resultb | No. (%) of samples with FISHa result:
|
Total no. of samples | |||
|---|---|---|---|---|---|
| 0 | + | ++ | +++ | ||
| 0 | 17 (28.8) | 2 (3.4) | 2 (3.4) | 21 (35.6) | |
| + | 1 (1.7) | 2 (3.4) | 2 (3.4) | 0 | 5 (8.5) |
| ++ | 0 | 6 (10.2) | 0 | 1 (1.7) | 7 (11.9) |
| +++ | 0 | 2 (3.4) | 8 (13.5) | 16 (27.1) | 26 (44.0) |
| Total | 1 (1.7) | 27 (45.8) | 12 (20.3) | 19 (32.2) | 59 (100.0) |
The number and percentage of samples identified as positive by FISH are given. FISH was graded as 0 (less than 103 CFU/ml), + (103 to 104 CFU/ml), ++ (105 to 106 CFU/ml), and +++ (107 to 108 CFU/ml).
Cultures were graded as 0 (no growth of GBS), + (low colonization), ++ (moderate colonization), +++ (heavy colonization).
GBS could be detected by FISH in 58 (22.5%) of the 258 investigated vaginal specimens, whereas 200 (77.5%) of the samples were negative (Table 1). Positive probes were identified by the presence of fluorochrome Cy3 (red signal)-labeled chains of cocci, which were specifically hybridized with the probe Saga 67 a/b for S. agalactiae. All the positive samples also hybridized with the FITC (green signal)-labeled eubacterial probe (EUB338-FITC), which was used as a positive control. In contrast to the probe Saga 67 a/b specific for S. agalactiae, probe EUB338-FITC revealed positive stainings in most of the 258 investigated samples. The quantitative evaluation of the 58 GBS-positive probes identified by FISH showed that 19 (32.8%) samples were heavily colonized (+++), 12 (20.7%) samples were moderately colonized (++), and 27 (46.5%) samples had a low level of colonization (+) (Table 2).
In 199 vaginal specimens (group I), no GBS were detected by any method, and in 37 specimens, the pathogen was identified by both methods, FISH and culture (group II) (Table 1). In the last group, a heavy colonization result (+++) by the culture method corresponded to 8 samples with a moderate colonization result (++) and 16 samples with a heavy colonization result (+++) determined by FISH. Only 2 of the samples, which were graded as heavily colonized by the culture technique showed only a small number (+) of GBS by FISH (Table 2). Group III comprises 1 vaginal specimen which was GBS positive by culture but GBS negative by FISH. In one woman with discordant results, the positive culture showed a low colonization (+) with GBS. However, in another 21 vaginal specimens, GBS were detected by FISH but not by the culture method (group IV). In this group, the vast majority of samples (17 samples) contained a low number (+) of GBS. Nevertheless, 4 samples demonstrated a moderate (++) or heavy colonization (+++) (Table 2).
Thus, the culture method showed a sensitivity of 64.4%, whereas FISH was the more accurate technique with a sensitivity of 98.3%. The time required for results was 3.5 h for the FISH technique and at least 36 h for the bacterial culture.
We evaluated the FISH technique for the detection of GBS in vaginal specimens as a rapid, highly specific and sensitive test to identify 16S rRNA of microbial pathogens.
Neonatal GBS sepsis can be prevented by identifying and treating pregnant women which carry GBS and have a risk of transmitting the bacteria to their newborns. The current recommendation is to screen pregnant women by using a culture of vaginal and anal secretions obtained at 35 to 37 weeks of gestation and to identify the presence of at least one of many risk factors associated with neonatal infection (7).
While implementation of screening-based antibiotic prophylaxis is not possible for women who are not receiving adequate prenatal care or whose prenatal records are not available at the time of delivery (2), it is extremely important to develop rapid and sensitive screening methods to accurately identify GBS colonization in pregnant women at the time of delivery.
Two PCR assays were recently described as rapid methods for identification of GBS colonization in pregnant women. Compared with the culture results, the sensitivity of both PCR assays was 97% (2).
In our study, the prevalence of colonization among the 258 pregnant women was 22.8%, which corresponds with that reported in the literature (4, 5, 9). Compared with the conventional culture method, GBS colonization was detected earlier (after 3.5 versus 36 h) and with a significantly higher sensitivity (98.3 versus 64.4%; P = 0.024) by FISH than by culture. From a total of 59 GBS-positive specimens, in 21 vaginal samples the culture results were negative, whereas the FISH results were positive (group IV). Most of them (17 samples) contained a low number (103 to 104 CFU/ml) of GBS. However, in 4 cases, a moderately high number of CFU per milliliter was identified (Table 2). Because of the high specificity of the oligonucleotide probe Saga 67 a/b evaluated in a recent study, we can exclude false-positive results (8). In this study, a set of fluorescently labeled 16S, 18S, or 23S rRNA-targeted oligonucleotide probes was developed for the detection of microorganisms in blood culture. Each probe, including that for S. agalactiae Saga 16/67, was tested by FISH for specificity by using the respective target strain as well as related microbial species. In our study, the discrepancy between the number of positive results by FISH and culture might be explained by several factors. (i) By analyzing the culture, usually only beta-hemolytic colonies were selected as suspect and further examined. However, the frequency of strains which are not hemolytic is estimated to be 2 to 5% (1). (ii) On the other hand, 12 (57%) of 21 false-negative cultures which were FISH positive showed a heavy colonization of the vaginal specimens with gram-negative rods or enterococci (data not shown). Their rapid growth may inhibit the slowly growing GBS. The fact that, in most samples which were FISH positive, a low number of GBS was found also supports this hypothesis. (iii) Moreover, the stability of S. agalactiae in vaginal swabs during transport may be a limiting factor for the culture method. In addition, the time between obtaining specimens and preparation of vaginal swabs in the microbiology laboratory varied between 30 min and 4 h. The above-mentioned advantage of FISH over culturing might be even more evident if larger transport distances or a delayed transport occurred. (iv) Another possibility might be the fact that bacteria which have been killed by antibiotic drugs and which cannot be grown by culture were still detectable by FISH (10). This advantage of the FISH technique for the detection of microorganisms from antibiotic-treated patient was also underlined in a recent study with clinical samples obtained from cystic fibrosis patients (6).
Group III comprises only one vaginal specimen positive for GBS in the standard culture but negative by FISH. A possible explanation might be that small amounts of rRNA can lead to the failure of FISH to detect bacteria, as recently reported in reference 10. In fact, in this single case, a very low number of bacteria might be the reason for the failure of FISH to detect GBS. Moreover, the limit of detection of bacteria by FISH corresponds to that of other microscopic methods and can be estimated to be 103 to 104 CFU/ml of sample.
In conclusion, the application of FISH significantly increases the sensitivity of detection of GBS in vaginal specimens of pregnant women. Such a rapid method allows for the identification of the bacteria even during the labor and delivery period, for example, for women with prolonged membrane rupture, and for the subsequent intrapartum antibiotic prophylaxis. Other methods like the recently described PCR assays (2) have no significantly higher sensitivity compared to culture (97%), are more expensive than both culture and FISH, and require special equipment. We recommend FISH as a rapid, specific, and cost-effective screening technique with a high sensitivity (98.3%).
Acknowledgments
We thank Lydia Häuβermann for expert technical assistance.
REFERENCES
- 1.Baker, C. J. 1977. Summary of the workshop on perinatal infections due to group B Streptococcus. J. Infect. Dis. 136:137-152. [DOI] [PubMed] [Google Scholar]
- 2.Bergeron, M. G., D. Ke, C. Menard, F. J. Picard, M. Gagnon, M. Bernier, M. Ouellette, P. H. Roy, S. Marcoux, and W. D. Fraser. 2000. Rapid detection of group B streptococci in pregnant women at delivery. N. Engl. J. Med. 343:175-179. [DOI] [PubMed] [Google Scholar]
- 3.Boyer, K. M. 1995. Neonatal group B streptococcal infections. Curr. Opin. Pediatr. 7:13-18. [DOI] [PubMed] [Google Scholar]
- 4.Boyer, K. M., C. A. Gadzala, P. D. Kelly, L. I. Burd, and S. P. Gotoff. 1983. Selective intrapartum chemoprophylaxis of neonatal group B streptococcal early-onset disease. II. Predictive value of prenatal cultures. J. Infect. Dis. 148:802-809. [DOI] [PubMed] [Google Scholar]
- 5.Glantz, J. C., and K. E. Kedley. 1998. Concepts and controversies in the management of group B streptococcus during pregnancy. Birth 25:45-53. [DOI] [PubMed] [Google Scholar]
- 6.Hogardt, M., K. Trebesius, A. M. Geiger, M. Hornef, J. Rosenecker, and J. Heesemann. 2000. Specific and rapid detection by fluorescent in situ hybridization of bacteria in clinical samples obtained from cystic fibrosis patients. J. Clin. Microbiol. 38:818-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keenan, C. 1998. Prevention of neonatal group B streptococcal infection. Am. Fam. Physician 57:2713-2720, 2725. [PubMed]
- 8.Kempf, V. A. J., K. Trebesius, and I. B. Autenrieth. 2000. Fluorescent in situ hybridization allows rapid identification of microorganisms in blood cultures. J. Clin. Microbiol. 38:830-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McKenna, D. S., and J. D. Iams. 1998. Group B streptococcal infections. Semin. Perinatol. 22:267-276. [DOI] [PubMed] [Google Scholar]
- 10.Rüssmann, H., V. A. J. Kempf, S. Koletzko, J. Heesemann, and I. B. Autenrieth. 2001. Comparison of fluorescent in situ hybridization and conventional culturing for detection of Helicobacter pylori in gastric biopsy specimens. J. Clin. Microbiol. 39:304-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schrag, S. J., S. Zywicki, M. M. Farley, A. L. Reingold, L. H. Harrison, L. L. Lefkowitz, J. L. Hadler, R. Danila, P. R. Cieslak, and A. Schuchat. 2000. Group B streptococcal disease in the era of intrapartum antibiotic prophylaxis. N. Engl. J. Med. 342:15-20. [DOI] [PubMed] [Google Scholar]
- 12.Siegel, J. D. 1998. Prophylaxis for neonatal group B streptococcus infection. Semin. Perinatol. 22:33-49. [DOI] [PubMed] [Google Scholar]