Abstract
Shigella sonnei is a significant cause of gastroenteritis in both developing and industrialized countries. Definition of the diversity and antimicrobial susceptibility of S. sonnei isolates may be helpful in the management of individual cases and outbreaks. Antimicrobial susceptibility testing and pulsed-field gel electrophoresis (PFGE) were performed with 67 isolates of S. sonnei predominantly (n = 59) from three counties in the west of Ireland. Phage typing (n = 17), plasmid profiling (n = 28), and integron analysis (n = 24) were performed with subsets of strains. PFGE typing permitted recognition of two major clusters: PFGE type A (n = 53) and PFGE type B (n = 14). PFGE type A was associated with resistance to ampicillin, streptomycin, and sulfonamides (51 of 53 isolates), and those that were phage typed (n = 6) were phage type 3. PFGE type B was associated with resistance to streptomycin, sulfonamides, tetracycline, and trimethoprim (11 of 14 isolates) and phage type 6 (9 of 11 isolates). Fifteen different plasmid profiles were identified among the 28 isolates analyzed. A class 2 integron was present in all 14 PFGE type B isolates. One of these isolates also contained a class 1 integron and showed a unique variant of the PFGE type B pattern. Sequence analysis of the gene cassette structures contained within these integrons identified distinct open reading frames that encoded determinants of resistance to trimethoprim, streptomycin, and streptothricin. Our data demonstrate two predominant PFGE types among S. sonnei isolates circulating in this region. The limited diversity of the S. sonnei isolates in this region means that detection of isolates indistinguishable by PFGE and according to their antibiograms in two or more patients is not persuasive evidence of a common-source food- or waterborne outbreak. Indistinguishable plasmid profiles in addition to indistinguishable PFGE and antibiogram types may be more suggestive of an epidemiologically relevant link between cases.
Shigella sonnei is typically associated with mild self-limiting infection. In recent years S. sonnei has become the most prevalent Shigella species in the developed world. The spread of S. sonnei is particularly problematic in institutional or crowded settings, such as day-care centers and prisons and in military field settings (11, 12). Shigellosis is the third leading bacterial gastrointestinal disease in the United States, with 25,000 cases reported in 1998 and 18,000 cases reported in 1999 (8). Approximately 900 cases of S. sonnei infection are reported annually in the United Kingdom, and 15 cases of S. sonnei infection were reported to the National Disease Surveillance Centre in Ireland in 2001. In our laboratory, which serves a population of approximately 350,000 people, 813 isolates of S. sonnei were isolated between 1988 and 2001. There was marked variation from year-to-year in the number of cases, as follows: 1988, 180 cases; 1989, 4 cases; 1990, 96 cases; 1991, 373 cases; 1992, 45 cases; 1993, 1 case; 1994, 28 cases; 1995, 6 cases; 1996, 15 cases; 1997, 0 cases; 1998, 16 cases; 1999, 47 cases; 2000, 1 case; and 2001, 1 case.
The routes of transmission and the degree of diversity of the S. sonnei strains circulating in Ireland at present have not been well studied. We wished to determine the degree of diversity that exists among S. sonnei strains and to identify the potential of a number of typing methods for application in investigations of outbreaks or sporadic cases of S. sonnei infection.
The techniques used to type S. sonnei include antimicrobial susceptibility testing, phage typing, biotyping (S. sonnei can be subdivided into five biotypes), and a variety of molecular methods, including pulsed-field gel electrophoresis (PFGE), ribotyping, plasmid profiling, and plasmid fingerprinting (18). PFGE is a broadly applicable typing method with a high degree of intra- and interlaboratory reproducibility when standardized protocols are followed (6). We selected PFGE as the primary typing technique for the present study.
Most S. sonnei infections are self-limiting; however, antibiotic treatment may be useful in some cases to prevent further transmission. Antimicrobial resistance patterns are valuable as a guide to empirical therapy (when required), as a typing method, and as an indicator of dissemination of antimicrobial resistance determinants. S. sonnei, unlike most enteric pathogens, has no natural reservoir other than humans; therefore, antimicrobial resistance in S. sonnei may be expected to predominantly reflect gene transfer and selective pressures in the human gastrointestinal tract.
Mobile genetic elements including plasmids, transposons, and gene cassettes in integrons are important in the dissemination of resistance determinants. Conjugative plasmids encoding resistance to multiple antibiotics have been detected in S. sonnei (3, 17); indeed, the first report of carriage of tetracycline resistance genes on conjugative R plasmids was reported for a Shigella dysenteriae isolate in Japan in 1953 (25).
Integrons are genetic elements that encode a DNA recombinase that promotes the site-specific integration of gene cassettes. Four classes of integrons have been identified to date (26). Class 1 and 2 integrons have been reported in S. sonnei (19). Class 1 integrons are frequently reported in clinical and veterinary isolates of the family Enterobacteriaceae and are associated with lateral transfer of antimicrobial resistance genes (5, 10, 13). The gene cassettes are integrated between two conserved segments at the 5′ and 3′ ends at a receptor attI1 site. Class 2 integrons are members of the Tn7 family of transposons and are defined as containing three integrated gene cassettes (dhfrI, sat, and aadA1) located near a defective intI2 gene (5). This class of integrons does not contain the sul1 or qacEΔ gene at the 3′ conserved segment, characteristic of class 1 integrons but, rather, contain genes which promote the function of Tn7 transposition. Recent studies suggest that class 2 integrons predominate in S. sonnei (19). Integrons in S. sonnei have been associated with the dfrA1 and oxa-1 genes, which confer resistance to trimethoprim and ampicillin, respectively, in a study of isolates from Tanzania (21), while class 2 integrons containing gene cassettes encoding dfrA1 and sat1 (streptomycin and spectinomycin resistance) were detected in all (n = 40) isolates examined in an Australian study (19). We have examined representative isolates from a collection of isolates predominantly from the west of Ireland for the presence of class 1 and class 2 integrons and the associated antimicrobial resistance genes by PCR and DNA sequence analysis.
MATERIALS AND METHODS
Bacterial strains.
The majority of isolates analyzed (59 of 67) were clinical isolates from three counties in the west of Ireland. Six isolates were received from a hospital in the east of Ireland, and two were received from a hospital in the south of Ireland. The collection included family or institutional clusters (13 isolates in five clusters). The isolates were stored at −70°C. The bacterial strains were confirmed to be S. sonnei with the API 20E system (Biomerieux, Marcy l'Etoile, France) and slide agglutination with standard antisera (Murex Biotech Ltd., Dartford, England).
Phenotypic testing.
Antimicrobial susceptibility testing was performed by the disk diffusion method of the National Committee of Clinical Laboratory Standards (20). The following antimicrobial agents (the disk content is indicated in parentheses) were tested: ampicillin (10 μg), chloramphenicol (30 μg), streptomycin (10 μg), sulfonamides (300 μg), tetracycline (30 μg) trimethoprim (5 μg) ciprofloxacin (5 μg), kanamycin (30 μg), naladixic acid (30 μg), and nitrofurantoin (300 μg). Escherichia coli ATCC 25922 was used as a control. The disks were obtained from Oxoid (Basingstoke, United Kingdom). The stability of the antimicrobial resistance of a number of the isolates was determined by subculturing representative isolates each day for 4 weeks. At the end of each week the isolates were retested against the same panel of antimicrobial agents. Phage typing was performed at the Laboratory of Enteric Pathogens, Public Health Laboratory Service, London, United Kingdom, by standard procedures on a subset (n = 17) of the S. sonnei isolates.
PFGE.
PFGE plugs were prepared and digested (with restriction enzyme Xba) as described previously (9). Macrorestriction fragment patterns were initially analyzed by use of the criteria of Tenover et al. (22). Computer-assisted analysis of the PFGE banding patterns was performed with Bionumerics software (Applied Maths, Kortrijk, Belgium). Tiff images of the gel were normalized by aligning the size standards located in the outer lanes of the gel with the reference standard of the database. Analysis of banding patterns was performed with the Dice coefficient by using a 1.2% tolerance for the band migration distance. Clustering of the patterns was performed by the unweighted pair group method with arithmetic averages. Ten isolates with the different patterns observed by PFGE on initial analysis were reexamined by the standardized PulseNet protocol (6) when it became available for use in our laboratory.
Plasmid profiles.
Plasmids were extracted from the isolates by a modification of the alkaline lysis method of Kado and Liu (14). Twenty-five microliters of plasmid extract was added to 2 μl of tracking buffer, and the mixture was added to a 0.7% agarose gel (Sigma-Aldrich, St. Louis, Mo.) with a Pasteur pipette. Horizontal gel electrophoresis was carried out in 1× TAE (Tris-acetate-EDTA) at 70 V for 4.5 h. Size controls consisted of Rts1 (84 kb), R1 (41 kb), R6k (17 kb), and a 2- to 10-kb size marker (Promega, Madison, Wis.). The gels were stained with 5 μg of ethidium bromide per ml and visualized under UV light. The plasmid size was estimated from plots of a standard curve on two-cycle log paper and extrapolation of the sizes from the distances traveled.
Plasmid conjugation.
A subset of isolates were examined for the presence of conjugative plasmids encoding antimicrobial resistance by the liquid mating method of Bradley et al. (4). The donor and the recipient (strain J-53, which is nalidixic acid resistant) were cultured overnight at 37°C in 4 ml of brain heart infusion broth. Conjugation was performed by adding aliquots of overnight cultures of the donor (0.1 ml) and the recipient (0.4 ml) to 0.5 ml of prewarmed brain heart infusion broth, and the mixture was incubated at 37°C for 4 h. An aliquot (100 μl) of the conjugation mixture was spread plated onto tryptone soy agar (BDH) made selective by the addition of antimicrobial agents (i.e., nalidixic acid and ampicillin, nalidixic acid and tetracycline, and nalidixic acid and streptomycin). The tryptone soy agar plates were incubated overnight at 37°C. Suspected transconjugants were subcultured for purity on a nonselective agar. Antimicrobial susceptibility testing and biochemical identification (API 20E) were performed with all transconjugants.
Integron analysis.
DNA was extracted from each isolate (24) and quantitated spectrophotometrically. Integron analysis was performed by PCR with specific primer pairs with a subset (n = 24) of isolates. To amplify class 1 integron structures, the primer pair Int1F (5′-GGCATCCAAGCAGCAAGC-3′) and Int1R (5′-AAGCAGACTTGACCTGAT-3′) was used (15), and to detect the class 2 integrons, primer pair hep74 (5′-CGGGATCCCGGACGGCATGCACGATTTGTA-3′) and hep51 (5′-GATGCCATCGCAAGTACGAG-3′) was used (26). To each reaction mixture was added 100 ng of template DNA, 1.5 mM MgCl2, each deoxynucleoside triphosphate at a concentration of 200 μM, 5 μl of 10× PCR buffer (50 mM Tris-HCl [pH 8.0], 100 mM NaCl, 0.1 mM EDTA, 1 mM dithiothreitol, 50% glycerol, 1% Triton X-100), 25 pmol of each primer, and 2.5 U of Taq DNA polymerase in a final reaction volume of 50 μl. Amplification was performed in a MiniCycler (MJ Research, Watertown, Mass.) by using the following temperature profile: predenaturation at 94°C for 10 min, followed by 35 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 5 min, with a final extension step at 72°C for 5 min. The amplified DNA products were analyzed by conventional 1.5% (wt/vol) agarose gel electrophoresis in 1× TAE buffer and stained with ethidium bromide as outlined above.
DNA sequencing.
Amplified DNA products of interest were initially gel extracted and purified (QIAGEN, West Sussex, United Kingdom). These were subsequently cloned by use of a TOPO TA cloning kit (Invitrogen BV, Groningen, The Netherlands). The cloned products were sequenced by automated methods (MWG-Biotech, Ebersberg, Germany), and the resulting DNA sequence was analyzed by use of the BLAST suite of programs. The sequences identified in the present databases available over the Internet were subsequently aligned by using the CLUSTALW program (23).
Nucleotide sequence accession numbers.
The sequences of two class 2 integron structures of 2.22 kbp (the GenBank accession number AY140652) and 1.37 kbp (GenBank accession no. AY090896) and a single class 1 integron structure of 1.95 kbp and their respective gene cassettes have been submitted to GenBank.
RESULTS
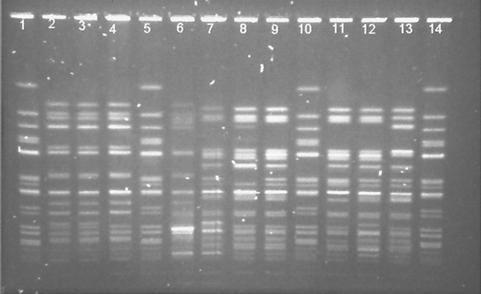
Two major clusters of S. sonnei were identified by PFGE: PFGE type A (n = 53) and PFGE type B (n = 14). Limited diversity was observed within the clusters (Fig. 1). The similarity index for the two groups was 55%, whereas similarity indices were 82% for isolates within cluster A and 75% for isolates within cluster B. Forty-nine of the PFGE type A isolates (92%) were indistinguishable, and the remaining four (designated A1) differed by a single band. All of the PFGE type A isolates and all but one of the PFGE type A1 variants were isolated from the western region of Ireland between February 1998 and May 1999. A single type A1 isolate was from the east coast area of Ireland. Five variants (B, B1, B2, B3, and B4) were observed among the PFGE type B strains (Fig. 1). These isolates originated from patients distributed more widely geographically. The five PFGE type B isolates from the eastern region comprised a single representative of each variant, while both isolates from the southern region were PFGE type B. The results of phage typing of a subset of isolates (n = 17) correlated with those of PFGE. PFGE type A strains (n = 6) were phage type 3 (PT3), while PFGE type B isolates were PT6 (n = 8) or PT50 (n = 1) or did not conform to a recognized phage type (n = 1).
FIG. 1.
PFGE patterns generated with XbaI of representative S. sonnei isolates from Ireland. Lane 1, strain F2353 (S. sonnei control); lane 2, isolate 3331 (PFGE type A); lane 3, isolate 3367 (type A); lane 4, isolate 3219 (type A); lane 5, F2353 control; lane 6, isolate Dublin 1 (type A1); lane 7, isolate CK1 (type B); lane 8, isolate Dublin 4 (type B1); lane 9, isolate 510 (type B); lane 10, F2353 control; lane 11, isolate Dublin 2 (type B4); lane 12, isolate 3601 (type B1); lane 13, isolate Dublin 3 (type B3); lane 14, F2353 control.
The antimicrobial resistance phenotypes of the isolates by PFGE type are summarized in Table 1. The stabilities of the resistance phenotypes in vitro were confirmed by continuous subculture of a representative isolate of each resistance phenotype for 4 weeks, followed by repeat susceptibility testing.
TABLE 1.
Antimicrobial resistance phenotypes of S. sonnei isolates by PFGE type
| PFGE type | No. of isolates | Antimicrobial resistance phenotypea
|
|||||
|---|---|---|---|---|---|---|---|
| AMP | STREP | SUL | TET | TRIM | NAL | ||
| A | 51b | R | R | R | S | S | S |
| A | 1b,c | R | S | S | S | S | S |
| A | 1b,d | R | S | R | S | R | S |
| B | 11b | S | R | R | R | R | S |
| B | 1b,e | R | R | R | R | R | S |
| B | 1b,f | S | R | R | R | R | R |
| B | 1b,g | S | R | S | S | R | S |
AMP, ampicillin; STREP, streptomycin; SUL, sulfonamide; TET, tetracycline, TRIM, trimethoprim; NAL, nalidixic acid; R, resistance; S, susceptible.
In addition, all isolates were susceptible to chloramphenicol, ciprofloxacin, kanamycin, and nitrofurantoin.
Isolate 3599.
Isolate 3219.
Isolates CK2.
Isolate 1778.
Isolate CK1.
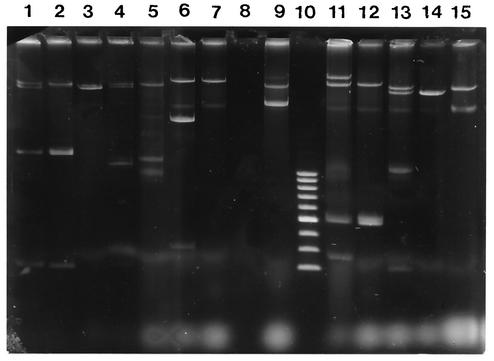
One or more plasmids ranging in size from 1 kb to greater than 90 kb were present in all 28 isolates examined for plasmids. Four distinct plasmid profiles were observed among the 14 PFGE type A isolates tested (Fig. 2). Five isolates were PFGE type A and plasmid profile a, including three isolates from three members of one family. Seven isolates were PFGE type A and plasmid profile b, including two isolates from the members of one family. Plasmid profiles a and b were not observed among the PFGE type B isolates; however, plasmid profile d occurred among the isolates in both PFGE clusters. Greater diversity of plasmid profiles was observed among PFGE type B isolates (12 different plasmid profiles) than among PFGE type A isolates.
FIG. 2.
Plasmid profiles of selected S. sonnei isolates along with their respective E. coli J-53 transconjugants. Lane 1, isolate 3331; lane 2, 3331 transconjugant resistant to ampicillin, streptomycin, and sulfonamides; lane 3, 3331 transconjugant A; lane 4, isolate 3599; lane 5, 3599 transconjugant A; lane 6, isolate Dublin 3; lane 7, Dublin 3 transconjugant resistant to streptomycin, sulfonamides, and tetracycline; lane 8, E. coli J-53 recipient; lane 9, controls Rts1 (84 kb), R1 (41 kb), and R6K (17 kb); lane 10, 2- to 10-kb ladder; lane 11, isolate CK2; lane 12, CK2 transconjugant resistant to ampicillin, sulfonamides, and tetracycline; lane 13, isolate 3219; lane 14, 3219 transconjugant A; lane 15, 3219 transconjugant resistant to ampicillin, sulfonamides, and trimethoprim.
Several isolates transferred resistance determinants to nalidixic acid-resistant strain J-53 when they were cocultivated (Table 2). Plasmids were extracted from the transconjugants and run in tandem with their source isolates. The transfer of isolated ampicillin resistance was associated with a single plasmid of 33 kb (isolate 3219) or 48 kb (isolate 3331) in two isolates; however, transfer of combined resistance to ampicillin, sulfonamides, and trimethoprim from isolate 3219 in association with a 47-kb plasmid was also observed. These results suggest the presence of a determinant of ampicillin resistance on two distinct plasmids in isolate 3219.
TABLE 2.
Results of liquid mating conjugation of S. sonnei isolatesa
| Isolate no.b | Resistance profilec | Transconjugant resistance profilec | Selective agard | Size (kb) of transconjugant plasmid(s) |
|---|---|---|---|---|
| 3331Ab | ASSu | ASSuNa | A + Na | 2,12, and 48 |
| 3331Ab | ASSu | ANa | A + Na | 48 |
| 3599Ab | A | ANa | A + Na | 11 and 45 |
| CK2Bi | ASSuTTm | ASuTNa | A + Na | 5 and 60 |
| 3219Ab | ASuTm | ANa | A + Na | 33 |
| 3219Ab | ASuTm | ASuTmNa | Tm + Na | 47 |
| D3B3k | SSuTTm | SSuTNa | T + Na | 60 |
No transconjugants were obtained with isolate 3331, CK2, or D3 when streptomycin (30 mg/ml) and nalidixic acid (30 μg/ml) were used. No transconjugants were obtained with isolate CK2 or D3 when Mueller-Hinton agar with nalidixic acid (30 μg/ml) and trimethoprim (5 μg/ml) were used.
Superscript letters indicate the PFGE type (upper case letters) and plasmid profile pattern (lowercase letters) for each strain.
The resistance profiles are represented by the abbreviations for the antimicrobial agents to which the isolate tested was resistant: A, ampicillin; S, streptomycin; Su, sulfonamides; T, tetracycline; Tm, trimethoprim.
Isolates were selected on Tryptone soya agar with nalidixic acid (30 μg/ml) and ampicillin (100 μg/ml) (A + Na) or tetracycline (20 μg/ml) (T + Na) or Mueller-Hinton agar with nalidixic acid (30 μg/ml) and trimethoprim (5 μg/ml) (Tm + Na).
Two different class 2 integron structures were identified in the S. sonnei isolates in this subset of the collection. Thirteen isolates contained a 2.22-kbp gene cassette identified by DNA sequence analysis as containing three open reading frames (including dhfr1, sat, and aad). This gene array is similar to that found in the promiscuous transposon Tn7 (19). A single isolate (isolate 1778) produced a smaller cassette structure of 1.37 kbp encoding only two genes within the cassette (dhfr1 and sat). All isolates containing a class 2 integron were phenotypically resistant to trimethoprim and streptomycin (S), which can be accounted for by the resistance genes encoded within the gene cassettes. Possession of a class 2 integron and streptomycin and trimethoprim resistance was associated with the PFGE type B pattern. Isolate 3219, the only PFGE type A isolate resistant to trimethoprim, did not contain a class 2 integron. It is possible that in this case the trimethoprim resistance may be due to an alterative dhfr gene other than dhfr1 present on a plasmid (7). A single isolate (isolate D3) had a class 1 integron (1.95 kbp) containing two open reading frames (sat and aad) within the gene cassette, in addition to a class 2 integron (dhfr1, sat, and aad). Noticeably, this isolate had a unique PFGE pattern (PFGE pattern B3) and a unique plasmid profile (plasmid profile k). Analysis of gene cassettes of both integron classes did not identify any gene associated with resistance to ampicillin. No class 3 integrons were detected. Alignments of the deduced amino acid sequences (data not shown) did not reveal any unusual sequence substitutions, indicating a high degree of sequence conservation.
DISCUSSION
PFGE indicated the presence of just two predominant clusters, clusters A (n = 53) and B (n = 14), among S. sonnei isolates circulating in Ireland in 1998 and 1999. Plasmid profiles and results of integron analysis are consistent with this interpretation. The results obtained by phenotypic typing methods (phage typing and antimicrobial susceptibility testing) also support the existence of two distinct groups. The larger group, group A (which correlated with PT3), appears to be very homogeneous, with only one minor variant detected by PFGE. Greater diversity was observed among group B isolates (which mainly correlate with PT6).
Our study was intended to provide baseline data for the application of PFGE to the routine typing of S. sonnei isolates and to the investigation of possible common-source outbreaks of S. sonnei in Ireland. The 53 isolates that made up group A were primarily isolated from patients with no apparent connection from three counties over a period for 16 months. There was no indication of a common-source outbreak during the period when the isolates were collected. The findings may suggest person-to-person transmission of a particular strain over an extended period of time, although this interpretation must be regarded as tentative, given the lack of detailed case studies. A practical consequence of the relative homogeneity of S. sonnei in this region is that the detection of isolates from two patients indistinguishable by PFGE has a high probability of occurrence by chance alone and is not strongly supportive of a common-source food or waterborne outbreak. Indistinguishable plasmid profiles, in addition to indistinguishable PFGE types, appear to be more suggestive of a link between cases. The application of the standardized PulseNet protocol to representative isolates from this collection may allow comparison with other data to determine if S. sonnei isolates with similar PFGE patterns also occur elsewhere in the world and over time should permit us to determine if the periodic increases in the occurrence of S. sonnei observed in Ireland are associated with the emergence of new strains.
It appears that greater diversity exists among S. sonnei isolates in the United States, and the recognition of a group of isolates indistinguishable by PFGE may have greater public health significance in the United States. The application of PFGE (PulseNet protocol) to S. sonnei isolates from 16 people who had eaten at two separate restaurants in Los Angeles, Calif., during the summer of 1998 showed that the isolates were virtually identical to the strain associated with infection in Minnesota, Massachusetts, and Canada. A common source was identified as parsley grown on a Mexican farm which had been contaminated from tainted irrigation water (8).
The correlation of PFGE type with the results of phenotypic typing methods is useful for epidemiological purposes. The results of phage typing suggest some similarity between strains circulating in Ireland and those circulating in England and Wales. PT3 (which correlates with PFGE type A) was the most common phage type observed in England and Wales until it was recently displaced by PT6 (which correlates with PFGE type B), which now accounts for 80% of the S. sonnei isolates in England and Wales (T. Cheasty, unpublished data).
The association of specific antimicrobial resistance patterns with isolates of PFGE type A (ampicillin, streptomycin, and sulfonamide resistant) and PFGE type B (streptomycin, sulfonamide, tetracycline, and trimethoprim resistant) may provide a rapid and inexpensive initial approach to the categorization of isolates in the same way that pentaresistance to ampicillin, chloramphenicol, streptomycin, sulfonamides, and tetracycline is a useful marker for the DT104 strain of Salmonella enterica serovar Typhimurium. We are aware of reports of isolates with the streptomycin, sulfonamide, tetracycline, and trimethoprim resistance phenotype from a number of countries (2, 19). The resistance of S. sonnei strains to commonly used antimicrobial agents has been noted in several countries (Brazil [16], Hong Kong [17], the United States [3], Israel [1], and the United Kingdom).
An efficient route of acquisition and dissemination of resistance determinants is through mobile elements including plasmids, transposons, and gene cassettes in integrons. Conjugative plasmids encoding resistance to antibiotics have been detected in numerous studies of S. sonnei. All isolates tested in this study contained plasmids, and several of the isolates transferred resistance determinants to E. coli in conjugation studies. The isolates included the predominant PFGE type A strain (ampicillin, streptomycin, and sulfonamide resistant), which transferred both ampicillin resistance alone (in association with a 48-kb plasmid) and ampicillin, streptomycin, and sulfonamide resistance (in association with the cotransfer of plasmids of 2, 12, and 48 kb). Ampicillin, sulfonamide, and tetracycline resistance was cotransferred with a 5- or 60-kb plasmid in one isolate (isolate CK2), while ampicillin, sulfonamide, and trimethoprim resistance was encoded by a 47-kb plasmid in isolate 3219. Interestingly, a 33-kb plasmid in this isolate transferred ampicillin resistance alone.
It was not possible to examine all isolates for plasmids and integrons, which limits the strength of our conclusions in this regard. Of the 24 isolates examined for the presence of integrons, only one (isolate D3) contained a class 1 integron structure. This isolate represented a unique variant within the PFGE type B cluster (PFGE type B3), was PT50, and also had a unique plasmid profile (plasmid profile k). All 14 PFGE type B isolates tested contained a class 2 integron; however, class 2 integrons were not detected in any of 10 PFGE type A isolates tested. In conjugation experiments, the cotransfer of streptomycin and trimethoprim resistance was not observed, which may suggest that the class 2 integron was not on a conjugative plasmid. Future work to confirm the location of the integron as the plasmid or chromosome is planned. Streptomycin and trimethoprim were introduced in the 1950s as alternative treatments for infections caused by sulfonamide-resistant Shigella species, and the presence of class 2 integrons may account for the rapid evolution of S. sonnei phenotypes resistant to these antimicrobials. Streptomycin resistance is strongly associated with integrons because of the high prevalence of aadA cassettes within both class 1 and class 2 integrons.
The majority of isolates studied belong to one PFGE group (PFGE type A), and there was a good correlation between PFGE, phage typing, and antimicrobial resistance patterns. There was a high rate of resistance to antimicrobial agents to which resistance is apparently associated with conjugative plasmids or integrons. It would be interesting to analyze isolates from other countries to extend our observations.
REFERENCES
- 1.Ashkenazi, S., M. May-Zahav, J. Sulkes, R. Zilberberg, and Z. Samra. 1995. Increasing antimicrobial resistance of Shigella isolates in Israel during the period 1984 to 1992. Antimicrob. Agents Chemother. 39:819-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aysev, A. D., and H. Guriz. 1998. Drug resistance of Shigella strains isolated in Ankara, Turkey, 1993-1996. Scand. J. Infect. Dis. 30:351-353. [DOI] [PubMed] [Google Scholar]
- 3.Barg, N. L., S. Register, C. Thomson, and S. Amyes. 1995. Sequence identity with type VIII and association with IS176 of type IIIc dihydrofolate reductase from Shigella sonnei. Antimicrob. Agents Chemother. 39:112-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradley, D. E., D. E. Taylor, and D. R. Cohen. 1980. Specification of surface mating systems among conjugative drug-resistant plasmids in Escherichia coli K-12. J. Bacteriol. 143:1466-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carattoli, A. 2001. Importance of integrons in the diffusion of resistance. Vet. Res. 32:243-259. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 2000. Standardized molecular subtyping of foodborne bacterial pathogens by pulsed-field gel electrophoresis, training manual. Centers for Disease Control and Prevention, Atlanta, Ga.
- 7.Chumg-Yu, C., C. Lin-Li, C. Yu-Hung, L. Tsong-Ming, L. Yuan-Hung, and C. Shui-Feng. 2000. Two new gene cassettes, dfr17 (for trimethoprim resistance) and aadA4 (for spectinomycin/streptomycin resistance), inserted in an Escherichia coli class 1 integron. J. Antimicrob. Chemother. 46:87-89. [DOI] [PubMed] [Google Scholar]
- 8.Cimmons, M. 2000. Rapid food-borne pathogen ID system is making a difference. ASM News 66:617. [Google Scholar]
- 9.Cormican, M. N. DeLappe, C. O'Hare, G. Doran, D. Morris, G. Corbett-Feeney, S. Fanning, M. Daly, M. Fitzgerald, and J. Moore. 2002. Salmonella enterica serotype Bredeney: antimicrobial susceptibility and molecular diversity of isolates from Ireland and Northern Ireland. Appl. Environ. Microbiol. 68:181-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daly, M., J. Buckley, E. Power, C. O'Hare, M. Cormican, B. Cryan, P. G. Wall, and S. Fanning. 2000. Molecular characterization of Irish Salmonella enterica serotype Typhimurium: detection of class 1 integrons and assessment of genetic relationships by DNA amplification fingerprinting. Appl. Environ. Microbiol. 66:614-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DuPont, H. L., M. M. Levine, and R. B. Hornick. 1989. Inoculum size in shigellosis and implications for expected mode of transmission. J. Infect. Dis. 159:1126.. [DOI] [PubMed] [Google Scholar]
- 12.Hyams, K. C., A. L. Bourgeois, B. R. Merrell, R. Rozmajzi, J. Escamilla, S. A. Thornton, G. M. Wasserman, A. Burke, P. Echeverria, K. Y. Green, A. Z. Kapikian, and J. N. Woody. 1991. Diarrheal disease during Operation Desert Shield. N. Engl. J. Med. 325:1423-1428. [DOI] [PubMed] [Google Scholar]
- 13.Guerra, B., S. Soto, S. Cal, and M. Carmen-Mendoza. 2000. Antimicrobial resistance and spread of class 1 integrons among Salmonella serotypes. Antimicrob. Agents Chemother. 44:2166-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kado, C. L., and S. T. Liu. 1981. Rapid procedure for detection of large and small plasmids. J. Bacteriol. 145:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levesque, C., L. Pyche. C. Larose, and P. H. Roy. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicron. Agents Chemother. 39:185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lima, A. A. M., N. L. Lima, M. C. N. Pinho, E. A. Barros, Jr., M. J. Teixeira, M. C. V. Martins, and R. L. Guerrant. 1995. High frequency of strains multiply resistant to ampicillin, trimethoprim-sulfamethoxazole, streptomycin, chloramphenicol, and tetracycline isolated from patients with shigellosis in northeastern Brazil during the period 1988 to 1993. Antimicrob. Agents Chemother. 39:256-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ling, J. M., P. C. Shaw, K. M. Kam, A. F. Cheng, and G. L. French. 1993. Molecular studies of plasmids of multiply-resistant Shigella spp. in Hong Kong. Epidemiol. Infect. 110:437-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu, P. Y., Y. J. Lau, B. S. Hu, J. M. Shyr, Z. Y. Shi, W. S. Tsai, Y. H. Lin, and C. Y. Tseng. 1995. Analysis of clonal relationships among isolates of Shigella sonnei by different molecular typing methods. J. Clin. Microbiol. 3:1779-1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McIver, C. J., P. A. White, L. A. Jones, T. Karagiannis, J. Harkness, D. Marriott, and W. D. Rawlinson. 2002. Epidemic strains of Shigella sonnei biotype g carrying integrons. J. Clin. Microbiol. 40:1538-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.National Committee for Clinical Laboratory Standards. 1997. Performance standards for antimicrobial disk susceptibility tests, 6th ed. Approved standard M2-A6. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 21.Navia, M. M., L. Capitano, J. Ruiz, M. Vargas, H. Urassa, D. Schellemberg, J. Gascon, and J. Vila. 1999. Typing and characterization of mechanisms of resistance of Shigella spp. isolated from feces of children under 5 years of age from Ifakara, Tanzania. J. Clin. Microbiol. 37:3113-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Pershing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Versalovic, J., T. Koeuth, and J. R. Lupski. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 24:6823-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wantabe, T. 1963. Infectious heredity of multiple drug resistance in bacteria. Bacteriol. Rev. 27:87-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.White, P. A., C. J. McIver, and W. D. Rawlinson. 2001. Integrons and gene cassettes in the Enterobacteriaceae. Antimicrob. Agents Chemother. 45:2658-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]