Abstract
One hundred ninety-three Streptococcus agalactiae isolates of neonatal origin and 146 isolates from adult women were analyzed for macrolide resistance and investigated for clonality. Among erythromycin-resistant isolates, serotype V turned out to be the most frequent. Comparative pulsed-field gel electrophoresis analysis revealed genetic clustering of resistant strains and predominance of a single clone family within an otherwise heterogeneous serotype V population.
Streptococcus agalactiae (group B Streptococcus [GBS]) is the leading cause of meningitis and early-onset sepsis in neonates (3, 21). Penicillin or ampicillin is the drug of choice for prophylaxis and treatment of GBS disease (1, 5). However, erythromycin and clindamycin are the recommended second-line drugs and the first alternative in case of a β-lactam allergy. In recent years, increasing macrolide resistance rates have been reported (9, 15, 16, 17, 19). Resistance mechanisms include target site modification mediated by erm genes leading to resistance to macrolides-lincosamides-streptogramin B (MLS phenotype) with either inducible or constitutive expression or an efflux mechanism encoded by mef genes mediating resistance only to 14- and 15-membered macrolides (M phenotype). After the first description of serotype V in 1977 (26), several studies reported a major increase of serotype V GBS among clinical isolates in the early 1990s (8, 13). To analyze the recent increase in macrolide resistance among GBS isolates in our region, the present study aimed to explore the issue of a clonal spread of resistant strains by means of randomly amplified polymorphic DNA (RAPD) analysis and restriction digest patterns (RDP) derived from pulsed-field gel electrophoresis (PFGE). Moreover, a possible correlation between the increase of a certain serotype and macrolide resistance was explored.
The bacterial strains used in this study have been presented previously (20). In brief, of 193 neonatal GBS strains, 26 invasive isolates were cultured from blood or cerebrospinal fluid while 167 isolates were grown from urine samples and swab cultures. A total of 146 GBS isolates from vaginal swabs of pregnant women were randomly collected. Serotyping was performed by using an enzymatic extraction method (2). All erythromycin-resistant isolates were screened by PCR analysis for erythromycin resistance genes (mefA or mefE, ermB, and ermTR) by using primers previously described (7, 23, 24). PCR assays were reproducibly repeated at least three times for every strain. Differentiation of specific MLS phenotypes was performed by use of a triple-disk test (10). For RAPD analysis, the core sequence of phage M13 and the previously described primer OPS16 were used (6, 12) and visual inspection of bands served to compare RAPD patterns for similarity. Different isolates with the same migration distances of all visible bands were considered highly similar. The presence or absence of two distinct bands was deemed to indicate a difference. Chromosomal DNA of GBS strains was prepared by modifications of a previously described method (22). Macrorestriction fragments were resolved by PFGE. Lambda ladder PFG marker and low-range PFG marker (New England Biolabs, Frankfurt, Germany) served as standards. Restriction digest patterns (RDPs) were visually analyzed by following the criteria offered by Tenover et al. (25).
Results of serotyping experiments are shown in Table 1. Serotype V was more frequently isolated in recent years. Among isolates of maternal origin, serotype V was found in 16 (16.7%) isolates from 1999 but in only 3 (6%) isolates from 1997. Among isolates of neonatal origin, serotype V was identified in 12 isolates (6.2%). Nine of these were isolated in the years 1998 and 1999, representing 19% of all isolates from this time period. Among all erythromycin-resistant isolates, GBS serotype V was predominant, accounting for 10 of 27 isolates (37%) (5 of 16 [31.6%] of maternal origin and 5 of 11 [45.5%] of neonatal origin). Obversely, 41.7% (5 of 12) of neonatal isolates and 26.3% (5 of 19) of maternal serotype V isolates were resistant to erythromycin.
TABLE 1.
Distribution of serotypes among GBS isolates
| Serotype | No. of isolates
|
|||
|---|---|---|---|---|
| Neonatal
|
Maternal
|
|||
| Erythromycin resistant | Total | Erythromycin resistant | Total | |
| Ia | 0 | 36 | 2 | 26 |
| Ib | 0 | 17 | 4 | 11 |
| II | 3 | 31 | 1 | 22 |
| III | 3 | 77 | 4 | 43 |
| IV | 0 | 2 | 0 | 4 |
| V | 5 | 12 | 5 | 19 |
| NTa | 0 | 18 | 0 | 21 |
| Total | 11 | 193 | 16 | 146 |
NT, nontypeable.
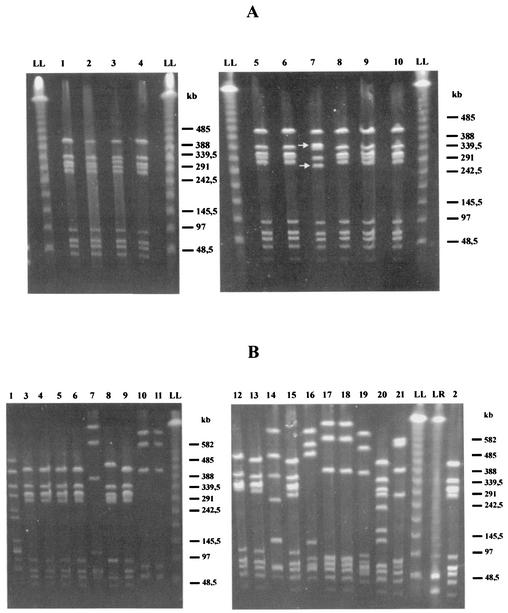
To determine the specific resistance mechanisms, PCR assays for all erythromycin-resistant isolates were performed, and the results showed predominance of erm genes in serotype V isolates (Table 2). Investigation of the erythromycin-resistant GBS strains by use of the M13 and OPS16 primers resulted in patterns that allowed the identification of genetically distinct subtypes. Using primer M13, visual differentiation resulted in 12 major fingerprinting patterns with 7 to 13 bands ranging from approximately 400 to 3,000 bp. Nine of 10 erythromycin-resistant serotype V isolates showed identical RAPD patterns, and 1 isolate had a single additional band. While serotype Ib isolates also revealed relatively homogenous patterns, serotype Ia, II, and III isolates could not be clustered. A PCR assay performed with primer OPS16 generated a lower number of amplification products and had less discriminatory power (data not shown). To confirm the above-mentioned RAPD results and for further exploration of the indicated clonal relatedness of the 10 erythromycin-resistant serotype V GBS strains, PFGE typing was performed. Analysis of SmaI-digested genomic DNA of all 10 erythromycin-resistant GBS serotype V isolates revealed nine identical PFGE patterns. One isolate showed a different size for two bands, indicating modification of a single SmaI restriction site on the chromosome only (Fig. 1A). The same isolate showed an additional amplification product in the M13 PCR. Thus, all resistant isolates turned out to be closely related.
TABLE 2.
Distribution of specific resistance genes among 27 erythromycin-resistant GBS isolatesa
| Serotype | No. of isolates carrying resistance gene:
|
||
|---|---|---|---|
| ermB | ermTR | mefA/mefB | |
| Ia | 0 | 0 | 2 |
| Ib | 3 | 3 | 1 |
| II | 1 | 2b | 1 |
| III | 2 | 1b | 4 |
| IV | 0 | 0 | 0 |
| V | 8 | 2b | 0 |
| Total | 13 | 8 | 8 |
Three serotype Ib isolates and one serotype III isolate harbored both the ermB and the ermTR genes.
Isolates revealing an inducible phenotype.
FIG. 1.
PFGE of genomic DNA of erythromycin-resistant (A) and sensitive (B) serotype V GBS isolates digested with SmaI. Lambda ladder PFG marker (LL) and low-range PFG marker (LR) were used as standards. (A) SmaI patterns of 10 erythromycin-resistant serotype V isolates, with the isolate in lane 7 showing different sizes for only two bands. (B) Lanes 1 to 21 represent SmaI digest patterns of 21 different erythromycin-sensitive serotype V GBS isolates of neonatal and maternal origins, respectively. Nine distinct RDPs were identified: pattern A (lane 1), pattern B (lanes 2 to 6, 8, 9, 12, 13, and 15), pattern C (lanes 7 and 19), pattern D (lanes 10 and 11), pattern E (lane 14), pattern F (lane 16), pattern G (lanes 17 and 18), pattern H (lane 20), and pattern I (lane 21).
To further investigate possible genetic clustering within the serotype V GBS population with regard to macrolide resistance, PFGE analysis was extended to 21 erythromycin-sensitive serotype V isolates of neonatal and maternal origins. SmaI digests of genomic DNA of these 21 sensitive isolates reproducibly gave rise to nine distinct RDPs (A to I) consisting of 8 to 11 SmaI fragments. This analysis revealed diversity within GBS serotype V as well as the striking predominance of one specific pattern (Fig. 1B). The most common PFGE pattern was B, accounting for 48% (10 of 21) of all isolates and showing two subtypes. RDPs C, D, and G were shared by two isolates each, whereas patterns A, E, F, H, and I were each represented by a single isolate. Compared to the RDPs found within the group of erythromycin-sensitive isolates, the patterns of the resistant bacteria could all be assigned to the most common B pattern of the sensitive strains. The differing resistant isolate added another subtype to this group.
The data obtained from the present study reveal that the frequency of resistant strains was related to the capsular serotype. Serotype V accounted for a high percentage of erythromycin-resistant isolates of maternal as well as neonatal origin. These observations confirm the findings of some recent studies reporting that 30 to 35% of erythromycin-resistant GBS isolates of different origins belong to serotype V (9, 15). Using two primers for arbitrarily primed-PCR-based fingerprinting of erythromycin-resistant GBS isolates, our study revealed that two major clusters were formed by isolates of serotype V and Ib whereas the remaining serotypes showed different patterns. Remarkably, 9 of 10 serotype V isolates showed identical fingerprinting patterns and the remaining isolate differed by a single band only. These results, indicating possible genetic clustering of a macrolide-resistant clone family within the serotype V population, led to the initiation of further typing analysis by PFGE. This method has been demonstrated by several investigators to be a powerful tool for genomic characterization, epidemiological analysis, and subtyping of GBS (4, 8, 11, 18). Different restriction endonucleases have been used, with SmaI becoming the best-established one for GBS typing (11, 18, 25). PFGE analysis of SmaI-digested genomic DNA of all erythromycin-resistant GBS serotype V isolates provided strong evidence for a very close molecular relatedness, too. Extension of PFGE analysis to 21 erythromycin-sensitive serotype V isolates revealed diversity within the GBS serotype V population as well as one predominant RDP (pattern B) accounting for 48% of all sensitive serotype V isolates (10 of 21). These findings are in good agreement with data published in previous studies (8, 14). Even though direct comparative analysis of our isolates with the predominant U.S. or French serotype V GBS strains was not performed in this work, the B pattern obtained from the majority of our isolates seems to be very similar to the pattern published as being predominant in the United States and France. Evaluating these findings, we suggest the emergence of a specific macrolide-resistant clone family that possibly acquired resistance at a certain point of evolution and then subsequently increased in numbers. This observation is of significant epidemiological impact. The emergence of a new clone family that carries resistance to an antimicrobial agent that is widely used in prophylaxis and therapy may have major consequences for clinical practice not only in a given region but also beyond regional frontiers.
REFERENCES
- 1.American Academy of Pediatrics. Committee on Infectious Disease and Committee on Fetus and Newborn. 1997. Revised guidelines for prevention of early-onset group B streptococcal (GBS) infection. Pediatrics 99:489-496. [DOI] [PubMed] [Google Scholar]
- 2.Berner, R., A. Bender, C. Rensing, J. Forster, and M. Brandis. 1999. Low prevalence of the immunoglobulin-A-binding β antigen of the C protein among Streptococcus agalactiae isolates causing neonatal sepsis. Eur. J. Clin. Microbiol. Infect. Dis. 18:545-550. [DOI] [PubMed] [Google Scholar]
- 3.Berner, R., R. F. Schumacher, S. Bartelt, J. Forster, and M. Brandis. 1998. Bacteremia in hospitalized children: predisposing conditions and case-related microorganisms. Eur. J. Clin. Microbiol. Infect. Dis. 17:337-340. [DOI] [PubMed] [Google Scholar]
- 4.Bohnsack, J. F., A. A. Whiting, R. D. Bradford, K. B. Van Frank, S. Takahashi, and E. E. Adderson. 2002. Long-range mapping of the Streptococcus agalactiae phylogenic lineage restriction digest pattern type III-3 reveals clustering of virulence genes. Infect. Immun. 70:134-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 1996. Prevention of perinatal group B streptococcal disease: a public health perspective. Morb. Mortal. Wkly. Rep. 45:1-24. [PubMed] [Google Scholar]
- 6.Chatellier, S., C. Ramanantsoa, P. Harriau, K. Rolland, A. Rosenau, and R. Quentin. 1997. Characterization of Streptococcus agalactiae strains by randomly amplified polymorphic DNA analysis. J. Clin. Microbiol. 35:2573-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clancy, J., J. Petitpas, F. Dib-Hajj, W. Yuan, M. Cronan, A. V. Kamath, J. Bergeron, and J. A. Retsema. 1998. Molecular cloning and functional analysis of a novel macrolide-resistance determinant, mefA, from Streptococcus pyogenes. Mol. Microbiol. 42:1493-1494. [DOI] [PubMed] [Google Scholar]
- 8.Elliot, J. A., K. D. Farmer, and R. R. Facklam. 1998. Sudden increase in isolation of group B streptococci, serotype V, is not due to emergence of a new pulsed-field gel electrophoresis type. J. Clin. Microbiol. 36:2115-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fernandez, M., M. E. Hickman, and C. J. Baker. 1998. Antimicrobial susceptibility of group B streptococci isolated between 1992 and 1996 from patients with bacteremia and meningitis. Antimicrob. Agents Chemother. 42:1517-1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giovanetti, E., M. P. Montanari, M. Mingoia, and P. E. Varaldo. 1999. Phenotypes and genotypes of erythromycin-resistant Streptococcus pyogenes strains in Italy and heterogeneity of inducibly resistant strains. Antimicrob. Agents Chemother. 43:1935-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gordillo, M. E., K. V. Singh, C. J. Baker, and B. E. Murray. 1993. Typing of group B streptococci: comparison of pulsed-field gel electrophoresis and conventional electrophoresis. J. Clin. Microbiol. 31:1430-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gräser, Y., I. Klare, E. Halle, R. Gantenberg, P. Buchholz, H. D. Jacobi, W. Presber, and G. Schönian. 1993. Epidemiological study of an Acinetobacter baumannii outbreak by using polymerase chain reaction fingerprinting. J. Clin. Microbiol. 31:2417-2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harrison, L. H., D. M. Dwyer, and J. A. Johnson. 1995. Emergence of serotype V group B streptococcal infection among infants and adults. J. Infect. Dis. 171:513.. [DOI] [PubMed] [Google Scholar]
- 14.Le Thomas-Bories, I., F. Fitoussi, P. Mariani-Kurkdjian, J. Raymond, N. Brahimi, P. Bidet, V. Lefranc, and E. Bingen. 2001. Clonal relationship between U.S. and French serotype V group B streptococcus isolates. J. Clin. Microbiol. 39:4526-4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin, F. Y. C., P. H. Azimi, L. E. Weisman, J. B. Philips III, J. Regan, P. Clark, C. G. Rhoads, J. Clemens, J. Troendle, E. Pratt, R. A. Brenner, and V. Gill. 2000. Antibiotic susceptibility profiles for group B streptococci isolated from neonates, 1995-1998. Clin. Infect. Dis. 31:76-79. [DOI] [PubMed] [Google Scholar]
- 16.Morales, W. J., S. S. Dickey, P. Bornick, and D. V. Lim. 1999. Change in antibiotic resistance of group B streptococcus: impact on intrapartum management. Am. J. Obstet. Gynecol. 181:310-314. [DOI] [PubMed] [Google Scholar]
- 17.Pearlman, M. D., C. L. Piersoni, and R. G. Faix. 1998. Frequent resistance of clinical group B streptococci isolates to clindamycin and erythromycin. Obstet. Gynecol. 92:258-261. [DOI] [PubMed] [Google Scholar]
- 18.Rolland, K., C. Marois, V. Siquier, B. Cattier, and R. Quentin. 1999. Genetic features of Streptococcus agalactiae strains causing neonatal infections, as revealed by pulsed-field gel electrophoresis and hlyB gene analysis. J. Clin. Microbiol. 37:1892-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rouse, D. J., W. W. Andrews, F. Y. C. Lin, C. W. Mott, J. C. Ware, and J. B. Philips III. 1998. Antibiotic susceptibility profile of group B streptococcus acquired vertically. Obstet. Gynecol. 92:931-934. [DOI] [PubMed] [Google Scholar]
- 20.Ruess, M., U. Müller, A. Sander, and R. Berner. 2000. Antimicrobial susceptibility patterns of Streptococcus agalactiae in a German university hospital. Scand. J. Infect. Dis. 32:623-626. [DOI] [PubMed] [Google Scholar]
- 21.Schrag, S. J., S. Zywicki, M. M. Farley, A. L. Reingold, L. H. Harrison, L. B. Lefkowitz, J. L. Hadler, R. Danila, P. R. Cieslak, and A. Schuchat. 2000. Group B streptococcal disease in the era of intrapartum prophylaxis. N. Engl. J. Med. 342:15-20. [DOI] [PubMed] [Google Scholar]
- 22.Schwarzkopf, A., C. Cuny, and W. Witte. 1995. Bestimmung der Fragmentmuster der genomischen DNA mittels Pulsfeld-Gelelektrophorese bei Staphylococcus aureus. Bundesgesundheitsblatt 38:215-219. [Google Scholar]
- 23.Seppälä, H., M. Skurnik, H. Soini, M. C. Roberts, and P. Huovinen. 1998. A novel erythromycin resistance methylase gene (ermTR) in Streptococcus pyogenes. Antimicrob. Agents Chemother. 42:257-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sutcliffe, J., T. Grebe, T. Tait-Kamradt, and L. Wondrack. 1996. Detection of erythromycin-resistant determinants by PCR. Antimicrob. Agents Chemother. 40:2562-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilkinson, H. W. 1977. Nontypable group B-streptococci isolated from human sources. J. Clin. Microbiol. 6:183-184. [DOI] [PMC free article] [PubMed] [Google Scholar]