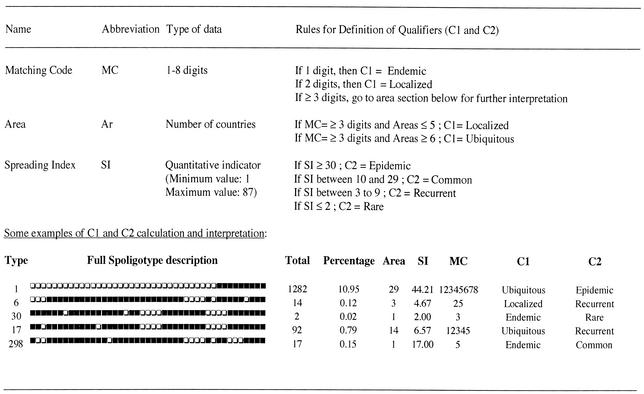
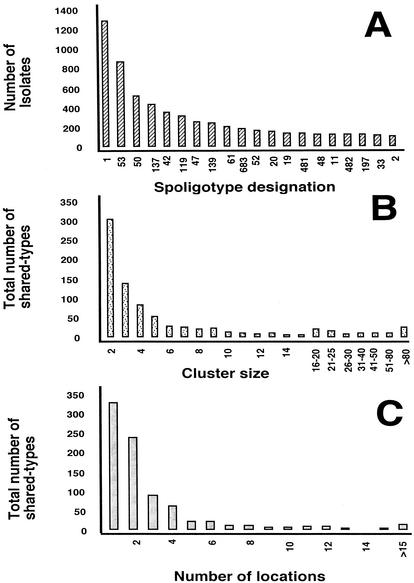
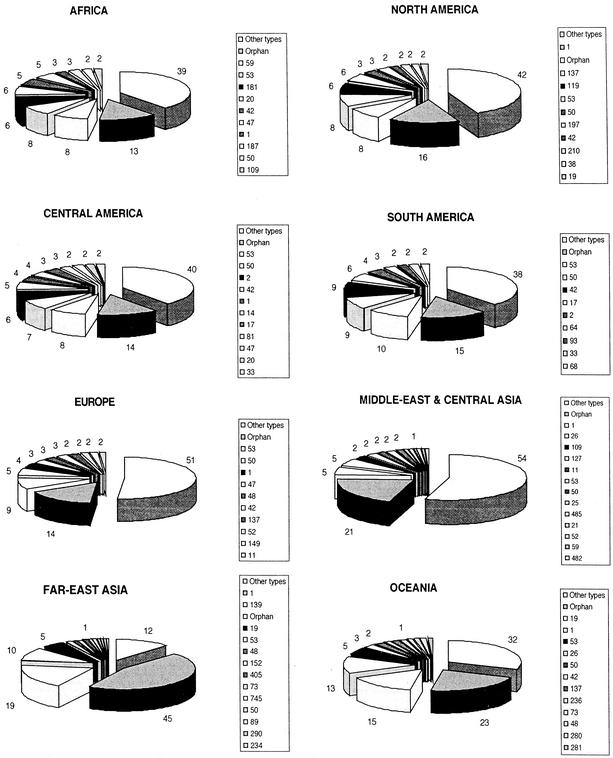
Ingrid Filliol
Ingrid Filliol
Unité de la Tuberculose et des Mycobactéries, Institut Pasteur de Guadeloupe, Pointe-à-Pitre, Guadeloupe,1 Wadsworth Center, New York State Department Of Health, Albany New York,2 Public Health Research Institute, Tuberculosis Center, Newark, New Jersey,4 Diagnosis Laboratory for Infectious Diseases and Perinatal Screening RIVM, Bilthoven, The Netherlands,3 National Institute of Hygiene and Epidemiology, Hanoi, Vietnam,5 Department of Microbiology, Leeds General Infirmary, Leeds,6 Medical School, St Georges Hospital,7 Mycobacterium Reference Unit, PHLS, Dulwich Hospital, London,13 University of Surrey, Guildford, Surrey, United Kingdom,11 Instituto de Biotecnologia INTA, Moron, Argentina,8 Tuberculosis Mycobacteriology Branch, Centers for Disease Control and Prevention, Atlanta, Georgia,9 Australian Reference Laboratory for Bovine Tuberculosis, Department of Agriculture, South Perth, Australia,10 Centro de Biotecnologia, Universidade Federal de Pelotas, Pelotas,12 Universidade Federal do Rio Grande do Sul, Porto Alegre,27 Department of Biochemistry and Molecular Biology, Fiocruz, Oswaldo Cruz Institute, Rio de Janeiro, Brazil,29 University Childrens Hospital, Heidelberg,14 Infektions Epidemiology, Robert Koch Institute, Berlin,16 Bundesinstitut für Gesundheitlichen Verbraucherschutz und Veterinärmedizin, Jena, Germany,20 Centre National de Référence des Mycobactéries, Institut Pasteur, Paris,15 Laboratoire de Bactériologie, CHU de Brest, Brest,22 Laboratoire de Bactériologie, CHU de Bordeaux, Bordeaux, France,31 Institut Pasteur de Bangui, Bangui, Central African Republic,17 Klinische Mikrobiologie, Hygiene-Institut der Universität,18 Bundesstaatliche Bakteriologisch-Serologische Untersuchungsanstalt Wien, Vienna, Austria,28 Department of Hygiene and Microbiology, University of Palermo, Palermo,19 Department of Public Health, University of Florence, Florence, Italy,23 Laboratory of Molecular Microbiology, Saint Petersburg Pasteur Institute, Saint Petersburg, Russia,21 Les Centres Gheskio, INLR, Port au Prince, Haïti,24 Institut Pasteur de Madagascar, Tananarive, Madagascar,25 Institute of Medical Microbiology and Immunology, Göteborg University, Göteborg, Sweden,26 IVIC, Centro de Microbiología y Biología Celular, Laboratorio de Genética Molecular,30 Tuberculosis Laboratory, Instituto de Biomedicina, Caracas, Venezuela32
1,
Jeffrey R Driscoll
Jeffrey R Driscoll
Unité de la Tuberculose et des Mycobactéries, Institut Pasteur de Guadeloupe, Pointe-à-Pitre, Guadeloupe,1 Wadsworth Center, New York State Department Of Health, Albany New York,2 Public Health Research Institute, Tuberculosis Center, Newark, New Jersey,4 Diagnosis Laboratory for Infectious Diseases and Perinatal Screening RIVM, Bilthoven, The Netherlands,3 National Institute of Hygiene and Epidemiology, Hanoi, Vietnam,5 Department of Microbiology, Leeds General Infirmary, Leeds,6 Medical School, St Georges Hospital,7 Mycobacterium Reference Unit, PHLS, Dulwich Hospital, London,13 University of Surrey, Guildford, Surrey, United Kingdom,11 Instituto de Biotecnologia INTA, Moron, Argentina,8 Tuberculosis Mycobacteriology Branch, Centers for Disease Control and Prevention, Atlanta, Georgia,9 Australian Reference Laboratory for Bovine Tuberculosis, Department of Agriculture, South Perth, Australia,10 Centro de Biotecnologia, Universidade Federal de Pelotas, Pelotas,12 Universidade Federal do Rio Grande do Sul, Porto Alegre,27 Department of Biochemistry and Molecular Biology, Fiocruz, Oswaldo Cruz Institute, Rio de Janeiro, Brazil,29 University Childrens Hospital, Heidelberg,14 Infektions Epidemiology, Robert Koch Institute, Berlin,16 Bundesinstitut für Gesundheitlichen Verbraucherschutz und Veterinärmedizin, Jena, Germany,20 Centre National de Référence des Mycobactéries, Institut Pasteur, Paris,15 Laboratoire de Bactériologie, CHU de Brest, Brest,22 Laboratoire de Bactériologie, CHU de Bordeaux, Bordeaux, France,31 Institut Pasteur de Bangui, Bangui, Central African Republic,17 Klinische Mikrobiologie, Hygiene-Institut der Universität,18 Bundesstaatliche Bakteriologisch-Serologische Untersuchungsanstalt Wien, Vienna, Austria,28 Department of Hygiene and Microbiology, University of Palermo, Palermo,19 Department of Public Health, University of Florence, Florence, Italy,23 Laboratory of Molecular Microbiology, Saint Petersburg Pasteur Institute, Saint Petersburg, Russia,21 Les Centres Gheskio, INLR, Port au Prince, Haïti,24 Institut Pasteur de Madagascar, Tananarive, Madagascar,25 Institute of Medical Microbiology and Immunology, Göteborg University, Göteborg, Sweden,26 IVIC, Centro de Microbiología y Biología Celular, Laboratorio de Genética Molecular,30 Tuberculosis Laboratory, Instituto de Biomedicina, Caracas, Venezuela32
2,
Dick van Soolingen
Dick van Soolingen
Unité de la Tuberculose et des Mycobactéries, Institut Pasteur de Guadeloupe, Pointe-à-Pitre, Guadeloupe,1 Wadsworth Center, New York State Department Of Health, Albany New York,2 Public Health Research Institute, Tuberculosis Center, Newark, New Jersey,4 Diagnosis Laboratory for Infectious Diseases and Perinatal Screening RIVM, Bilthoven, The Netherlands,3 National Institute of Hygiene and Epidemiology, Hanoi, Vietnam,5 Department of Microbiology, Leeds General Infirmary, Leeds,6 Medical School, St Georges Hospital,7 Mycobacterium Reference Unit, PHLS, Dulwich Hospital, London,13 University of Surrey, Guildford, Surrey, United Kingdom,11 Instituto de Biotecnologia INTA, Moron, Argentina,8 Tuberculosis Mycobacteriology Branch, Centers for Disease Control and Prevention, Atlanta, Georgia,9 Australian Reference Laboratory for Bovine Tuberculosis, Department of Agriculture, South Perth, Australia,10 Centro de Biotecnologia, Universidade Federal de Pelotas, Pelotas,12 Universidade Federal do Rio Grande do Sul, Porto Alegre,27 Department of Biochemistry and Molecular Biology, Fiocruz, Oswaldo Cruz Institute, Rio de Janeiro, Brazil,29 University Childrens Hospital, Heidelberg,14 Infektions Epidemiology, Robert Koch Institute, Berlin,16 Bundesinstitut für Gesundheitlichen Verbraucherschutz und Veterinärmedizin, Jena, Germany,20 Centre National de Référence des Mycobactéries, Institut Pasteur, Paris,15 Laboratoire de Bactériologie, CHU de Brest, Brest,22 Laboratoire de Bactériologie, CHU de Bordeaux, Bordeaux, France,31 Institut Pasteur de Bangui, Bangui, Central African Republic,17 Klinische Mikrobiologie, Hygiene-Institut der Universität,18 Bundesstaatliche Bakteriologisch-Serologische Untersuchungsanstalt Wien, Vienna, Austria,28 Department of Hygiene and Microbiology, University of Palermo, Palermo,19 Department of Public Health, University of Florence, Florence, Italy,23 Laboratory of Molecular Microbiology, Saint Petersburg Pasteur Institute, Saint Petersburg, Russia,21 Les Centres Gheskio, INLR, Port au Prince, Haïti,24 Institut Pasteur de Madagascar, Tananarive, Madagascar,25 Institute of Medical Microbiology and Immunology, Göteborg University, Göteborg, Sweden,26 IVIC, Centro de Microbiología y Biología Celular, Laboratorio de Genética Molecular,30 Tuberculosis Laboratory, Instituto de Biomedicina, Caracas, Venezuela32
3,
Barry N Kreiswirth
Barry N Kreiswirth
Unité de la Tuberculose et des Mycobactéries, Institut Pasteur de Guadeloupe, Pointe-à-Pitre, Guadeloupe,1 Wadsworth Center, New York State Department Of Health, Albany New York,2 Public Health Research Institute, Tuberculosis Center, Newark, New Jersey,4 Diagnosis Laboratory for Infectious Diseases and Perinatal Screening RIVM, Bilthoven, The Netherlands,3 National Institute of Hygiene and Epidemiology, Hanoi, Vietnam,5 Department of Microbiology, Leeds General Infirmary, Leeds,6 Medical School, St Georges Hospital,7 Mycobacterium Reference Unit, PHLS, Dulwich Hospital, London,13 University of Surrey, Guildford, Surrey, United Kingdom,11 Instituto de Biotecnologia INTA, Moron, Argentina,8 Tuberculosis Mycobacteriology Branch, Centers for Disease Control and Prevention, Atlanta, Georgia,9 Australian Reference Laboratory for Bovine Tuberculosis, Department of Agriculture, South Perth, Australia,10 Centro de Biotecnologia, Universidade Federal de Pelotas, Pelotas,12 Universidade Federal do Rio Grande do Sul, Porto Alegre,27 Department of Biochemistry and Molecular Biology, Fiocruz, Oswaldo Cruz Institute, Rio de Janeiro, Brazil,29 University Childrens Hospital, Heidelberg,14 Infektions Epidemiology, Robert Koch Institute, Berlin,16 Bundesinstitut für Gesundheitlichen Verbraucherschutz und Veterinärmedizin, Jena, Germany,20 Centre National de Référence des Mycobactéries, Institut Pasteur, Paris,15 Laboratoire de Bactériologie, CHU de Brest, Brest,22 Laboratoire de Bactériologie, CHU de Bordeaux, Bordeaux, France,31 Institut Pasteur de Bangui, Bangui, Central African Republic,17 Klinische Mikrobiologie, Hygiene-Institut der Universität,18 Bundesstaatliche Bakteriologisch-Serologische Untersuchungsanstalt Wien, Vienna, Austria,28 Department of Hygiene and Microbiology, University of Palermo, Palermo,19 Department of Public Health, University of Florence, Florence, Italy,23 Laboratory of Molecular Microbiology, Saint Petersburg Pasteur Institute, Saint Petersburg, Russia,21 Les Centres Gheskio, INLR, Port au Prince, Haïti,24 Institut Pasteur de Madagascar, Tananarive, Madagascar,25 Institute of Medical Microbiology and Immunology, Göteborg University, Göteborg, Sweden,26 IVIC, Centro de Microbiología y Biología Celular, Laboratorio de Genética Molecular,30 Tuberculosis Laboratory, Instituto de Biomedicina, Caracas, Venezuela32
4,
Kristin Kremer
Kristin Kremer
Unité de la Tuberculose et des Mycobactéries, Institut Pasteur de Guadeloupe, Pointe-à-Pitre, Guadeloupe,1 Wadsworth Center, New York State Department Of Health, Albany New York,2 Public Health Research Institute, Tuberculosis Center, Newark, New Jersey,4 Diagnosis Laboratory for Infectious Diseases and Perinatal Screening RIVM, Bilthoven, The Netherlands,3 National Institute of Hygiene and Epidemiology, Hanoi, Vietnam,5 Department of Microbiology, Leeds General Infirmary, Leeds,6 Medical School, St Georges Hospital,7 Mycobacterium Reference Unit, PHLS, Dulwich Hospital, London,13 University of Surrey, Guildford, Surrey, United Kingdom,11 Instituto de Biotecnologia INTA, Moron, Argentina,8 Tuberculosis Mycobacteriology Branch, Centers for Disease Control and Prevention, Atlanta, Georgia,9 Australian Reference Laboratory for Bovine Tuberculosis, Department of Agriculture, South Perth, Australia,10 Centro de Biotecnologia, Universidade Federal de Pelotas, Pelotas,12 Universidade Federal do Rio Grande do Sul, Porto Alegre,27 Department of Biochemistry and Molecular Biology, Fiocruz, Oswaldo Cruz Institute, Rio de Janeiro, Brazil,29 University Childrens Hospital, Heidelberg,14 Infektions Epidemiology, Robert Koch Institute, Berlin,16 Bundesinstitut für Gesundheitlichen Verbraucherschutz und Veterinärmedizin, Jena, Germany,20 Centre National de Référence des Mycobactéries, Institut Pasteur, Paris,15 Laboratoire de Bactériologie, CHU de Brest, Brest,22 Laboratoire de Bactériologie, CHU de Bordeaux, Bordeaux, France,31 Institut Pasteur de Bangui, Bangui, Central African Republic,17 Klinische Mikrobiologie, Hygiene-Institut der Universität,18 Bundesstaatliche Bakteriologisch-Serologische Untersuchungsanstalt Wien, Vienna, Austria,28 Department of Hygiene and Microbiology, University of Palermo, Palermo,19 Department of Public Health, University of Florence, Florence, Italy,23 Laboratory of Molecular Microbiology, Saint Petersburg Pasteur Institute, Saint Petersburg, Russia,21 Les Centres Gheskio, INLR, Port au Prince, Haïti,24 Institut Pasteur de Madagascar, Tananarive, Madagascar,25 Institute of Medical Microbiology and Immunology, Göteborg University, Göteborg, Sweden,26 IVIC, Centro de Microbiología y Biología Celular, Laboratorio de Genética Molecular,30 Tuberculosis Laboratory, Instituto de Biomedicina, Caracas, Venezuela32
3,
Georges Valétudie
Georges Valétudie
Unité de la Tuberculose et des Mycobactéries, Institut Pasteur de Guadeloupe, Pointe-à-Pitre, Guadeloupe,1 Wadsworth Center, New York State Department Of Health, Albany New York,2 Public Health Research Institute, Tuberculosis Center, Newark, New Jersey,4 Diagnosis Laboratory for Infectious Diseases and Perinatal Screening RIVM, Bilthoven, The Netherlands,3 National Institute of Hygiene and Epidemiology, Hanoi, Vietnam,5 Department of Microbiology, Leeds General Infirmary, Leeds,6 Medical School, St Georges Hospital,7 Mycobacterium Reference Unit, PHLS, Dulwich Hospital, London,13 University of Surrey, Guildford, Surrey, United Kingdom,11 Instituto de Biotecnologia INTA, Moron, Argentina,8 Tuberculosis Mycobacteriology Branch, Centers for Disease Control and Prevention, Atlanta, Georgia,9 Australian Reference Laboratory for Bovine Tuberculosis, Department of Agriculture, South Perth, Australia,10 Centro de Biotecnologia, Universidade Federal de Pelotas, Pelotas,12 Universidade Federal do Rio Grande do Sul, Porto Alegre,27 Department of Biochemistry and Molecular Biology, Fiocruz, Oswaldo Cruz Institute, Rio de Janeiro, Brazil,29 University Childrens Hospital, Heidelberg,14 Infektions Epidemiology, Robert Koch Institute, Berlin,16 Bundesinstitut für Gesundheitlichen Verbraucherschutz und Veterinärmedizin, Jena, Germany,20 Centre National de Référence des Mycobactéries, Institut Pasteur, Paris,15 Laboratoire de Bactériologie, CHU de Brest, Brest,22 Laboratoire de Bactériologie, CHU de Bordeaux, Bordeaux, France,31 Institut Pasteur de Bangui, Bangui, Central African Republic,17 Klinische Mikrobiologie, Hygiene-Institut der Universität,18 Bundesstaatliche Bakteriologisch-Serologische Untersuchungsanstalt Wien, Vienna, Austria,28 Department of Hygiene and Microbiology, University of Palermo, Palermo,19 Department of Public Health, University of Florence, Florence, Italy,23 Laboratory of Molecular Microbiology, Saint Petersburg Pasteur Institute, Saint Petersburg, Russia,21 Les Centres Gheskio, INLR, Port au Prince, Haïti,24 Institut Pasteur de Madagascar, Tananarive, Madagascar,25 Institute of Medical Microbiology and Immunology, Göteborg University, Göteborg, Sweden,26 IVIC, Centro de Microbiología y Biología Celular, Laboratorio de Genética Molecular,30 Tuberculosis Laboratory, Instituto de Biomedicina, Caracas, Venezuela32
1,
Dang Duc Anh
Dang Duc Anh
Unité de la Tuberculose et des Mycobactéries, Institut Pasteur de Guadeloupe, Pointe-à-Pitre, Guadeloupe,1 Wadsworth Center, New York State Department Of Health, Albany New York,2 Public Health Research Institute, Tuberculosis Center, Newark, New Jersey,4 Diagnosis Laboratory for Infectious Diseases and Perinatal Screening RIVM, Bilthoven, The Netherlands,3 National Institute of Hygiene and Epidemiology, Hanoi, Vietnam,5 Department of Microbiology, Leeds General Infirmary, Leeds,6 Medical School, St Georges Hospital,7 Mycobacterium Reference Unit, PHLS, Dulwich Hospital, London,13 University of Surrey, Guildford, Surrey, United Kingdom,11 Instituto de Biotecnologia INTA, Moron, Argentina,8 Tuberculosis Mycobacteriology Branch, Centers for Disease Control and Prevention, Atlanta, Georgia,9 Australian Reference Laboratory for Bovine Tuberculosis, Department of Agriculture, South Perth, Australia,10 Centro de Biotecnologia, Universidade Federal de Pelotas, Pelotas,12 Universidade Federal do Rio Grande do Sul, Porto Alegre,27 Department of Biochemistry and Molecular Biology, Fiocruz, Oswaldo Cruz Institute, Rio de Janeiro, Brazil,29 University Childrens Hospital, Heidelberg,14 Infektions Epidemiology, Robert Koch Institute, Berlin,16 Bundesinstitut für Gesundheitlichen Verbraucherschutz und Veterinärmedizin, Jena, Germany,20 Centre National de Référence des Mycobactéries, Institut Pasteur, Paris,15 Laboratoire de Bactériologie, CHU de Brest, Brest,22 Laboratoire de Bactériologie, CHU de Bordeaux, Bordeaux, France,31 Institut Pasteur de Bangui, Bangui, Central African Republic,17 Klinische Mikrobiologie, Hygiene-Institut der Universität,18 Bundesstaatliche Bakteriologisch-Serologische Untersuchungsanstalt Wien, Vienna, Austria,28 Department of Hygiene and Microbiology, University of Palermo, Palermo,19 Department of Public Health, University of Florence, Florence, Italy,23 Laboratory of Molecular Microbiology, Saint Petersburg Pasteur Institute, Saint Petersburg, Russia,21 Les Centres Gheskio, INLR, Port au Prince, Haïti,24 Institut Pasteur de Madagascar, Tananarive, Madagascar,25 Institute of Medical Microbiology and Immunology, Göteborg University, Göteborg, Sweden,26 IVIC, Centro de Microbiología y Biología Celular, Laboratorio de Genética Molecular,30 Tuberculosis Laboratory, Instituto de Biomedicina, Caracas, Venezuela32
5,
Rachael Barlow
Rachael Barlow
Unité de la Tuberculose et des Mycobactéries, Institut Pasteur de Guadeloupe, Pointe-à-Pitre, Guadeloupe,1 Wadsworth Center, New York State Department Of Health, Albany New York,2 Public Health Research Institute, Tuberculosis Center, Newark, New Jersey,4 Diagnosis Laboratory for Infectious Diseases and Perinatal Screening RIVM, Bilthoven, The Netherlands,3 National Institute of Hygiene and Epidemiology, Hanoi, Vietnam,5 Department of Microbiology, Leeds General Infirmary, Leeds,6 Medical School, St Georges Hospital,7 Mycobacterium Reference Unit, PHLS, Dulwich Hospital, London,13 University of Surrey, Guildford, Surrey, United Kingdom,11 Instituto de Biotecnologia INTA, Moron, Argentina,8 Tuberculosis Mycobacteriology Branch, Centers for Disease Control and Prevention, Atlanta, Georgia,9 Australian Reference Laboratory for Bovine Tuberculosis, Department of Agriculture, South Perth, Australia,10 Centro de Biotecnologia, Universidade Federal de Pelotas, Pelotas,12 Universidade Federal do Rio Grande do Sul, Porto Alegre,27 Department of Biochemistry and Molecular Biology, Fiocruz, Oswaldo Cruz Institute, Rio de Janeiro, Brazil,29 University Childrens Hospital, Heidelberg,14 Infektions Epidemiology, Robert Koch Institute, Berlin,16 Bundesinstitut für Gesundheitlichen Verbraucherschutz und Veterinärmedizin, Jena, Germany,20 Centre National de Référence des Mycobactéries, Institut Pasteur, Paris,15 Laboratoire de Bactériologie, CHU de Brest, Brest,22 Laboratoire de Bactériologie, CHU de Bordeaux, Bordeaux, France,31 Institut Pasteur de Bangui, Bangui, Central African Republic,17 Klinische Mikrobiologie, Hygiene-Institut der Universität,18 Bundesstaatliche Bakteriologisch-Serologische Untersuchungsanstalt Wien, Vienna, Austria,28 Department of Hygiene and Microbiology, University of Palermo, Palermo,19 Department of Public Health, University of Florence, Florence, Italy,23 Laboratory of Molecular Microbiology, Saint Petersburg Pasteur Institute, Saint Petersburg, Russia,21 Les Centres Gheskio, INLR, Port au Prince, Haïti,24 Institut Pasteur de Madagascar, Tananarive, Madagascar,25 Institute of Medical Microbiology and Immunology, Göteborg University, Göteborg, Sweden,26 IVIC, Centro de Microbiología y Biología Celular, Laboratorio de Genética Molecular,30 Tuberculosis Laboratory, Instituto de Biomedicina, Caracas, Venezuela32
6,
Dilip Banerjee
Dilip Banerjee
Unité de la Tuberculose et des Mycobactéries, Institut Pasteur de Guadeloupe, Pointe-à-Pitre, Guadeloupe,1 Wadsworth Center, New York State Department Of Health, Albany New York,2 Public Health Research Institute, Tuberculosis Center, Newark, New Jersey,4 Diagnosis Laboratory for Infectious Diseases and Perinatal Screening RIVM, Bilthoven, The Netherlands,3 National Institute of Hygiene and Epidemiology, Hanoi, Vietnam,5 Department of Microbiology, Leeds General Infirmary, Leeds,6 Medical School, St Georges Hospital,7 Mycobacterium Reference Unit, PHLS, Dulwich Hospital, London,13 University of Surrey, Guildford, Surrey, United Kingdom,11 Instituto de Biotecnologia INTA, Moron, Argentina,8 Tuberculosis Mycobacteriology Branch, Centers for Disease Control and Prevention, Atlanta, Georgia,9 Australian Reference Laboratory for Bovine Tuberculosis, Department of Agriculture, South Perth, Australia,10 Centro de Biotecnologia, Universidade Federal de Pelotas, Pelotas,12 Universidade Federal do Rio Grande do Sul, Porto Alegre,27 Department of Biochemistry and Molecular Biology, Fiocruz, Oswaldo Cruz Institute, Rio de Janeiro, Brazil,29 University Childrens Hospital, Heidelberg,14 Infektions Epidemiology, Robert Koch Institute, Berlin,16 Bundesinstitut für Gesundheitlichen Verbraucherschutz und Veterinärmedizin, Jena, Germany,20 Centre National de Référence des Mycobactéries, Institut Pasteur, Paris,15 Laboratoire de Bactériologie, CHU de Brest, Brest,22 Laboratoire de Bactériologie, CHU de Bordeaux, Bordeaux, France,31 Institut Pasteur de Bangui, Bangui, Central African Republic,17 Klinische Mikrobiologie, Hygiene-Institut der Universität,18 Bundesstaatliche Bakteriologisch-Serologische Untersuchungsanstalt Wien, Vienna, Austria,28 Department of Hygiene and Microbiology, University of Palermo, Palermo,19 Department of Public Health, University of Florence, Florence, Italy,23 Laboratory of Molecular Microbiology, Saint Petersburg Pasteur Institute, Saint Petersburg, Russia,21 Les Centres Gheskio, INLR, Port au Prince, Haïti,24 Institut Pasteur de Madagascar, Tananarive, Madagascar,25 Institute of Medical Microbiology and Immunology, Göteborg University, Göteborg, Sweden,26 IVIC, Centro de Microbiología y Biología Celular, Laboratorio de Genética Molecular,30 Tuberculosis Laboratory, Instituto de Biomedicina, Caracas, Venezuela32
7,
Pablo J Bifani
Pablo J Bifani
Unité de la Tuberculose et des Mycobactéries, Institut Pasteur de Guadeloupe, Pointe-à-Pitre, Guadeloupe,1 Wadsworth Center, New York State Department Of Health, Albany New York,2 Public Health Research Institute, Tuberculosis Center, Newark, New Jersey,4 Diagnosis Laboratory for Infectious Diseases and Perinatal Screening RIVM, Bilthoven, The Netherlands,3 National Institute of Hygiene and Epidemiology, Hanoi, Vietnam,5 Department of Microbiology, Leeds General Infirmary, Leeds,6 Medical School, St Georges Hospital,7 Mycobacterium Reference Unit, PHLS, Dulwich Hospital, London,13 University of Surrey, Guildford, Surrey, United Kingdom,11 Instituto de Biotecnologia INTA, Moron, Argentina,8 Tuberculosis Mycobacteriology Branch, Centers for Disease Control and Prevention, Atlanta, Georgia,9 Australian Reference Laboratory for Bovine Tuberculosis, Department of Agriculture, South Perth, Australia,10 Centro de Biotecnologia, Universidade Federal de Pelotas, Pelotas,12 Universidade Federal do Rio Grande do Sul, Porto Alegre,27 Department of Biochemistry and Molecular Biology, Fiocruz, Oswaldo Cruz Institute, Rio de Janeiro, Brazil,29 University Childrens Hospital, Heidelberg,14 Infektions Epidemiology, Robert Koch Institute, Berlin,16 Bundesinstitut für Gesundheitlichen Verbraucherschutz und Veterinärmedizin, Jena, Germany,20 Centre National de Référence des Mycobactéries, Institut Pasteur, Paris,15 Laboratoire de Bactériologie, CHU de Brest, Brest,22 Laboratoire de Bactériologie, CHU de Bordeaux, Bordeaux, France,31 Institut Pasteur de Bangui, Bangui, Central African Republic,17 Klinische Mikrobiologie, Hygiene-Institut der Universität,18 Bundesstaatliche Bakteriologisch-Serologische Untersuchungsanstalt Wien, Vienna, Austria,28 Department of Hygiene and Microbiology, University of Palermo, Palermo,19 Department of Public Health, University of Florence, Florence, Italy,23 Laboratory of Molecular Microbiology, Saint Petersburg Pasteur Institute, Saint Petersburg, Russia,21 Les Centres Gheskio, INLR, Port au Prince, Haïti,24 Institut Pasteur de Madagascar, Tananarive, Madagascar,25 Institute of Medical Microbiology and Immunology, Göteborg University, Göteborg, Sweden,26 IVIC, Centro de Microbiología y Biología Celular, Laboratorio de Genética Molecular,30 Tuberculosis Laboratory, Instituto de Biomedicina, Caracas, Venezuela32
4,
Karine Brudey
Karine Brudey
Unité de la Tuberculose et des Mycobactéries, Institut Pasteur de Guadeloupe, Pointe-à-Pitre, Guadeloupe,1 Wadsworth Center, New York State Department Of Health, Albany New York,2 Public Health Research Institute, Tuberculosis Center, Newark, New Jersey,4 Diagnosis Laboratory for Infectious Diseases and Perinatal Screening RIVM, Bilthoven, The Netherlands,3 National Institute of Hygiene and Epidemiology, Hanoi, Vietnam,5 Department of Microbiology, Leeds General Infirmary, Leeds,6 Medical School, St Georges Hospital,7 Mycobacterium Reference Unit, PHLS, Dulwich Hospital, London,13 University of Surrey, Guildford, Surrey, United Kingdom,11 Instituto de Biotecnologia INTA, Moron, Argentina,8 Tuberculosis Mycobacteriology Branch, Centers for Disease Control and Prevention, Atlanta, Georgia,9 Australian Reference Laboratory for Bovine Tuberculosis, Department of Agriculture, South Perth, Australia,10 Centro de Biotecnologia, Universidade Federal de Pelotas, Pelotas,12 Universidade Federal do Rio Grande do Sul, Porto Alegre,27 Department of Biochemistry and Molecular Biology, Fiocruz, Oswaldo Cruz Institute, Rio de Janeiro, Brazil,29 University Childrens Hospital, Heidelberg,14 Infektions Epidemiology, Robert Koch Institute, Berlin,16 Bundesinstitut für Gesundheitlichen Verbraucherschutz und Veterinärmedizin, Jena, Germany,20 Centre National de Référence des Mycobactéries, Institut Pasteur, Paris,15 Laboratoire de Bactériologie, CHU de Brest, Brest,22 Laboratoire de Bactériologie, CHU de Bordeaux, Bordeaux, France,31 Institut Pasteur de Bangui, Bangui, Central African Republic,17 Klinische Mikrobiologie, Hygiene-Institut der Universität,18 Bundesstaatliche Bakteriologisch-Serologische Untersuchungsanstalt Wien, Vienna, Austria,28 Department of Hygiene and Microbiology, University of Palermo, Palermo,19 Department of Public Health, University of Florence, Florence, Italy,23 Laboratory of Molecular Microbiology, Saint Petersburg Pasteur Institute, Saint Petersburg, Russia,21 Les Centres Gheskio, INLR, Port au Prince, Haïti,24 Institut Pasteur de Madagascar, Tananarive, Madagascar,25 Institute of Medical Microbiology and Immunology, Göteborg University, Göteborg, Sweden,26 IVIC, Centro de Microbiología y Biología Celular, Laboratorio de Genética Molecular,30 Tuberculosis Laboratory, Instituto de Biomedicina, Caracas, Venezuela32
1,
Angel Cataldi
Angel Cataldi
Unité de la Tuberculose et des Mycobactéries, Institut Pasteur de Guadeloupe, Pointe-à-Pitre, Guadeloupe,1 Wadsworth Center, New York State Department Of Health, Albany New York,2 Public Health Research Institute, Tuberculosis Center, Newark, New Jersey,4 Diagnosis Laboratory for Infectious Diseases and Perinatal Screening RIVM, Bilthoven, The Netherlands,3 National Institute of Hygiene and Epidemiology, Hanoi, Vietnam,5 Department of Microbiology, Leeds General Infirmary, Leeds,6 Medical School, St Georges Hospital,7 Mycobacterium Reference Unit, PHLS, Dulwich Hospital, London,13 University of Surrey, Guildford, Surrey, United Kingdom,11 Instituto de Biotecnologia INTA, Moron, Argentina,8 Tuberculosis Mycobacteriology Branch, Centers for Disease Control and Prevention, Atlanta, Georgia,9 Australian Reference Laboratory for Bovine Tuberculosis, Department of Agriculture, South Perth, Australia,10 Centro de Biotecnologia, Universidade Federal de Pelotas, Pelotas,12 Universidade Federal do Rio Grande do Sul, Porto Alegre,27 Department of Biochemistry and Molecular Biology, Fiocruz, Oswaldo Cruz Institute, Rio de Janeiro, Brazil,29 University Childrens Hospital, Heidelberg,14 Infektions Epidemiology, Robert Koch Institute, Berlin,16 Bundesinstitut für Gesundheitlichen Verbraucherschutz und Veterinärmedizin, Jena, Germany,20 Centre National de Référence des Mycobactéries, Institut Pasteur, Paris,15 Laboratoire de Bactériologie, CHU de Brest, Brest,22 Laboratoire de Bactériologie, CHU de Bordeaux, Bordeaux, France,31 Institut Pasteur de Bangui, Bangui, Central African Republic,17 Klinische Mikrobiologie, Hygiene-Institut der Universität,18 Bundesstaatliche Bakteriologisch-Serologische Untersuchungsanstalt Wien, Vienna, Austria,28 Department of Hygiene and Microbiology, University of Palermo, Palermo,19 Department of Public Health, University of Florence, Florence, Italy,23 Laboratory of Molecular Microbiology, Saint Petersburg Pasteur Institute, Saint Petersburg, Russia,21 Les Centres Gheskio, INLR, Port au Prince, Haïti,24 Institut Pasteur de Madagascar, Tananarive, Madagascar,25 Institute of Medical Microbiology and Immunology, Göteborg University, Göteborg, Sweden,26 IVIC, Centro de Microbiología y Biología Celular, Laboratorio de Genética Molecular,30 Tuberculosis Laboratory, Instituto de Biomedicina, Caracas, Venezuela32
8,
Robert C Cooksey
Robert C Cooksey
Unité de la Tuberculose et des Mycobactéries, Institut Pasteur de Guadeloupe, Pointe-à-Pitre, Guadeloupe,1 Wadsworth Center, New York State Department Of Health, Albany New York,2 Public Health Research Institute, Tuberculosis Center, Newark, New Jersey,4 Diagnosis Laboratory for Infectious Diseases and Perinatal Screening RIVM, Bilthoven, The Netherlands,3 National Institute of Hygiene and Epidemiology, Hanoi, Vietnam,5 Department of Microbiology, Leeds General Infirmary, Leeds,6 Medical School, St Georges Hospital,7 Mycobacterium Reference Unit, PHLS, Dulwich Hospital, London,13 University of Surrey, Guildford, Surrey, United Kingdom,11 Instituto de Biotecnologia INTA, Moron, Argentina,8 Tuberculosis Mycobacteriology Branch, Centers for Disease Control and Prevention, Atlanta, Georgia,9 Australian Reference Laboratory for Bovine Tuberculosis, Department of Agriculture, South Perth, Australia,10 Centro de Biotecnologia, Universidade Federal de Pelotas, Pelotas,12 Universidade Federal do Rio Grande do Sul, Porto Alegre,27 Department of Biochemistry and Molecular Biology, Fiocruz, Oswaldo Cruz Institute, Rio de Janeiro, Brazil,29 University Childrens Hospital, Heidelberg,14 Infektions Epidemiology, Robert Koch Institute, Berlin,16 Bundesinstitut für Gesundheitlichen Verbraucherschutz und Veterinärmedizin, Jena, Germany,20 Centre National de Référence des Mycobactéries, Institut Pasteur, Paris,15 Laboratoire de Bactériologie, CHU de Brest, Brest,22 Laboratoire de Bactériologie, CHU de Bordeaux, Bordeaux, France,31 Institut Pasteur de Bangui, Bangui, Central African Republic,17 Klinische Mikrobiologie, Hygiene-Institut der Universität,18 Bundesstaatliche Bakteriologisch-Serologische Untersuchungsanstalt Wien, Vienna, Austria,28 Department of Hygiene and Microbiology, University of Palermo, Palermo,19 Department of Public Health, University of Florence, Florence, Italy,23 Laboratory of Molecular Microbiology, Saint Petersburg Pasteur Institute, Saint Petersburg, Russia,21 Les Centres Gheskio, INLR, Port au Prince, Haïti,24 Institut Pasteur de Madagascar, Tananarive, Madagascar,25 Institute of Medical Microbiology and Immunology, Göteborg University, Göteborg, Sweden,26 IVIC, Centro de Microbiología y Biología Celular, Laboratorio de Genética Molecular,30 Tuberculosis Laboratory, Instituto de Biomedicina, Caracas, Venezuela32
9,
Debby V Cousins
Debby V Cousins
Unité de la Tuberculose et des Mycobactéries, Institut Pasteur de Guadeloupe, Pointe-à-Pitre, Guadeloupe,1 Wadsworth Center, New York State Department Of Health, Albany New York,2 Public Health Research Institute, Tuberculosis Center, Newark, New Jersey,4 Diagnosis Laboratory for Infectious Diseases and Perinatal Screening RIVM, Bilthoven, The Netherlands,3 National Institute of Hygiene and Epidemiology, Hanoi, Vietnam,5 Department of Microbiology, Leeds General Infirmary, Leeds,6 Medical School, St Georges Hospital,7 Mycobacterium Reference Unit, PHLS, Dulwich Hospital, London,13 University of Surrey, Guildford, Surrey, United Kingdom,11 Instituto de Biotecnologia INTA, Moron, Argentina,8 Tuberculosis Mycobacteriology Branch, Centers for Disease Control and Prevention, Atlanta, Georgia,9 Australian Reference Laboratory for Bovine Tuberculosis, Department of Agriculture, South Perth, Australia,10 Centro de Biotecnologia, Universidade Federal de Pelotas, Pelotas,12 Universidade Federal do Rio Grande do Sul, Porto Alegre,27 Department of Biochemistry and Molecular Biology, Fiocruz, Oswaldo Cruz Institute, Rio de Janeiro, Brazil,29 University Childrens Hospital, Heidelberg,14 Infektions Epidemiology, Robert Koch Institute, Berlin,16 Bundesinstitut für Gesundheitlichen Verbraucherschutz und Veterinärmedizin, Jena, Germany,20 Centre National de Référence des Mycobactéries, Institut Pasteur, Paris,15 Laboratoire de Bactériologie, CHU de Brest, Brest,22 Laboratoire de Bactériologie, CHU de Bordeaux, Bordeaux, France,31 Institut Pasteur de Bangui, Bangui, Central African Republic,17 Klinische Mikrobiologie, Hygiene-Institut der Universität,18 Bundesstaatliche Bakteriologisch-Serologische Untersuchungsanstalt Wien, Vienna, Austria,28 Department of Hygiene and Microbiology, University of Palermo, Palermo,19 Department of Public Health, University of Florence, Florence, Italy,23 Laboratory of Molecular Microbiology, Saint Petersburg Pasteur Institute, Saint Petersburg, Russia,21 Les Centres Gheskio, INLR, Port au Prince, Haïti,24 Institut Pasteur de Madagascar, Tananarive, Madagascar,25 Institute of Medical Microbiology and Immunology, Göteborg University, Göteborg, Sweden,26 IVIC, Centro de Microbiología y Biología Celular, Laboratorio de Genética Molecular,30 Tuberculosis Laboratory, Instituto de Biomedicina, Caracas, Venezuela32
10,
Jeremy W Dale
Jeremy W Dale
Unité de la Tuberculose et des Mycobactéries, Institut Pasteur de Guadeloupe, Pointe-à-Pitre, Guadeloupe,1 Wadsworth Center, New York State Department Of Health, Albany New York,2 Public Health Research Institute, Tuberculosis Center, Newark, New Jersey,4 Diagnosis Laboratory for Infectious Diseases and Perinatal Screening RIVM, Bilthoven, The Netherlands,3 National Institute of Hygiene and Epidemiology, Hanoi, Vietnam,5 Department of Microbiology, Leeds General Infirmary, Leeds,6 Medical School, St Georges Hospital,7 Mycobacterium Reference Unit, PHLS, Dulwich Hospital, London,13 University of Surrey, Guildford, Surrey, United Kingdom,11 Instituto de Biotecnologia INTA, Moron, Argentina,8 Tuberculosis Mycobacteriology Branch, Centers for Disease Control and Prevention, Atlanta, Georgia,9 Australian Reference Laboratory for Bovine Tuberculosis, Department of Agriculture, South Perth, Australia,10 Centro de Biotecnologia, Universidade Federal de Pelotas, Pelotas,12 Universidade Federal do Rio Grande do Sul, Porto Alegre,27 Department of Biochemistry and Molecular Biology, Fiocruz, Oswaldo Cruz Institute, Rio de Janeiro, Brazil,29 University Childrens Hospital, Heidelberg,14 Infektions Epidemiology, Robert Koch Institute, Berlin,16 Bundesinstitut für Gesundheitlichen Verbraucherschutz und Veterinärmedizin, Jena, Germany,20 Centre National de Référence des Mycobactéries, Institut Pasteur, Paris,15 Laboratoire de Bactériologie, CHU de Brest, Brest,22 Laboratoire de Bactériologie, CHU de Bordeaux, Bordeaux, France,31 Institut Pasteur de Bangui, Bangui, Central African Republic,17 Klinische Mikrobiologie, Hygiene-Institut der Universität,18 Bundesstaatliche Bakteriologisch-Serologische Untersuchungsanstalt Wien, Vienna, Austria,28 Department of Hygiene and Microbiology, University of Palermo, Palermo,19 Department of Public Health, University of Florence, Florence, Italy,23 Laboratory of Molecular Microbiology, Saint Petersburg Pasteur Institute, Saint Petersburg, Russia,21 Les Centres Gheskio, INLR, Port au Prince, Haïti,24 Institut Pasteur de Madagascar, Tananarive, Madagascar,25 Institute of Medical Microbiology and Immunology, Göteborg University, Göteborg, Sweden,26 IVIC, Centro de Microbiología y Biología Celular, Laboratorio de Genética Molecular,30 Tuberculosis Laboratory, Instituto de Biomedicina, Caracas, Venezuela32
11,
Odir A Dellagostin
Odir A Dellagostin
Unité de la Tuberculose et des Mycobactéries, Institut Pasteur de Guadeloupe, Pointe-à-Pitre, Guadeloupe,1 Wadsworth Center, New York State Department Of Health, Albany New York,2 Public Health Research Institute, Tuberculosis Center, Newark, New Jersey,4 Diagnosis Laboratory for Infectious Diseases and Perinatal Screening RIVM, Bilthoven, The Netherlands,3 National Institute of Hygiene and Epidemiology, Hanoi, Vietnam,5 Department of Microbiology, Leeds General Infirmary, Leeds,6 Medical School, St Georges Hospital,7 Mycobacterium Reference Unit, PHLS, Dulwich Hospital, London,13 University of Surrey, Guildford, Surrey, United Kingdom,11 Instituto de Biotecnologia INTA, Moron, Argentina,8 Tuberculosis Mycobacteriology Branch, Centers for Disease Control and Prevention, Atlanta, Georgia,9 Australian Reference Laboratory for Bovine Tuberculosis, Department of Agriculture, South Perth, Australia,10 Centro de Biotecnologia, Universidade Federal de Pelotas, Pelotas,12 Universidade Federal do Rio Grande do Sul, Porto Alegre,27 Department of Biochemistry and Molecular Biology, Fiocruz, Oswaldo Cruz Institute, Rio de Janeiro, Brazil,29 University Childrens Hospital, Heidelberg,14 Infektions Epidemiology, Robert Koch Institute, Berlin,16 Bundesinstitut für Gesundheitlichen Verbraucherschutz und Veterinärmedizin, Jena, Germany,20 Centre National de Référence des Mycobactéries, Institut Pasteur, Paris,15 Laboratoire de Bactériologie, CHU de Brest, Brest,22 Laboratoire de Bactériologie, CHU de Bordeaux, Bordeaux, France,31 Institut Pasteur de Bangui, Bangui, Central African Republic,17 Klinische Mikrobiologie, Hygiene-Institut der Universität,18 Bundesstaatliche Bakteriologisch-Serologische Untersuchungsanstalt Wien, Vienna, Austria,28 Department of Hygiene and Microbiology, University of Palermo, Palermo,19 Department of Public Health, University of Florence, Florence, Italy,23 Laboratory of Molecular Microbiology, Saint Petersburg Pasteur Institute, Saint Petersburg, Russia,21 Les Centres Gheskio, INLR, Port au Prince, Haïti,24 Institut Pasteur de Madagascar, Tananarive, Madagascar,25 Institute of Medical Microbiology and Immunology, Göteborg University, Göteborg, Sweden,26 IVIC, Centro de Microbiología y Biología Celular, Laboratorio de Genética Molecular,30 Tuberculosis Laboratory, Instituto de Biomedicina, Caracas, Venezuela32
12,
Francis Drobniewski
Francis Drobniewski
Unité de la Tuberculose et des Mycobactéries, Institut Pasteur de Guadeloupe, Pointe-à-Pitre, Guadeloupe,1 Wadsworth Center, New York State Department Of Health, Albany New York,2 Public Health Research Institute, Tuberculosis Center, Newark, New Jersey,4 Diagnosis Laboratory for Infectious Diseases and Perinatal Screening RIVM, Bilthoven, The Netherlands,3 National Institute of Hygiene and Epidemiology, Hanoi, Vietnam,5 Department of Microbiology, Leeds General Infirmary, Leeds,6 Medical School, St Georges Hospital,7 Mycobacterium Reference Unit, PHLS, Dulwich Hospital, London,13 University of Surrey, Guildford, Surrey, United Kingdom,11 Instituto de Biotecnologia INTA, Moron, Argentina,8 Tuberculosis Mycobacteriology Branch, Centers for Disease Control and Prevention, Atlanta, Georgia,9 Australian Reference Laboratory for Bovine Tuberculosis, Department of Agriculture, South Perth, Australia,10 Centro de Biotecnologia, Universidade Federal de Pelotas, Pelotas,12 Universidade Federal do Rio Grande do Sul, Porto Alegre,27 Department of Biochemistry and Molecular Biology, Fiocruz, Oswaldo Cruz Institute, Rio de Janeiro, Brazil,29 University Childrens Hospital, Heidelberg,14 Infektions Epidemiology, Robert Koch Institute, Berlin,16 Bundesinstitut für Gesundheitlichen Verbraucherschutz und Veterinärmedizin, Jena, Germany,20 Centre National de Référence des Mycobactéries, Institut Pasteur, Paris,15 Laboratoire de Bactériologie, CHU de Brest, Brest,22 Laboratoire de Bactériologie, CHU de Bordeaux, Bordeaux, France,31 Institut Pasteur de Bangui, Bangui, Central African Republic,17 Klinische Mikrobiologie, Hygiene-Institut der Universität,18 Bundesstaatliche Bakteriologisch-Serologische Untersuchungsanstalt Wien, Vienna, Austria,28 Department of Hygiene and Microbiology, University of Palermo, Palermo,19 Department of Public Health, University of Florence, Florence, Italy,23 Laboratory of Molecular Microbiology, Saint Petersburg Pasteur Institute, Saint Petersburg, Russia,21 Les Centres Gheskio, INLR, Port au Prince, Haïti,24 Institut Pasteur de Madagascar, Tananarive, Madagascar,25 Institute of Medical Microbiology and Immunology, Göteborg University, Göteborg, Sweden,26 IVIC, Centro de Microbiología y Biología Celular, Laboratorio de Genética Molecular,30 Tuberculosis Laboratory, Instituto de Biomedicina, Caracas, Venezuela32
13,
Guido Engelmann
Guido Engelmann
Unité de la Tuberculose et des Mycobactéries, Institut Pasteur de Guadeloupe, Pointe-à-Pitre, Guadeloupe,1 Wadsworth Center, New York State Department Of Health, Albany New York,2 Public Health Research Institute, Tuberculosis Center, Newark, New Jersey,4 Diagnosis Laboratory for Infectious Diseases and Perinatal Screening RIVM, Bilthoven, The Netherlands,3 National Institute of Hygiene and Epidemiology, Hanoi, Vietnam,5 Department of Microbiology, Leeds General Infirmary, Leeds,6 Medical School, St Georges Hospital,7 Mycobacterium Reference Unit, PHLS, Dulwich Hospital, London,13 University of Surrey, Guildford, Surrey, United Kingdom,11 Instituto de Biotecnologia INTA, Moron, Argentina,8 Tuberculosis Mycobacteriology Branch, Centers for Disease Control and Prevention, Atlanta, Georgia,9 Australian Reference Laboratory for Bovine Tuberculosis, Department of Agriculture, South Perth, Australia,10 Centro de Biotecnologia, Universidade Federal de Pelotas, Pelotas,12 Universidade Federal do Rio Grande do Sul, Porto Alegre,27 Department of Biochemistry and Molecular Biology, Fiocruz, Oswaldo Cruz Institute, Rio de Janeiro, Brazil,29 University Childrens Hospital, Heidelberg,14 Infektions Epidemiology, Robert Koch Institute, Berlin,16 Bundesinstitut für Gesundheitlichen Verbraucherschutz und Veterinärmedizin, Jena, Germany,20 Centre National de Référence des Mycobactéries, Institut Pasteur, Paris,15 Laboratoire de Bactériologie, CHU de Brest, Brest,22 Laboratoire de Bactériologie, CHU de Bordeaux, Bordeaux, France,31 Institut Pasteur de Bangui, Bangui, Central African Republic,17 Klinische Mikrobiologie, Hygiene-Institut der Universität,18 Bundesstaatliche Bakteriologisch-Serologische Untersuchungsanstalt Wien, Vienna, Austria,28 Department of Hygiene and Microbiology, University of Palermo, Palermo,19 Department of Public Health, University of Florence, Florence, Italy,23 Laboratory of Molecular Microbiology, Saint Petersburg Pasteur Institute, Saint Petersburg, Russia,21 Les Centres Gheskio, INLR, Port au Prince, Haïti,24 Institut Pasteur de Madagascar, Tananarive, Madagascar,25 Institute of Medical Microbiology and Immunology, Göteborg University, Göteborg, Sweden,26 IVIC, Centro de Microbiología y Biología Celular, Laboratorio de Genética Molecular,30 Tuberculosis Laboratory, Instituto de Biomedicina, Caracas, Venezuela32
14,
Séverine Ferdinand
Séverine Ferdinand
Unité de la Tuberculose et des Mycobactéries, Institut Pasteur de Guadeloupe, Pointe-à-Pitre, Guadeloupe,1 Wadsworth Center, New York State Department Of Health, Albany New York,2 Public Health Research Institute, Tuberculosis Center, Newark, New Jersey,4 Diagnosis Laboratory for Infectious Diseases and Perinatal Screening RIVM, Bilthoven, The Netherlands,3 National Institute of Hygiene and Epidemiology, Hanoi, Vietnam,5 Department of Microbiology, Leeds General Infirmary, Leeds,6 Medical School, St Georges Hospital,7 Mycobacterium Reference Unit, PHLS, Dulwich Hospital, London,13 University of Surrey, Guildford, Surrey, United Kingdom,11 Instituto de Biotecnologia INTA, Moron, Argentina,8 Tuberculosis Mycobacteriology Branch, Centers for Disease Control and Prevention, Atlanta, Georgia,9 Australian Reference Laboratory for Bovine Tuberculosis, Department of Agriculture, South Perth, Australia,10 Centro de Biotecnologia, Universidade Federal de Pelotas, Pelotas,12 Universidade Federal do Rio Grande do Sul, Porto Alegre,27 Department of Biochemistry and Molecular Biology, Fiocruz, Oswaldo Cruz Institute, Rio de Janeiro, Brazil,29 University Childrens Hospital, Heidelberg,14 Infektions Epidemiology, Robert Koch Institute, Berlin,16 Bundesinstitut für Gesundheitlichen Verbraucherschutz und Veterinärmedizin, Jena, Germany,20 Centre National de Référence des Mycobactéries, Institut Pasteur, Paris,15 Laboratoire de Bactériologie, CHU de Brest, Brest,22 Laboratoire de Bactériologie, CHU de Bordeaux, Bordeaux, France,31 Institut Pasteur de Bangui, Bangui, Central African Republic,17 Klinische Mikrobiologie, Hygiene-Institut der Universität,18 Bundesstaatliche Bakteriologisch-Serologische Untersuchungsanstalt Wien, Vienna, Austria,28 Department of Hygiene and Microbiology, University of Palermo, Palermo,19 Department of Public Health, University of Florence, Florence, Italy,23 Laboratory of Molecular Microbiology, Saint Petersburg Pasteur Institute, Saint Petersburg, Russia,21 Les Centres Gheskio, INLR, Port au Prince, Haïti,24 Institut Pasteur de Madagascar, Tananarive, Madagascar,25 Institute of Medical Microbiology and Immunology, Göteborg University, Göteborg, Sweden,26 IVIC, Centro de Microbiología y Biología Celular, Laboratorio de Genética Molecular,30 Tuberculosis Laboratory, Instituto de Biomedicina, Caracas, Venezuela32
1,
Deborah Gascoyne-Binzi
Deborah Gascoyne-Binzi
Unité de la Tuberculose et des Mycobactéries, Institut Pasteur de Guadeloupe, Pointe-à-Pitre, Guadeloupe,1 Wadsworth Center, New York State Department Of Health, Albany New York,2 Public Health Research Institute, Tuberculosis Center, Newark, New Jersey,4 Diagnosis Laboratory for Infectious Diseases and Perinatal Screening RIVM, Bilthoven, The Netherlands,3 National Institute of Hygiene and Epidemiology, Hanoi, Vietnam,5 Department of Microbiology, Leeds General Infirmary, Leeds,6 Medical School, St Georges Hospital,7 Mycobacterium Reference Unit, PHLS, Dulwich Hospital, London,13 University of Surrey, Guildford, Surrey, United Kingdom,11 Instituto de Biotecnologia INTA, Moron, Argentina,8 Tuberculosis Mycobacteriology Branch, Centers for Disease Control and Prevention, Atlanta, Georgia,9 Australian Reference Laboratory for Bovine Tuberculosis, Department of Agriculture, South Perth, Australia,10 Centro de Biotecnologia, Universidade Federal de Pelotas, Pelotas,12 Universidade Federal do Rio Grande do Sul, Porto Alegre,27 Department of Biochemistry and Molecular Biology, Fiocruz, Oswaldo Cruz Institute, Rio de Janeiro, Brazil,29 University Childrens Hospital, Heidelberg,14 Infektions Epidemiology, Robert Koch Institute, Berlin,16 Bundesinstitut für Gesundheitlichen Verbraucherschutz und Veterinärmedizin, Jena, Germany,20 Centre National de Référence des Mycobactéries, Institut Pasteur, Paris,15 Laboratoire de Bactériologie, CHU de Brest, Brest,22 Laboratoire de Bactériologie, CHU de Bordeaux, Bordeaux, France,31 Institut Pasteur de Bangui, Bangui, Central African Republic,17 Klinische Mikrobiologie, Hygiene-Institut der Universität,18 Bundesstaatliche Bakteriologisch-Serologische Untersuchungsanstalt Wien, Vienna, Austria,28 Department of Hygiene and Microbiology, University of Palermo, Palermo,19 Department of Public Health, University of Florence, Florence, Italy,23 Laboratory of Molecular Microbiology, Saint Petersburg Pasteur Institute, Saint Petersburg, Russia,21 Les Centres Gheskio, INLR, Port au Prince, Haïti,24 Institut Pasteur de Madagascar, Tananarive, Madagascar,25 Institute of Medical Microbiology and Immunology, Göteborg University, Göteborg, Sweden,26 IVIC, Centro de Microbiología y Biología Celular, Laboratorio de Genética Molecular,30 Tuberculosis Laboratory, Instituto de Biomedicina, Caracas, Venezuela32
6,
Max Gordon
Max Gordon
Unité de la Tuberculose et des Mycobactéries, Institut Pasteur de Guadeloupe, Pointe-à-Pitre, Guadeloupe,1 Wadsworth Center, New York State Department Of Health, Albany New York,2 Public Health Research Institute, Tuberculosis Center, Newark, New Jersey,4 Diagnosis Laboratory for Infectious Diseases and Perinatal Screening RIVM, Bilthoven, The Netherlands,3 National Institute of Hygiene and Epidemiology, Hanoi, Vietnam,5 Department of Microbiology, Leeds General Infirmary, Leeds,6 Medical School, St Georges Hospital,7 Mycobacterium Reference Unit, PHLS, Dulwich Hospital, London,13 University of Surrey, Guildford, Surrey, United Kingdom,11 Instituto de Biotecnologia INTA, Moron, Argentina,8 Tuberculosis Mycobacteriology Branch, Centers for Disease Control and Prevention, Atlanta, Georgia,9 Australian Reference Laboratory for Bovine Tuberculosis, Department of Agriculture, South Perth, Australia,10 Centro de Biotecnologia, Universidade Federal de Pelotas, Pelotas,12 Universidade Federal do Rio Grande do Sul, Porto Alegre,27 Department of Biochemistry and Molecular Biology, Fiocruz, Oswaldo Cruz Institute, Rio de Janeiro, Brazil,29 University Childrens Hospital, Heidelberg,14 Infektions Epidemiology, Robert Koch Institute, Berlin,16 Bundesinstitut für Gesundheitlichen Verbraucherschutz und Veterinärmedizin, Jena, Germany,20 Centre National de Référence des Mycobactéries, Institut Pasteur, Paris,15 Laboratoire de Bactériologie, CHU de Brest, Brest,22 Laboratoire de Bactériologie, CHU de Bordeaux, Bordeaux, France,31 Institut Pasteur de Bangui, Bangui, Central African Republic,17 Klinische Mikrobiologie, Hygiene-Institut der Universität,18 Bundesstaatliche Bakteriologisch-Serologische Untersuchungsanstalt Wien, Vienna, Austria,28 Department of Hygiene and Microbiology, University of Palermo, Palermo,19 Department of Public Health, University of Florence, Florence, Italy,23 Laboratory of Molecular Microbiology, Saint Petersburg Pasteur Institute, Saint Petersburg, Russia,21 Les Centres Gheskio, INLR, Port au Prince, Haïti,24 Institut Pasteur de Madagascar, Tananarive, Madagascar,25 Institute of Medical Microbiology and Immunology, Göteborg University, Göteborg, Sweden,26 IVIC, Centro de Microbiología y Biología Celular, Laboratorio de Genética Molecular,30 Tuberculosis Laboratory, Instituto de Biomedicina, Caracas, Venezuela32
1,
M Cristina Gutierrez
M Cristina Gutierrez
Unité de la Tuberculose et des Mycobactéries, Institut Pasteur de Guadeloupe, Pointe-à-Pitre, Guadeloupe,1 Wadsworth Center, New York State Department Of Health, Albany New York,2 Public Health Research Institute, Tuberculosis Center, Newark, New Jersey,4 Diagnosis Laboratory for Infectious Diseases and Perinatal Screening RIVM, Bilthoven, The Netherlands,3 National Institute of Hygiene and Epidemiology, Hanoi, Vietnam,5 Department of Microbiology, Leeds General Infirmary, Leeds,6 Medical School, St Georges Hospital,7 Mycobacterium Reference Unit, PHLS, Dulwich Hospital, London,13 University of Surrey, Guildford, Surrey, United Kingdom,11 Instituto de Biotecnologia INTA, Moron, Argentina,8 Tuberculosis Mycobacteriology Branch, Centers for Disease Control and Prevention, Atlanta, Georgia,9 Australian Reference Laboratory for Bovine Tuberculosis, Department of Agriculture, South Perth, Australia,10 Centro de Biotecnologia, Universidade Federal de Pelotas, Pelotas,12 Universidade Federal do Rio Grande do Sul, Porto Alegre,27 Department of Biochemistry and Molecular Biology, Fiocruz, Oswaldo Cruz Institute, Rio de Janeiro, Brazil,29 University Childrens Hospital, Heidelberg,14 Infektions Epidemiology, Robert Koch Institute, Berlin,16 Bundesinstitut für Gesundheitlichen Verbraucherschutz und Veterinärmedizin, Jena, Germany,20 Centre National de Référence des Mycobactéries, Institut Pasteur, Paris,15 Laboratoire de Bactériologie, CHU de Brest, Brest,22 Laboratoire de Bactériologie, CHU de Bordeaux, Bordeaux, France,31 Institut Pasteur de Bangui, Bangui, Central African Republic,17 Klinische Mikrobiologie, Hygiene-Institut der Universität,18 Bundesstaatliche Bakteriologisch-Serologische Untersuchungsanstalt Wien, Vienna, Austria,28 Department of Hygiene and Microbiology, University of Palermo, Palermo,19 Department of Public Health, University of Florence, Florence, Italy,23 Laboratory of Molecular Microbiology, Saint Petersburg Pasteur Institute, Saint Petersburg, Russia,21 Les Centres Gheskio, INLR, Port au Prince, Haïti,24 Institut Pasteur de Madagascar, Tananarive, Madagascar,25 Institute of Medical Microbiology and Immunology, Göteborg University, Göteborg, Sweden,26 IVIC, Centro de Microbiología y Biología Celular, Laboratorio de Genética Molecular,30 Tuberculosis Laboratory, Instituto de Biomedicina, Caracas, Venezuela32
15,
Walter H Haas
Walter H Haas
Unité de la Tuberculose et des Mycobactéries, Institut Pasteur de Guadeloupe, Pointe-à-Pitre, Guadeloupe,1 Wadsworth Center, New York State Department Of Health, Albany New York,2 Public Health Research Institute, Tuberculosis Center, Newark, New Jersey,4 Diagnosis Laboratory for Infectious Diseases and Perinatal Screening RIVM, Bilthoven, The Netherlands,3 National Institute of Hygiene and Epidemiology, Hanoi, Vietnam,5 Department of Microbiology, Leeds General Infirmary, Leeds,6 Medical School, St Georges Hospital,7 Mycobacterium Reference Unit, PHLS, Dulwich Hospital, London,13 University of Surrey, Guildford, Surrey, United Kingdom,11 Instituto de Biotecnologia INTA, Moron, Argentina,8 Tuberculosis Mycobacteriology Branch, Centers for Disease Control and Prevention, Atlanta, Georgia,9 Australian Reference Laboratory for Bovine Tuberculosis, Department of Agriculture, South Perth, Australia,10 Centro de Biotecnologia, Universidade Federal de Pelotas, Pelotas,12 Universidade Federal do Rio Grande do Sul, Porto Alegre,27 Department of Biochemistry and Molecular Biology, Fiocruz, Oswaldo Cruz Institute, Rio de Janeiro, Brazil,29 University Childrens Hospital, Heidelberg,14 Infektions Epidemiology, Robert Koch Institute, Berlin,16 Bundesinstitut für Gesundheitlichen Verbraucherschutz und Veterinärmedizin, Jena, Germany,20 Centre National de Référence des Mycobactéries, Institut Pasteur, Paris,15 Laboratoire de Bactériologie, CHU de Brest, Brest,22 Laboratoire de Bactériologie, CHU de Bordeaux, Bordeaux, France,31 Institut Pasteur de Bangui, Bangui, Central African Republic,17 Klinische Mikrobiologie, Hygiene-Institut der Universität,18 Bundesstaatliche Bakteriologisch-Serologische Untersuchungsanstalt Wien, Vienna, Austria,28 Department of Hygiene and Microbiology, University of Palermo, Palermo,19 Department of Public Health, University of Florence, Florence, Italy,23 Laboratory of Molecular Microbiology, Saint Petersburg Pasteur Institute, Saint Petersburg, Russia,21 Les Centres Gheskio, INLR, Port au Prince, Haïti,24 Institut Pasteur de Madagascar, Tananarive, Madagascar,25 Institute of Medical Microbiology and Immunology, Göteborg University, Göteborg, Sweden,26 IVIC, Centro de Microbiología y Biología Celular, Laboratorio de Genética Molecular,30 Tuberculosis Laboratory, Instituto de Biomedicina, Caracas, Venezuela32
16,
Herre Heersma
Herre Heersma
Unité de la Tuberculose et des Mycobactéries, Institut Pasteur de Guadeloupe, Pointe-à-Pitre, Guadeloupe,1 Wadsworth Center, New York State Department Of Health, Albany New York,2 Public Health Research Institute, Tuberculosis Center, Newark, New Jersey,4 Diagnosis Laboratory for Infectious Diseases and Perinatal Screening RIVM, Bilthoven, The Netherlands,3 National Institute of Hygiene and Epidemiology, Hanoi, Vietnam,5 Department of Microbiology, Leeds General Infirmary, Leeds,6 Medical School, St Georges Hospital,7 Mycobacterium Reference Unit, PHLS, Dulwich Hospital, London,13 University of Surrey, Guildford, Surrey, United Kingdom,11 Instituto de Biotecnologia INTA, Moron, Argentina,8 Tuberculosis Mycobacteriology Branch, Centers for Disease Control and Prevention, Atlanta, Georgia,9 Australian Reference Laboratory for Bovine Tuberculosis, Department of Agriculture, South Perth, Australia,10 Centro de Biotecnologia, Universidade Federal de Pelotas, Pelotas,12 Universidade Federal do Rio Grande do Sul, Porto Alegre,27 Department of Biochemistry and Molecular Biology, Fiocruz, Oswaldo Cruz Institute, Rio de Janeiro, Brazil,29 University Childrens Hospital, Heidelberg,14 Infektions Epidemiology, Robert Koch Institute, Berlin,16 Bundesinstitut für Gesundheitlichen Verbraucherschutz und Veterinärmedizin, Jena, Germany,20 Centre National de Référence des Mycobactéries, Institut Pasteur, Paris,15 Laboratoire de Bactériologie, CHU de Brest, Brest,22 Laboratoire de Bactériologie, CHU de Bordeaux, Bordeaux, France,31 Institut Pasteur de Bangui, Bangui, Central African Republic,17 Klinische Mikrobiologie, Hygiene-Institut der Universität,18 Bundesstaatliche Bakteriologisch-Serologische Untersuchungsanstalt Wien, Vienna, Austria,28 Department of Hygiene and Microbiology, University of Palermo, Palermo,19 Department of Public Health, University of Florence, Florence, Italy,23 Laboratory of Molecular Microbiology, Saint Petersburg Pasteur Institute, Saint Petersburg, Russia,21 Les Centres Gheskio, INLR, Port au Prince, Haïti,24 Institut Pasteur de Madagascar, Tananarive, Madagascar,25 Institute of Medical Microbiology and Immunology, Göteborg University, Göteborg, Sweden,26 IVIC, Centro de Microbiología y Biología Celular, Laboratorio de Genética Molecular,30 Tuberculosis Laboratory, Instituto de Biomedicina, Caracas, Venezuela32
3,
Eric Kassa-Kelembho
Eric Kassa-Kelembho
Unité de la Tuberculose et des Mycobactéries, Institut Pasteur de Guadeloupe, Pointe-à-Pitre, Guadeloupe,1 Wadsworth Center, New York State Department Of Health, Albany New York,2 Public Health Research Institute, Tuberculosis Center, Newark, New Jersey,4 Diagnosis Laboratory for Infectious Diseases and Perinatal Screening RIVM, Bilthoven, The Netherlands,3 National Institute of Hygiene and Epidemiology, Hanoi, Vietnam,5 Department of Microbiology, Leeds General Infirmary, Leeds,6 Medical School, St Georges Hospital,7 Mycobacterium Reference Unit, PHLS, Dulwich Hospital, London,13 University of Surrey, Guildford, Surrey, United Kingdom,11 Instituto de Biotecnologia INTA, Moron, Argentina,8 Tuberculosis Mycobacteriology Branch, Centers for Disease Control and Prevention, Atlanta, Georgia,9 Australian Reference Laboratory for Bovine Tuberculosis, Department of Agriculture, South Perth, Australia,10 Centro de Biotecnologia, Universidade Federal de Pelotas, Pelotas,12 Universidade Federal do Rio Grande do Sul, Porto Alegre,27 Department of Biochemistry and Molecular Biology, Fiocruz, Oswaldo Cruz Institute, Rio de Janeiro, Brazil,29 University Childrens Hospital, Heidelberg,14 Infektions Epidemiology, Robert Koch Institute, Berlin,16 Bundesinstitut für Gesundheitlichen Verbraucherschutz und Veterinärmedizin, Jena, Germany,20 Centre National de Référence des Mycobactéries, Institut Pasteur, Paris,15 Laboratoire de Bactériologie, CHU de Brest, Brest,22 Laboratoire de Bactériologie, CHU de Bordeaux, Bordeaux, France,31 Institut Pasteur de Bangui, Bangui, Central African Republic,17 Klinische Mikrobiologie, Hygiene-Institut der Universität,18 Bundesstaatliche Bakteriologisch-Serologische Untersuchungsanstalt Wien, Vienna, Austria,28 Department of Hygiene and Microbiology, University of Palermo, Palermo,19 Department of Public Health, University of Florence, Florence, Italy,23 Laboratory of Molecular Microbiology, Saint Petersburg Pasteur Institute, Saint Petersburg, Russia,21 Les Centres Gheskio, INLR, Port au Prince, Haïti,24 Institut Pasteur de Madagascar, Tananarive, Madagascar,25 Institute of Medical Microbiology and Immunology, Göteborg University, Göteborg, Sweden,26 IVIC, Centro de Microbiología y Biología Celular, Laboratorio de Genética Molecular,30 Tuberculosis Laboratory, Instituto de Biomedicina, Caracas, Venezuela32
17,
Ho Minh Ly
Ho Minh Ly
Unité de la Tuberculose et des Mycobactéries, Institut Pasteur de Guadeloupe, Pointe-à-Pitre, Guadeloupe,1 Wadsworth Center, New York State Department Of Health, Albany New York,2 Public Health Research Institute, Tuberculosis Center, Newark, New Jersey,4 Diagnosis Laboratory for Infectious Diseases and Perinatal Screening RIVM, Bilthoven, The Netherlands,3 National Institute of Hygiene and Epidemiology, Hanoi, Vietnam,5 Department of Microbiology, Leeds General Infirmary, Leeds,6 Medical School, St Georges Hospital,7 Mycobacterium Reference Unit, PHLS, Dulwich Hospital, London,13 University of Surrey, Guildford, Surrey, United Kingdom,11 Instituto de Biotecnologia INTA, Moron, Argentina,8 Tuberculosis Mycobacteriology Branch, Centers for Disease Control and Prevention, Atlanta, Georgia,9 Australian Reference Laboratory for Bovine Tuberculosis, Department of Agriculture, South Perth, Australia,10 Centro de Biotecnologia, Universidade Federal de Pelotas, Pelotas,12 Universidade Federal do Rio Grande do Sul, Porto Alegre,27 Department of Biochemistry and Molecular Biology, Fiocruz, Oswaldo Cruz Institute, Rio de Janeiro, Brazil,29 University Childrens Hospital, Heidelberg,14 Infektions Epidemiology, Robert Koch Institute, Berlin,16 Bundesinstitut für Gesundheitlichen Verbraucherschutz und Veterinärmedizin, Jena, Germany,20 Centre National de Référence des Mycobactéries, Institut Pasteur, Paris,15 Laboratoire de Bactériologie, CHU de Brest, Brest,22 Laboratoire de Bactériologie, CHU de Bordeaux, Bordeaux, France,31 Institut Pasteur de Bangui, Bangui, Central African Republic,17 Klinische Mikrobiologie, Hygiene-Institut der Universität,18 Bundesstaatliche Bakteriologisch-Serologische Untersuchungsanstalt Wien, Vienna, Austria,28 Department of Hygiene and Microbiology, University of Palermo, Palermo,19 Department of Public Health, University of Florence, Florence, Italy,23 Laboratory of Molecular Microbiology, Saint Petersburg Pasteur Institute, Saint Petersburg, Russia,21 Les Centres Gheskio, INLR, Port au Prince, Haïti,24 Institut Pasteur de Madagascar, Tananarive, Madagascar,25 Institute of Medical Microbiology and Immunology, Göteborg University, Göteborg, Sweden,26 IVIC, Centro de Microbiología y Biología Celular, Laboratorio de Genética Molecular,30 Tuberculosis Laboratory, Instituto de Biomedicina, Caracas, Venezuela32
5,
Athanasios Makristathis
Athanasios Makristathis
Unité de la Tuberculose et des Mycobactéries, Institut Pasteur de Guadeloupe, Pointe-à-Pitre, Guadeloupe,1 Wadsworth Center, New York State Department Of Health, Albany New York,2 Public Health Research Institute, Tuberculosis Center, Newark, New Jersey,4 Diagnosis Laboratory for Infectious Diseases and Perinatal Screening RIVM, Bilthoven, The Netherlands,3 National Institute of Hygiene and Epidemiology, Hanoi, Vietnam,5 Department of Microbiology, Leeds General Infirmary, Leeds,6 Medical School, St Georges Hospital,7 Mycobacterium Reference Unit, PHLS, Dulwich Hospital, London,13 University of Surrey, Guildford, Surrey, United Kingdom,11 Instituto de Biotecnologia INTA, Moron, Argentina,8 Tuberculosis Mycobacteriology Branch, Centers for Disease Control and Prevention, Atlanta, Georgia,9 Australian Reference Laboratory for Bovine Tuberculosis, Department of Agriculture, South Perth, Australia,10 Centro de Biotecnologia, Universidade Federal de Pelotas, Pelotas,12 Universidade Federal do Rio Grande do Sul, Porto Alegre,27 Department of Biochemistry and Molecular Biology, Fiocruz, Oswaldo Cruz Institute, Rio de Janeiro, Brazil,29 University Childrens Hospital, Heidelberg,14 Infektions Epidemiology, Robert Koch Institute, Berlin,16 Bundesinstitut für Gesundheitlichen Verbraucherschutz und Veterinärmedizin, Jena, Germany,20 Centre National de Référence des Mycobactéries, Institut Pasteur, Paris,15 Laboratoire de Bactériologie, CHU de Brest, Brest,22 Laboratoire de Bactériologie, CHU de Bordeaux, Bordeaux, France,31 Institut Pasteur de Bangui, Bangui, Central African Republic,17 Klinische Mikrobiologie, Hygiene-Institut der Universität,18 Bundesstaatliche Bakteriologisch-Serologische Untersuchungsanstalt Wien, Vienna, Austria,28 Department of Hygiene and Microbiology, University of Palermo, Palermo,19 Department of Public Health, University of Florence, Florence, Italy,23 Laboratory of Molecular Microbiology, Saint Petersburg Pasteur Institute, Saint Petersburg, Russia,21 Les Centres Gheskio, INLR, Port au Prince, Haïti,24 Institut Pasteur de Madagascar, Tananarive, Madagascar,25 Institute of Medical Microbiology and Immunology, Göteborg University, Göteborg, Sweden,26 IVIC, Centro de Microbiología y Biología Celular, Laboratorio de Genética Molecular,30 Tuberculosis Laboratory, Instituto de Biomedicina, Caracas, Venezuela32
18,
Caterina Mammina
Caterina Mammina
Unité de la Tuberculose et des Mycobactéries, Institut Pasteur de Guadeloupe, Pointe-à-Pitre, Guadeloupe,1 Wadsworth Center, New York State Department Of Health, Albany New York,2 Public Health Research Institute, Tuberculosis Center, Newark, New Jersey,4 Diagnosis Laboratory for Infectious Diseases and Perinatal Screening RIVM, Bilthoven, The Netherlands,3 National Institute of Hygiene and Epidemiology, Hanoi, Vietnam,5 Department of Microbiology, Leeds General Infirmary, Leeds,6 Medical School, St Georges Hospital,7 Mycobacterium Reference Unit, PHLS, Dulwich Hospital, London,13 University of Surrey, Guildford, Surrey, United Kingdom,11 Instituto de Biotecnologia INTA, Moron, Argentina,8 Tuberculosis Mycobacteriology Branch, Centers for Disease Control and Prevention, Atlanta, Georgia,9 Australian Reference Laboratory for Bovine Tuberculosis, Department of Agriculture, South Perth, Australia,10 Centro de Biotecnologia, Universidade Federal de Pelotas, Pelotas,12 Universidade Federal do Rio Grande do Sul, Porto Alegre,27 Department of Biochemistry and Molecular Biology, Fiocruz, Oswaldo Cruz Institute, Rio de Janeiro, Brazil,29 University Childrens Hospital, Heidelberg,14 Infektions Epidemiology, Robert Koch Institute, Berlin,16 Bundesinstitut für Gesundheitlichen Verbraucherschutz und Veterinärmedizin, Jena, Germany,20 Centre National de Référence des Mycobactéries, Institut Pasteur, Paris,15 Laboratoire de Bactériologie, CHU de Brest, Brest,22 Laboratoire de Bactériologie, CHU de Bordeaux, Bordeaux, France,31 Institut Pasteur de Bangui, Bangui, Central African Republic,17 Klinische Mikrobiologie, Hygiene-Institut der Universität,18 Bundesstaatliche Bakteriologisch-Serologische Untersuchungsanstalt Wien, Vienna, Austria,28 Department of Hygiene and Microbiology, University of Palermo, Palermo,19 Department of Public Health, University of Florence, Florence, Italy,23 Laboratory of Molecular Microbiology, Saint Petersburg Pasteur Institute, Saint Petersburg, Russia,21 Les Centres Gheskio, INLR, Port au Prince, Haïti,24 Institut Pasteur de Madagascar, Tananarive, Madagascar,25 Institute of Medical Microbiology and Immunology, Göteborg University, Göteborg, Sweden,26 IVIC, Centro de Microbiología y Biología Celular, Laboratorio de Genética Molecular,30 Tuberculosis Laboratory, Instituto de Biomedicina, Caracas, Venezuela32
19,
Gerald Martin
Gerald Martin
Unité de la Tuberculose et des Mycobactéries, Institut Pasteur de Guadeloupe, Pointe-à-Pitre, Guadeloupe,1 Wadsworth Center, New York State Department Of Health, Albany New York,2 Public Health Research Institute, Tuberculosis Center, Newark, New Jersey,4 Diagnosis Laboratory for Infectious Diseases and Perinatal Screening RIVM, Bilthoven, The Netherlands,3 National Institute of Hygiene and Epidemiology, Hanoi, Vietnam,5 Department of Microbiology, Leeds General Infirmary, Leeds,6 Medical School, St Georges Hospital,7 Mycobacterium Reference Unit, PHLS, Dulwich Hospital, London,13 University of Surrey, Guildford, Surrey, United Kingdom,11 Instituto de Biotecnologia INTA, Moron, Argentina,8 Tuberculosis Mycobacteriology Branch, Centers for Disease Control and Prevention, Atlanta, Georgia,9 Australian Reference Laboratory for Bovine Tuberculosis, Department of Agriculture, South Perth, Australia,10 Centro de Biotecnologia, Universidade Federal de Pelotas, Pelotas,12 Universidade Federal do Rio Grande do Sul, Porto Alegre,27 Department of Biochemistry and Molecular Biology, Fiocruz, Oswaldo Cruz Institute, Rio de Janeiro, Brazil,29 University Childrens Hospital, Heidelberg,14 Infektions Epidemiology, Robert Koch Institute, Berlin,16 Bundesinstitut für Gesundheitlichen Verbraucherschutz und Veterinärmedizin, Jena, Germany,20 Centre National de Référence des Mycobactéries, Institut Pasteur, Paris,15 Laboratoire de Bactériologie, CHU de Brest, Brest,22 Laboratoire de Bactériologie, CHU de Bordeaux, Bordeaux, France,31 Institut Pasteur de Bangui, Bangui, Central African Republic,17 Klinische Mikrobiologie, Hygiene-Institut der Universität,18 Bundesstaatliche Bakteriologisch-Serologische Untersuchungsanstalt Wien, Vienna, Austria,28 Department of Hygiene and Microbiology, University of Palermo, Palermo,19 Department of Public Health, University of Florence, Florence, Italy,23 Laboratory of Molecular Microbiology, Saint Petersburg Pasteur Institute, Saint Petersburg, Russia,21 Les Centres Gheskio, INLR, Port au Prince, Haïti,24 Institut Pasteur de Madagascar, Tananarive, Madagascar,25 Institute of Medical Microbiology and Immunology, Göteborg University, Göteborg, Sweden,26 IVIC, Centro de Microbiología y Biología Celular, Laboratorio de Genética Molecular,30 Tuberculosis Laboratory, Instituto de Biomedicina, Caracas, Venezuela32
20,
Peter Moström
Peter Moström
Unité de la Tuberculose et des Mycobactéries, Institut Pasteur de Guadeloupe, Pointe-à-Pitre, Guadeloupe,1 Wadsworth Center, New York State Department Of Health, Albany New York,2 Public Health Research Institute, Tuberculosis Center, Newark, New Jersey,4 Diagnosis Laboratory for Infectious Diseases and Perinatal Screening RIVM, Bilthoven, The Netherlands,3 National Institute of Hygiene and Epidemiology, Hanoi, Vietnam,5 Department of Microbiology, Leeds General Infirmary, Leeds,6 Medical School, St Georges Hospital,7 Mycobacterium Reference Unit, PHLS, Dulwich Hospital, London,13 University of Surrey, Guildford, Surrey, United Kingdom,11 Instituto de Biotecnologia INTA, Moron, Argentina,8 Tuberculosis Mycobacteriology Branch, Centers for Disease Control and Prevention, Atlanta, Georgia,9 Australian Reference Laboratory for Bovine Tuberculosis, Department of Agriculture, South Perth, Australia,10 Centro de Biotecnologia, Universidade Federal de Pelotas, Pelotas,12 Universidade Federal do Rio Grande do Sul, Porto Alegre,27 Department of Biochemistry and Molecular Biology, Fiocruz, Oswaldo Cruz Institute, Rio de Janeiro, Brazil,29 University Childrens Hospital, Heidelberg,14 Infektions Epidemiology, Robert Koch Institute, Berlin,16 Bundesinstitut für Gesundheitlichen Verbraucherschutz und Veterinärmedizin, Jena, Germany,20 Centre National de Référence des Mycobactéries, Institut Pasteur, Paris,15 Laboratoire de Bactériologie, CHU de Brest, Brest,22 Laboratoire de Bactériologie, CHU de Bordeaux, Bordeaux, France,31 Institut Pasteur de Bangui, Bangui, Central African Republic,17 Klinische Mikrobiologie, Hygiene-Institut der Universität,18 Bundesstaatliche Bakteriologisch-Serologische Untersuchungsanstalt Wien, Vienna, Austria,28 Department of Hygiene and Microbiology, University of Palermo, Palermo,19 Department of Public Health, University of Florence, Florence, Italy,23 Laboratory of Molecular Microbiology, Saint Petersburg Pasteur Institute, Saint Petersburg, Russia,21 Les Centres Gheskio, INLR, Port au Prince, Haïti,24 Institut Pasteur de Madagascar, Tananarive, Madagascar,25 Institute of Medical Microbiology and Immunology, Göteborg University, Göteborg, Sweden,26 IVIC, Centro de Microbiología y Biología Celular, Laboratorio de Genética Molecular,30 Tuberculosis Laboratory, Instituto de Biomedicina, Caracas, Venezuela32
1,
Igor Mokrousov
Igor Mokrousov
Unité de la Tuberculose et des Mycobactéries, Institut Pasteur de Guadeloupe, Pointe-à-Pitre, Guadeloupe,1 Wadsworth Center, New York State Department Of Health, Albany New York,2 Public Health Research Institute, Tuberculosis Center, Newark, New Jersey,4 Diagnosis Laboratory for Infectious Diseases and Perinatal Screening RIVM, Bilthoven, The Netherlands,3 National Institute of Hygiene and Epidemiology, Hanoi, Vietnam,5 Department of Microbiology, Leeds General Infirmary, Leeds,6 Medical School, St Georges Hospital,7 Mycobacterium Reference Unit, PHLS, Dulwich Hospital, London,13 University of Surrey, Guildford, Surrey, United Kingdom,11 Instituto de Biotecnologia INTA, Moron, Argentina,8 Tuberculosis Mycobacteriology Branch, Centers for Disease Control and Prevention, Atlanta, Georgia,9 Australian Reference Laboratory for Bovine Tuberculosis, Department of Agriculture, South Perth, Australia,10 Centro de Biotecnologia, Universidade Federal de Pelotas, Pelotas,12 Universidade Federal do Rio Grande do Sul, Porto Alegre,27 Department of Biochemistry and Molecular Biology, Fiocruz, Oswaldo Cruz Institute, Rio de Janeiro, Brazil,29 University Childrens Hospital, Heidelberg,14 Infektions Epidemiology, Robert Koch Institute, Berlin,16 Bundesinstitut für Gesundheitlichen Verbraucherschutz und Veterinärmedizin, Jena, Germany,20 Centre National de Référence des Mycobactéries, Institut Pasteur, Paris,15 Laboratoire de Bactériologie, CHU de Brest, Brest,22 Laboratoire de Bactériologie, CHU de Bordeaux, Bordeaux, France,31 Institut Pasteur de Bangui, Bangui, Central African Republic,17 Klinische Mikrobiologie, Hygiene-Institut der Universität,18 Bundesstaatliche Bakteriologisch-Serologische Untersuchungsanstalt Wien, Vienna, Austria,28 Department of Hygiene and Microbiology, University of Palermo, Palermo,19 Department of Public Health, University of Florence, Florence, Italy,23 Laboratory of Molecular Microbiology, Saint Petersburg Pasteur Institute, Saint Petersburg, Russia,21 Les Centres Gheskio, INLR, Port au Prince, Haïti,24 Institut Pasteur de Madagascar, Tananarive, Madagascar,25 Institute of Medical Microbiology and Immunology, Göteborg University, Göteborg, Sweden,26 IVIC, Centro de Microbiología y Biología Celular, Laboratorio de Genética Molecular,30 Tuberculosis Laboratory, Instituto de Biomedicina, Caracas, Venezuela32
21,
Valérie Narbonne
Valérie Narbonne
Unité de la Tuberculose et des Mycobactéries, Institut Pasteur de Guadeloupe, Pointe-à-Pitre, Guadeloupe,1 Wadsworth Center, New York State Department Of Health, Albany New York,2 Public Health Research Institute, Tuberculosis Center, Newark, New Jersey,4 Diagnosis Laboratory for Infectious Diseases and Perinatal Screening RIVM, Bilthoven, The Netherlands,3 National Institute of Hygiene and Epidemiology, Hanoi, Vietnam,5 Department of Microbiology, Leeds General Infirmary, Leeds,6 Medical School, St Georges Hospital,7 Mycobacterium Reference Unit, PHLS, Dulwich Hospital, London,13 University of Surrey, Guildford, Surrey, United Kingdom,11 Instituto de Biotecnologia INTA, Moron, Argentina,8 Tuberculosis Mycobacteriology Branch, Centers for Disease Control and Prevention, Atlanta, Georgia,9 Australian Reference Laboratory for Bovine Tuberculosis, Department of Agriculture, South Perth, Australia,10 Centro de Biotecnologia, Universidade Federal de Pelotas, Pelotas,12 Universidade Federal do Rio Grande do Sul, Porto Alegre,27 Department of Biochemistry and Molecular Biology, Fiocruz, Oswaldo Cruz Institute, Rio de Janeiro, Brazil,29 University Childrens Hospital, Heidelberg,14 Infektions Epidemiology, Robert Koch Institute, Berlin,16 Bundesinstitut für Gesundheitlichen Verbraucherschutz und Veterinärmedizin, Jena, Germany,20 Centre National de Référence des Mycobactéries, Institut Pasteur, Paris,15 Laboratoire de Bactériologie, CHU de Brest, Brest,22 Laboratoire de Bactériologie, CHU de Bordeaux, Bordeaux, France,31 Institut Pasteur de Bangui, Bangui, Central African Republic,17 Klinische Mikrobiologie, Hygiene-Institut der Universität,18 Bundesstaatliche Bakteriologisch-Serologische Untersuchungsanstalt Wien, Vienna, Austria,28 Department of Hygiene and Microbiology, University of Palermo, Palermo,19 Department of Public Health, University of Florence, Florence, Italy,23 Laboratory of Molecular Microbiology, Saint Petersburg Pasteur Institute, Saint Petersburg, Russia,21 Les Centres Gheskio, INLR, Port au Prince, Haïti,24 Institut Pasteur de Madagascar, Tananarive, Madagascar,25 Institute of Medical Microbiology and Immunology, Göteborg University, Göteborg, Sweden,26 IVIC, Centro de Microbiología y Biología Celular, Laboratorio de Genética Molecular,30 Tuberculosis Laboratory, Instituto de Biomedicina, Caracas, Venezuela32
22,
Olga Narvskaya
Olga Narvskaya
Unité de la Tuberculose et des Mycobactéries, Institut Pasteur de Guadeloupe, Pointe-à-Pitre, Guadeloupe,1 Wadsworth Center, New York State Department Of Health, Albany New York,2 Public Health Research Institute, Tuberculosis Center, Newark, New Jersey,4 Diagnosis Laboratory for Infectious Diseases and Perinatal Screening RIVM, Bilthoven, The Netherlands,3 National Institute of Hygiene and Epidemiology, Hanoi, Vietnam,5 Department of Microbiology, Leeds General Infirmary, Leeds,6 Medical School, St Georges Hospital,7 Mycobacterium Reference Unit, PHLS, Dulwich Hospital, London,13 University of Surrey, Guildford, Surrey, United Kingdom,11 Instituto de Biotecnologia INTA, Moron, Argentina,8 Tuberculosis Mycobacteriology Branch, Centers for Disease Control and Prevention, Atlanta, Georgia,9 Australian Reference Laboratory for Bovine Tuberculosis, Department of Agriculture, South Perth, Australia,10 Centro de Biotecnologia, Universidade Federal de Pelotas, Pelotas,12 Universidade Federal do Rio Grande do Sul, Porto Alegre,27 Department of Biochemistry and Molecular Biology, Fiocruz, Oswaldo Cruz Institute, Rio de Janeiro, Brazil,29 University Childrens Hospital, Heidelberg,14 Infektions Epidemiology, Robert Koch Institute, Berlin,16 Bundesinstitut für Gesundheitlichen Verbraucherschutz und Veterinärmedizin, Jena, Germany,20 Centre National de Référence des Mycobactéries, Institut Pasteur, Paris,15 Laboratoire de Bactériologie, CHU de Brest, Brest,22 Laboratoire de Bactériologie, CHU de Bordeaux, Bordeaux, France,31 Institut Pasteur de Bangui, Bangui, Central African Republic,17 Klinische Mikrobiologie, Hygiene-Institut der Universität,18 Bundesstaatliche Bakteriologisch-Serologische Untersuchungsanstalt Wien, Vienna, Austria,28 Department of Hygiene and Microbiology, University of Palermo, Palermo,19 Department of Public Health, University of Florence, Florence, Italy,23 Laboratory of Molecular Microbiology, Saint Petersburg Pasteur Institute, Saint Petersburg, Russia,21 Les Centres Gheskio, INLR, Port au Prince, Haïti,24 Institut Pasteur de Madagascar, Tananarive, Madagascar,25 Institute of Medical Microbiology and Immunology, Göteborg University, Göteborg, Sweden,26 IVIC, Centro de Microbiología y Biología Celular, Laboratorio de Genética Molecular,30 Tuberculosis Laboratory, Instituto de Biomedicina, Caracas, Venezuela32
21,
Antonino Nastasi
Antonino Nastasi
Unité de la Tuberculose et des Mycobactéries, Institut Pasteur de Guadeloupe, Pointe-à-Pitre, Guadeloupe,1 Wadsworth Center, New York State Department Of Health, Albany New York,2 Public Health Research Institute, Tuberculosis Center, Newark, New Jersey,4 Diagnosis Laboratory for Infectious Diseases and Perinatal Screening RIVM, Bilthoven, The Netherlands,3 National Institute of Hygiene and Epidemiology, Hanoi, Vietnam,5 Department of Microbiology, Leeds General Infirmary, Leeds,6 Medical School, St Georges Hospital,7 Mycobacterium Reference Unit, PHLS, Dulwich Hospital, London,13 University of Surrey, Guildford, Surrey, United Kingdom,11 Instituto de Biotecnologia INTA, Moron, Argentina,8 Tuberculosis Mycobacteriology Branch, Centers for Disease Control and Prevention, Atlanta, Georgia,9 Australian Reference Laboratory for Bovine Tuberculosis, Department of Agriculture, South Perth, Australia,10 Centro de Biotecnologia, Universidade Federal de Pelotas, Pelotas,12 Universidade Federal do Rio Grande do Sul, Porto Alegre,27 Department of Biochemistry and Molecular Biology, Fiocruz, Oswaldo Cruz Institute, Rio de Janeiro, Brazil,29 University Childrens Hospital, Heidelberg,14 Infektions Epidemiology, Robert Koch Institute, Berlin,16 Bundesinstitut für Gesundheitlichen Verbraucherschutz und Veterinärmedizin, Jena, Germany,20 Centre National de Référence des Mycobactéries, Institut Pasteur, Paris,15 Laboratoire de Bactériologie, CHU de Brest, Brest,22 Laboratoire de Bactériologie, CHU de Bordeaux, Bordeaux, France,31 Institut Pasteur de Bangui, Bangui, Central African Republic,17 Klinische Mikrobiologie, Hygiene-Institut der Universität,18 Bundesstaatliche Bakteriologisch-Serologische Untersuchungsanstalt Wien, Vienna, Austria,28 Department of Hygiene and Microbiology, University of Palermo, Palermo,19 Department of Public Health, University of Florence, Florence, Italy,23 Laboratory of Molecular Microbiology, Saint Petersburg Pasteur Institute, Saint Petersburg, Russia,21 Les Centres Gheskio, INLR, Port au Prince, Haïti,24 Institut Pasteur de Madagascar, Tananarive, Madagascar,25 Institute of Medical Microbiology and Immunology, Göteborg University, Göteborg, Sweden,26 IVIC, Centro de Microbiología y Biología Celular, Laboratorio de Genética Molecular,30 Tuberculosis Laboratory, Instituto de Biomedicina, Caracas, Venezuela32
23,
Sara Ngo Niobe-Eyangoh
Sara Ngo Niobe-Eyangoh
Unité de la Tuberculose et des Mycobactéries, Institut Pasteur de Guadeloupe, Pointe-à-Pitre, Guadeloupe,1 Wadsworth Center, New York State Department Of Health, Albany New York,2 Public Health Research Institute, Tuberculosis Center, Newark, New Jersey,4 Diagnosis Laboratory for Infectious Diseases and Perinatal Screening RIVM, Bilthoven, The Netherlands,3 National Institute of Hygiene and Epidemiology, Hanoi, Vietnam,5 Department of Microbiology, Leeds General Infirmary, Leeds,6 Medical School, St Georges Hospital,7 Mycobacterium Reference Unit, PHLS, Dulwich Hospital, London,13 University of Surrey, Guildford, Surrey, United Kingdom,11 Instituto de Biotecnologia INTA, Moron, Argentina,8 Tuberculosis Mycobacteriology Branch, Centers for Disease Control and Prevention, Atlanta, Georgia,9 Australian Reference Laboratory for Bovine Tuberculosis, Department of Agriculture, South Perth, Australia,10 Centro de Biotecnologia, Universidade Federal de Pelotas, Pelotas,12 Universidade Federal do Rio Grande do Sul, Porto Alegre,27 Department of Biochemistry and Molecular Biology, Fiocruz, Oswaldo Cruz Institute, Rio de Janeiro, Brazil,29 University Childrens Hospital, Heidelberg,14 Infektions Epidemiology, Robert Koch Institute, Berlin,16 Bundesinstitut für Gesundheitlichen Verbraucherschutz und Veterinärmedizin, Jena, Germany,20 Centre National de Référence des Mycobactéries, Institut Pasteur, Paris,15 Laboratoire de Bactériologie, CHU de Brest, Brest,22 Laboratoire de Bactériologie, CHU de Bordeaux, Bordeaux, France,31 Institut Pasteur de Bangui, Bangui, Central African Republic,17 Klinische Mikrobiologie, Hygiene-Institut der Universität,18 Bundesstaatliche Bakteriologisch-Serologische Untersuchungsanstalt Wien, Vienna, Austria,28 Department of Hygiene and Microbiology, University of Palermo, Palermo,19 Department of Public Health, University of Florence, Florence, Italy,23 Laboratory of Molecular Microbiology, Saint Petersburg Pasteur Institute, Saint Petersburg, Russia,21 Les Centres Gheskio, INLR, Port au Prince, Haïti,24 Institut Pasteur de Madagascar, Tananarive, Madagascar,25 Institute of Medical Microbiology and Immunology, Göteborg University, Göteborg, Sweden,26 IVIC, Centro de Microbiología y Biología Celular, Laboratorio de Genética Molecular,30 Tuberculosis Laboratory, Instituto de Biomedicina, Caracas, Venezuela32
15,
Jean W Pape
Jean W Pape
Unité de la Tuberculose et des Mycobactéries, Institut Pasteur de Guadeloupe, Pointe-à-Pitre, Guadeloupe,1 Wadsworth Center, New York State Department Of Health, Albany New York,2 Public Health Research Institute, Tuberculosis Center, Newark, New Jersey,4 Diagnosis Laboratory for Infectious Diseases and Perinatal Screening RIVM, Bilthoven, The Netherlands,3 National Institute of Hygiene and Epidemiology, Hanoi, Vietnam,5 Department of Microbiology, Leeds General Infirmary, Leeds,6 Medical School, St Georges Hospital,7 Mycobacterium Reference Unit, PHLS, Dulwich Hospital, London,13 University of Surrey, Guildford, Surrey, United Kingdom,11 Instituto de Biotecnologia INTA, Moron, Argentina,8 Tuberculosis Mycobacteriology Branch, Centers for Disease Control and Prevention, Atlanta, Georgia,9 Australian Reference Laboratory for Bovine Tuberculosis, Department of Agriculture, South Perth, Australia,10 Centro de Biotecnologia, Universidade Federal de Pelotas, Pelotas,12 Universidade Federal do Rio Grande do Sul, Porto Alegre,27 Department of Biochemistry and Molecular Biology, Fiocruz, Oswaldo Cruz Institute, Rio de Janeiro, Brazil,29 University Childrens Hospital, Heidelberg,14 Infektions Epidemiology, Robert Koch Institute, Berlin,16 Bundesinstitut für Gesundheitlichen Verbraucherschutz und Veterinärmedizin, Jena, Germany,20 Centre National de Référence des Mycobactéries, Institut Pasteur, Paris,15 Laboratoire de Bactériologie, CHU de Brest, Brest,22 Laboratoire de Bactériologie, CHU de Bordeaux, Bordeaux, France,31 Institut Pasteur de Bangui, Bangui, Central African Republic,17 Klinische Mikrobiologie, Hygiene-Institut der Universität,18 Bundesstaatliche Bakteriologisch-Serologische Untersuchungsanstalt Wien, Vienna, Austria,28 Department of Hygiene and Microbiology, University of Palermo, Palermo,19 Department of Public Health, University of Florence, Florence, Italy,23 Laboratory of Molecular Microbiology, Saint Petersburg Pasteur Institute, Saint Petersburg, Russia,21 Les Centres Gheskio, INLR, Port au Prince, Haïti,24 Institut Pasteur de Madagascar, Tananarive, Madagascar,25 Institute of Medical Microbiology and Immunology, Göteborg University, Göteborg, Sweden,26 IVIC, Centro de Microbiología y Biología Celular, Laboratorio de Genética Molecular,30 Tuberculosis Laboratory, Instituto de Biomedicina, Caracas, Venezuela32
24,
Voahangy Rasolofo-Razanamparany
Voahangy Rasolofo-Razanamparany
Unité de la Tuberculose et des Mycobactéries, Institut Pasteur de Guadeloupe, Pointe-à-Pitre, Guadeloupe,1 Wadsworth Center, New York State Department Of Health, Albany New York,2 Public Health Research Institute, Tuberculosis Center, Newark, New Jersey,4 Diagnosis Laboratory for Infectious Diseases and Perinatal Screening RIVM, Bilthoven, The Netherlands,3 National Institute of Hygiene and Epidemiology, Hanoi, Vietnam,5 Department of Microbiology, Leeds General Infirmary, Leeds,6 Medical School, St Georges Hospital,7 Mycobacterium Reference Unit, PHLS, Dulwich Hospital, London,13 University of Surrey, Guildford, Surrey, United Kingdom,11 Instituto de Biotecnologia INTA, Moron, Argentina,8 Tuberculosis Mycobacteriology Branch, Centers for Disease Control and Prevention, Atlanta, Georgia,9 Australian Reference Laboratory for Bovine Tuberculosis, Department of Agriculture, South Perth, Australia,10 Centro de Biotecnologia, Universidade Federal de Pelotas, Pelotas,12 Universidade Federal do Rio Grande do Sul, Porto Alegre,27 Department of Biochemistry and Molecular Biology, Fiocruz, Oswaldo Cruz Institute, Rio de Janeiro, Brazil,29 University Childrens Hospital, Heidelberg,14 Infektions Epidemiology, Robert Koch Institute, Berlin,16 Bundesinstitut für Gesundheitlichen Verbraucherschutz und Veterinärmedizin, Jena, Germany,20 Centre National de Référence des Mycobactéries, Institut Pasteur, Paris,15 Laboratoire de Bactériologie, CHU de Brest, Brest,22 Laboratoire de Bactériologie, CHU de Bordeaux, Bordeaux, France,31 Institut Pasteur de Bangui, Bangui, Central African Republic,17 Klinische Mikrobiologie, Hygiene-Institut der Universität,18 Bundesstaatliche Bakteriologisch-Serologische Untersuchungsanstalt Wien, Vienna, Austria,28 Department of Hygiene and Microbiology, University of Palermo, Palermo,19 Department of Public Health, University of Florence, Florence, Italy,23 Laboratory of Molecular Microbiology, Saint Petersburg Pasteur Institute, Saint Petersburg, Russia,21 Les Centres Gheskio, INLR, Port au Prince, Haïti,24 Institut Pasteur de Madagascar, Tananarive, Madagascar,25 Institute of Medical Microbiology and Immunology, Göteborg University, Göteborg, Sweden,26 IVIC, Centro de Microbiología y Biología Celular, Laboratorio de Genética Molecular,30 Tuberculosis Laboratory, Instituto de Biomedicina, Caracas, Venezuela32
25,
Malin Ridell
Malin Ridell
Unité de la Tuberculose et des Mycobactéries, Institut Pasteur de Guadeloupe, Pointe-à-Pitre, Guadeloupe,1 Wadsworth Center, New York State Department Of Health, Albany New York,2 Public Health Research Institute, Tuberculosis Center, Newark, New Jersey,4 Diagnosis Laboratory for Infectious Diseases and Perinatal Screening RIVM, Bilthoven, The Netherlands,3 National Institute of Hygiene and Epidemiology, Hanoi, Vietnam,5 Department of Microbiology, Leeds General Infirmary, Leeds,6 Medical School, St Georges Hospital,7 Mycobacterium Reference Unit, PHLS, Dulwich Hospital, London,13 University of Surrey, Guildford, Surrey, United Kingdom,11 Instituto de Biotecnologia INTA, Moron, Argentina,8 Tuberculosis Mycobacteriology Branch, Centers for Disease Control and Prevention, Atlanta, Georgia,9 Australian Reference Laboratory for Bovine Tuberculosis, Department of Agriculture, South Perth, Australia,10 Centro de Biotecnologia, Universidade Federal de Pelotas, Pelotas,12 Universidade Federal do Rio Grande do Sul, Porto Alegre,27 Department of Biochemistry and Molecular Biology, Fiocruz, Oswaldo Cruz Institute, Rio de Janeiro, Brazil,29 University Childrens Hospital, Heidelberg,14 Infektions Epidemiology, Robert Koch Institute, Berlin,16 Bundesinstitut für Gesundheitlichen Verbraucherschutz und Veterinärmedizin, Jena, Germany,20 Centre National de Référence des Mycobactéries, Institut Pasteur, Paris,15 Laboratoire de Bactériologie, CHU de Brest, Brest,22 Laboratoire de Bactériologie, CHU de Bordeaux, Bordeaux, France,31 Institut Pasteur de Bangui, Bangui, Central African Republic,17 Klinische Mikrobiologie, Hygiene-Institut der Universität,18 Bundesstaatliche Bakteriologisch-Serologische Untersuchungsanstalt Wien, Vienna, Austria,28 Department of Hygiene and Microbiology, University of Palermo, Palermo,19 Department of Public Health, University of Florence, Florence, Italy,23 Laboratory of Molecular Microbiology, Saint Petersburg Pasteur Institute, Saint Petersburg, Russia,21 Les Centres Gheskio, INLR, Port au Prince, Haïti,24 Institut Pasteur de Madagascar, Tananarive, Madagascar,25 Institute of Medical Microbiology and Immunology, Göteborg University, Göteborg, Sweden,26 IVIC, Centro de Microbiología y Biología Celular, Laboratorio de Genética Molecular,30 Tuberculosis Laboratory, Instituto de Biomedicina, Caracas, Venezuela32
26,
M Lucia Rossetti
M Lucia Rossetti
Unité de la Tuberculose et des Mycobactéries, Institut Pasteur de Guadeloupe, Pointe-à-Pitre, Guadeloupe,1 Wadsworth Center, New York State Department Of Health, Albany New York,2 Public Health Research Institute, Tuberculosis Center, Newark, New Jersey,4 Diagnosis Laboratory for Infectious Diseases and Perinatal Screening RIVM, Bilthoven, The Netherlands,3 National Institute of Hygiene and Epidemiology, Hanoi, Vietnam,5 Department of Microbiology, Leeds General Infirmary, Leeds,6 Medical School, St Georges Hospital,7 Mycobacterium Reference Unit, PHLS, Dulwich Hospital, London,13 University of Surrey, Guildford, Surrey, United Kingdom,11 Instituto de Biotecnologia INTA, Moron, Argentina,8 Tuberculosis Mycobacteriology Branch, Centers for Disease Control and Prevention, Atlanta, Georgia,9 Australian Reference Laboratory for Bovine Tuberculosis, Department of Agriculture, South Perth, Australia,10 Centro de Biotecnologia, Universidade Federal de Pelotas, Pelotas,12 Universidade Federal do Rio Grande do Sul, Porto Alegre,27 Department of Biochemistry and Molecular Biology, Fiocruz, Oswaldo Cruz Institute, Rio de Janeiro, Brazil,29 University Childrens Hospital, Heidelberg,14 Infektions Epidemiology, Robert Koch Institute, Berlin,16 Bundesinstitut für Gesundheitlichen Verbraucherschutz und Veterinärmedizin, Jena, Germany,20 Centre National de Référence des Mycobactéries, Institut Pasteur, Paris,15 Laboratoire de Bactériologie, CHU de Brest, Brest,22 Laboratoire de Bactériologie, CHU de Bordeaux, Bordeaux, France,31 Institut Pasteur de Bangui, Bangui, Central African Republic,17 Klinische Mikrobiologie, Hygiene-Institut der Universität,18 Bundesstaatliche Bakteriologisch-Serologische Untersuchungsanstalt Wien, Vienna, Austria,28 Department of Hygiene and Microbiology, University of Palermo, Palermo,19 Department of Public Health, University of Florence, Florence, Italy,23 Laboratory of Molecular Microbiology, Saint Petersburg Pasteur Institute, Saint Petersburg, Russia,21 Les Centres Gheskio, INLR, Port au Prince, Haïti,24 Institut Pasteur de Madagascar, Tananarive, Madagascar,25 Institute of Medical Microbiology and Immunology, Göteborg University, Göteborg, Sweden,26 IVIC, Centro de Microbiología y Biología Celular, Laboratorio de Genética Molecular,30 Tuberculosis Laboratory, Instituto de Biomedicina, Caracas, Venezuela32
27,
Fritz Stauffer
Fritz Stauffer
Unité de la Tuberculose et des Mycobactéries, Institut Pasteur de Guadeloupe, Pointe-à-Pitre, Guadeloupe,1 Wadsworth Center, New York State Department Of Health, Albany New York,2 Public Health Research Institute, Tuberculosis Center, Newark, New Jersey,4 Diagnosis Laboratory for Infectious Diseases and Perinatal Screening RIVM, Bilthoven, The Netherlands,3 National Institute of Hygiene and Epidemiology, Hanoi, Vietnam,5 Department of Microbiology, Leeds General Infirmary, Leeds,6 Medical School, St Georges Hospital,7 Mycobacterium Reference Unit, PHLS, Dulwich Hospital, London,13 University of Surrey, Guildford, Surrey, United Kingdom,11 Instituto de Biotecnologia INTA, Moron, Argentina,8 Tuberculosis Mycobacteriology Branch, Centers for Disease Control and Prevention, Atlanta, Georgia,9 Australian Reference Laboratory for Bovine Tuberculosis, Department of Agriculture, South Perth, Australia,10 Centro de Biotecnologia, Universidade Federal de Pelotas, Pelotas,12 Universidade Federal do Rio Grande do Sul, Porto Alegre,27 Department of Biochemistry and Molecular Biology, Fiocruz, Oswaldo Cruz Institute, Rio de Janeiro, Brazil,29 University Childrens Hospital, Heidelberg,14 Infektions Epidemiology, Robert Koch Institute, Berlin,16 Bundesinstitut für Gesundheitlichen Verbraucherschutz und Veterinärmedizin, Jena, Germany,20 Centre National de Référence des Mycobactéries, Institut Pasteur, Paris,15 Laboratoire de Bactériologie, CHU de Brest, Brest,22 Laboratoire de Bactériologie, CHU de Bordeaux, Bordeaux, France,31 Institut Pasteur de Bangui, Bangui, Central African Republic,17 Klinische Mikrobiologie, Hygiene-Institut der Universität,18 Bundesstaatliche Bakteriologisch-Serologische Untersuchungsanstalt Wien, Vienna, Austria,28 Department of Hygiene and Microbiology, University of Palermo, Palermo,19 Department of Public Health, University of Florence, Florence, Italy,23 Laboratory of Molecular Microbiology, Saint Petersburg Pasteur Institute, Saint Petersburg, Russia,21 Les Centres Gheskio, INLR, Port au Prince, Haïti,24 Institut Pasteur de Madagascar, Tananarive, Madagascar,25 Institute of Medical Microbiology and Immunology, Göteborg University, Göteborg, Sweden,26 IVIC, Centro de Microbiología y Biología Celular, Laboratorio de Genética Molecular,30 Tuberculosis Laboratory, Instituto de Biomedicina, Caracas, Venezuela32
28,
Philip N Suffys
Philip N Suffys
Unité de la Tuberculose et des Mycobactéries, Institut Pasteur de Guadeloupe, Pointe-à-Pitre, Guadeloupe,1 Wadsworth Center, New York State Department Of Health, Albany New York,2 Public Health Research Institute, Tuberculosis Center, Newark, New Jersey,4 Diagnosis Laboratory for Infectious Diseases and Perinatal Screening RIVM, Bilthoven, The Netherlands,3 National Institute of Hygiene and Epidemiology, Hanoi, Vietnam,5 Department of Microbiology, Leeds General Infirmary, Leeds,6 Medical School, St Georges Hospital,7 Mycobacterium Reference Unit, PHLS, Dulwich Hospital, London,13 University of Surrey, Guildford, Surrey, United Kingdom,11 Instituto de Biotecnologia INTA, Moron, Argentina,8 Tuberculosis Mycobacteriology Branch, Centers for Disease Control and Prevention, Atlanta, Georgia,9 Australian Reference Laboratory for Bovine Tuberculosis, Department of Agriculture, South Perth, Australia,10 Centro de Biotecnologia, Universidade Federal de Pelotas, Pelotas,12 Universidade Federal do Rio Grande do Sul, Porto Alegre,27 Department of Biochemistry and Molecular Biology, Fiocruz, Oswaldo Cruz Institute, Rio de Janeiro, Brazil,29 University Childrens Hospital, Heidelberg,14 Infektions Epidemiology, Robert Koch Institute, Berlin,16 Bundesinstitut für Gesundheitlichen Verbraucherschutz und Veterinärmedizin, Jena, Germany,20 Centre National de Référence des Mycobactéries, Institut Pasteur, Paris,15 Laboratoire de Bactériologie, CHU de Brest, Brest,22 Laboratoire de Bactériologie, CHU de Bordeaux, Bordeaux, France,31 Institut Pasteur de Bangui, Bangui, Central African Republic,17 Klinische Mikrobiologie, Hygiene-Institut der Universität,18 Bundesstaatliche Bakteriologisch-Serologische Untersuchungsanstalt Wien, Vienna, Austria,28 Department of Hygiene and Microbiology, University of Palermo, Palermo,19 Department of Public Health, University of Florence, Florence, Italy,23 Laboratory of Molecular Microbiology, Saint Petersburg Pasteur Institute, Saint Petersburg, Russia,21 Les Centres Gheskio, INLR, Port au Prince, Haïti,24 Institut Pasteur de Madagascar, Tananarive, Madagascar,25 Institute of Medical Microbiology and Immunology, Göteborg University, Göteborg, Sweden,26 IVIC, Centro de Microbiología y Biología Celular, Laboratorio de Genética Molecular,30 Tuberculosis Laboratory, Instituto de Biomedicina, Caracas, Venezuela32
29,
Howard Takiff
Howard Takiff
Unité de la Tuberculose et des Mycobactéries, Institut Pasteur de Guadeloupe, Pointe-à-Pitre, Guadeloupe,1 Wadsworth Center, New York State Department Of Health, Albany New York,2 Public Health Research Institute, Tuberculosis Center, Newark, New Jersey,4 Diagnosis Laboratory for Infectious Diseases and Perinatal Screening RIVM, Bilthoven, The Netherlands,3 National Institute of Hygiene and Epidemiology, Hanoi, Vietnam,5 Department of Microbiology, Leeds General Infirmary, Leeds,6 Medical School, St Georges Hospital,7 Mycobacterium Reference Unit, PHLS, Dulwich Hospital, London,13 University of Surrey, Guildford, Surrey, United Kingdom,11 Instituto de Biotecnologia INTA, Moron, Argentina,8 Tuberculosis Mycobacteriology Branch, Centers for Disease Control and Prevention, Atlanta, Georgia,9 Australian Reference Laboratory for Bovine Tuberculosis, Department of Agriculture, South Perth, Australia,10 Centro de Biotecnologia, Universidade Federal de Pelotas, Pelotas,12 Universidade Federal do Rio Grande do Sul, Porto Alegre,27 Department of Biochemistry and Molecular Biology, Fiocruz, Oswaldo Cruz Institute, Rio de Janeiro, Brazil,29 University Childrens Hospital, Heidelberg,14 Infektions Epidemiology, Robert Koch Institute, Berlin,16 Bundesinstitut für Gesundheitlichen Verbraucherschutz und Veterinärmedizin, Jena, Germany,20 Centre National de Référence des Mycobactéries, Institut Pasteur, Paris,15 Laboratoire de Bactériologie, CHU de Brest, Brest,22 Laboratoire de Bactériologie, CHU de Bordeaux, Bordeaux, France,31 Institut Pasteur de Bangui, Bangui, Central African Republic,17 Klinische Mikrobiologie, Hygiene-Institut der Universität,18 Bundesstaatliche Bakteriologisch-Serologische Untersuchungsanstalt Wien, Vienna, Austria,28 Department of Hygiene and Microbiology, University of Palermo, Palermo,19 Department of Public Health, University of Florence, Florence, Italy,23 Laboratory of Molecular Microbiology, Saint Petersburg Pasteur Institute, Saint Petersburg, Russia,21 Les Centres Gheskio, INLR, Port au Prince, Haïti,24 Institut Pasteur de Madagascar, Tananarive, Madagascar,25 Institute of Medical Microbiology and Immunology, Göteborg University, Göteborg, Sweden,26 IVIC, Centro de Microbiología y Biología Celular, Laboratorio de Genética Molecular,30 Tuberculosis Laboratory, Instituto de Biomedicina, Caracas, Venezuela32
30,
Jeanne Texier-Maugein
Jeanne Texier-Maugein
Unité de la Tuberculose et des Mycobactéries, Institut Pasteur de Guadeloupe, Pointe-à-Pitre, Guadeloupe,1 Wadsworth Center, New York State Department Of Health, Albany New York,2 Public Health Research Institute, Tuberculosis Center, Newark, New Jersey,4 Diagnosis Laboratory for Infectious Diseases and Perinatal Screening RIVM, Bilthoven, The Netherlands,3 National Institute of Hygiene and Epidemiology, Hanoi, Vietnam,5 Department of Microbiology, Leeds General Infirmary, Leeds,6 Medical School, St Georges Hospital,7 Mycobacterium Reference Unit, PHLS, Dulwich Hospital, London,13 University of Surrey, Guildford, Surrey, United Kingdom,11 Instituto de Biotecnologia INTA, Moron, Argentina,8 Tuberculosis Mycobacteriology Branch, Centers for Disease Control and Prevention, Atlanta, Georgia,9 Australian Reference Laboratory for Bovine Tuberculosis, Department of Agriculture, South Perth, Australia,10 Centro de Biotecnologia, Universidade Federal de Pelotas, Pelotas,12 Universidade Federal do Rio Grande do Sul, Porto Alegre,27 Department of Biochemistry and Molecular Biology, Fiocruz, Oswaldo Cruz Institute, Rio de Janeiro, Brazil,29 University Childrens Hospital, Heidelberg,14 Infektions Epidemiology, Robert Koch Institute, Berlin,16 Bundesinstitut für Gesundheitlichen Verbraucherschutz und Veterinärmedizin, Jena, Germany,20 Centre National de Référence des Mycobactéries, Institut Pasteur, Paris,15 Laboratoire de Bactériologie, CHU de Brest, Brest,22 Laboratoire de Bactériologie, CHU de Bordeaux, Bordeaux, France,31 Institut Pasteur de Bangui, Bangui, Central African Republic,17 Klinische Mikrobiologie, Hygiene-Institut der Universität,18 Bundesstaatliche Bakteriologisch-Serologische Untersuchungsanstalt Wien, Vienna, Austria,28 Department of Hygiene and Microbiology, University of Palermo, Palermo,19 Department of Public Health, University of Florence, Florence, Italy,23 Laboratory of Molecular Microbiology, Saint Petersburg Pasteur Institute, Saint Petersburg, Russia,21 Les Centres Gheskio, INLR, Port au Prince, Haïti,24 Institut Pasteur de Madagascar, Tananarive, Madagascar,25 Institute of Medical Microbiology and Immunology, Göteborg University, Göteborg, Sweden,26 IVIC, Centro de Microbiología y Biología Celular, Laboratorio de Genética Molecular,30 Tuberculosis Laboratory, Instituto de Biomedicina, Caracas, Venezuela32
31,
Véronique Vincent
Véronique Vincent
Unité de la Tuberculose et des Mycobactéries, Institut Pasteur de Guadeloupe, Pointe-à-Pitre, Guadeloupe,1 Wadsworth Center, New York State Department Of Health, Albany New York,2 Public Health Research Institute, Tuberculosis Center, Newark, New Jersey,4 Diagnosis Laboratory for Infectious Diseases and Perinatal Screening RIVM, Bilthoven, The Netherlands,3 National Institute of Hygiene and Epidemiology, Hanoi, Vietnam,5 Department of Microbiology, Leeds General Infirmary, Leeds,6 Medical School, St Georges Hospital,7 Mycobacterium Reference Unit, PHLS, Dulwich Hospital, London,13 University of Surrey, Guildford, Surrey, United Kingdom,11 Instituto de Biotecnologia INTA, Moron, Argentina,8 Tuberculosis Mycobacteriology Branch, Centers for Disease Control and Prevention, Atlanta, Georgia,9 Australian Reference Laboratory for Bovine Tuberculosis, Department of Agriculture, South Perth, Australia,10 Centro de Biotecnologia, Universidade Federal de Pelotas, Pelotas,12 Universidade Federal do Rio Grande do Sul, Porto Alegre,27 Department of Biochemistry and Molecular Biology, Fiocruz, Oswaldo Cruz Institute, Rio de Janeiro, Brazil,29 University Childrens Hospital, Heidelberg,14 Infektions Epidemiology, Robert Koch Institute, Berlin,16 Bundesinstitut für Gesundheitlichen Verbraucherschutz und Veterinärmedizin, Jena, Germany,20 Centre National de Référence des Mycobactéries, Institut Pasteur, Paris,15 Laboratoire de Bactériologie, CHU de Brest, Brest,22 Laboratoire de Bactériologie, CHU de Bordeaux, Bordeaux, France,31 Institut Pasteur de Bangui, Bangui, Central African Republic,17 Klinische Mikrobiologie, Hygiene-Institut der Universität,18 Bundesstaatliche Bakteriologisch-Serologische Untersuchungsanstalt Wien, Vienna, Austria,28 Department of Hygiene and Microbiology, University of Palermo, Palermo,19 Department of Public Health, University of Florence, Florence, Italy,23 Laboratory of Molecular Microbiology, Saint Petersburg Pasteur Institute, Saint Petersburg, Russia,21 Les Centres Gheskio, INLR, Port au Prince, Haïti,24 Institut Pasteur de Madagascar, Tananarive, Madagascar,25 Institute of Medical Microbiology and Immunology, Göteborg University, Göteborg, Sweden,26 IVIC, Centro de Microbiología y Biología Celular, Laboratorio de Genética Molecular,30 Tuberculosis Laboratory, Instituto de Biomedicina, Caracas, Venezuela32
15,
Jacobus H de Waard
Jacobus H de Waard
Unité de la Tuberculose et des Mycobactéries, Institut Pasteur de Guadeloupe, Pointe-à-Pitre, Guadeloupe,1 Wadsworth Center, New York State Department Of Health, Albany New York,2 Public Health Research Institute, Tuberculosis Center, Newark, New Jersey,4 Diagnosis Laboratory for Infectious Diseases and Perinatal Screening RIVM, Bilthoven, The Netherlands,3 National Institute of Hygiene and Epidemiology, Hanoi, Vietnam,5 Department of Microbiology, Leeds General Infirmary, Leeds,6 Medical School, St Georges Hospital,7 Mycobacterium Reference Unit, PHLS, Dulwich Hospital, London,13 University of Surrey, Guildford, Surrey, United Kingdom,11 Instituto de Biotecnologia INTA, Moron, Argentina,8 Tuberculosis Mycobacteriology Branch, Centers for Disease Control and Prevention, Atlanta, Georgia,9 Australian Reference Laboratory for Bovine Tuberculosis, Department of Agriculture, South Perth, Australia,10 Centro de Biotecnologia, Universidade Federal de Pelotas, Pelotas,12 Universidade Federal do Rio Grande do Sul, Porto Alegre,27 Department of Biochemistry and Molecular Biology, Fiocruz, Oswaldo Cruz Institute, Rio de Janeiro, Brazil,29 University Childrens Hospital, Heidelberg,14 Infektions Epidemiology, Robert Koch Institute, Berlin,16 Bundesinstitut für Gesundheitlichen Verbraucherschutz und Veterinärmedizin, Jena, Germany,20 Centre National de Référence des Mycobactéries, Institut Pasteur, Paris,15 Laboratoire de Bactériologie, CHU de Brest, Brest,22 Laboratoire de Bactériologie, CHU de Bordeaux, Bordeaux, France,31 Institut Pasteur de Bangui, Bangui, Central African Republic,17 Klinische Mikrobiologie, Hygiene-Institut der Universität,18 Bundesstaatliche Bakteriologisch-Serologische Untersuchungsanstalt Wien, Vienna, Austria,28 Department of Hygiene and Microbiology, University of Palermo, Palermo,19 Department of Public Health, University of Florence, Florence, Italy,23 Laboratory of Molecular Microbiology, Saint Petersburg Pasteur Institute, Saint Petersburg, Russia,21 Les Centres Gheskio, INLR, Port au Prince, Haïti,24 Institut Pasteur de Madagascar, Tananarive, Madagascar,25 Institute of Medical Microbiology and Immunology, Göteborg University, Göteborg, Sweden,26 IVIC, Centro de Microbiología y Biología Celular, Laboratorio de Genética Molecular,30 Tuberculosis Laboratory, Instituto de Biomedicina, Caracas, Venezuela32
32,
Christophe Sola
Christophe Sola
Unité de la Tuberculose et des Mycobactéries, Institut Pasteur de Guadeloupe, Pointe-à-Pitre, Guadeloupe,1 Wadsworth Center, New York State Department Of Health, Albany New York,2 Public Health Research Institute, Tuberculosis Center, Newark, New Jersey,4 Diagnosis Laboratory for Infectious Diseases and Perinatal Screening RIVM, Bilthoven, The Netherlands,3 National Institute of Hygiene and Epidemiology, Hanoi, Vietnam,5 Department of Microbiology, Leeds General Infirmary, Leeds,6 Medical School, St Georges Hospital,7 Mycobacterium Reference Unit, PHLS, Dulwich Hospital, London,13 University of Surrey, Guildford, Surrey, United Kingdom,11 Instituto de Biotecnologia INTA, Moron, Argentina,8 Tuberculosis Mycobacteriology Branch, Centers for Disease Control and Prevention, Atlanta, Georgia,9 Australian Reference Laboratory for Bovine Tuberculosis, Department of Agriculture, South Perth, Australia,10 Centro de Biotecnologia, Universidade Federal de Pelotas, Pelotas,12 Universidade Federal do Rio Grande do Sul, Porto Alegre,27 Department of Biochemistry and Molecular Biology, Fiocruz, Oswaldo Cruz Institute, Rio de Janeiro, Brazil,29 University Childrens Hospital, Heidelberg,14 Infektions Epidemiology, Robert Koch Institute, Berlin,16 Bundesinstitut für Gesundheitlichen Verbraucherschutz und Veterinärmedizin, Jena, Germany,20 Centre National de Référence des Mycobactéries, Institut Pasteur, Paris,15 Laboratoire de Bactériologie, CHU de Brest, Brest,22 Laboratoire de Bactériologie, CHU de Bordeaux, Bordeaux, France,31 Institut Pasteur de Bangui, Bangui, Central African Republic,17 Klinische Mikrobiologie, Hygiene-Institut der Universität,18 Bundesstaatliche Bakteriologisch-Serologische Untersuchungsanstalt Wien, Vienna, Austria,28 Department of Hygiene and Microbiology, University of Palermo, Palermo,19 Department of Public Health, University of Florence, Florence, Italy,23 Laboratory of Molecular Microbiology, Saint Petersburg Pasteur Institute, Saint Petersburg, Russia,21 Les Centres Gheskio, INLR, Port au Prince, Haïti,24 Institut Pasteur de Madagascar, Tananarive, Madagascar,25 Institute of Medical Microbiology and Immunology, Göteborg University, Göteborg, Sweden,26 IVIC, Centro de Microbiología y Biología Celular, Laboratorio de Genética Molecular,30 Tuberculosis Laboratory, Instituto de Biomedicina, Caracas, Venezuela32
1,*,
Nalin Rastogi
Nalin Rastogi
Unité de la Tuberculose et des Mycobactéries, Institut Pasteur de Guadeloupe, Pointe-à-Pitre, Guadeloupe,1 Wadsworth Center, New York State Department Of Health, Albany New York,2 Public Health Research Institute, Tuberculosis Center, Newark, New Jersey,4 Diagnosis Laboratory for Infectious Diseases and Perinatal Screening RIVM, Bilthoven, The Netherlands,3 National Institute of Hygiene and Epidemiology, Hanoi, Vietnam,5 Department of Microbiology, Leeds General Infirmary, Leeds,6 Medical School, St Georges Hospital,7 Mycobacterium Reference Unit, PHLS, Dulwich Hospital, London,13 University of Surrey, Guildford, Surrey, United Kingdom,11 Instituto de Biotecnologia INTA, Moron, Argentina,8 Tuberculosis Mycobacteriology Branch, Centers for Disease Control and Prevention, Atlanta, Georgia,9 Australian Reference Laboratory for Bovine Tuberculosis, Department of Agriculture, South Perth, Australia,10 Centro de Biotecnologia, Universidade Federal de Pelotas, Pelotas,12 Universidade Federal do Rio Grande do Sul, Porto Alegre,27 Department of Biochemistry and Molecular Biology, Fiocruz, Oswaldo Cruz Institute, Rio de Janeiro, Brazil,29 University Childrens Hospital, Heidelberg,14 Infektions Epidemiology, Robert Koch Institute, Berlin,16 Bundesinstitut für Gesundheitlichen Verbraucherschutz und Veterinärmedizin, Jena, Germany,20 Centre National de Référence des Mycobactéries, Institut Pasteur, Paris,15 Laboratoire de Bactériologie, CHU de Brest, Brest,22 Laboratoire de Bactériologie, CHU de Bordeaux, Bordeaux, France,31 Institut Pasteur de Bangui, Bangui, Central African Republic,17 Klinische Mikrobiologie, Hygiene-Institut der Universität,18 Bundesstaatliche Bakteriologisch-Serologische Untersuchungsanstalt Wien, Vienna, Austria,28 Department of Hygiene and Microbiology, University of Palermo, Palermo,19 Department of Public Health, University of Florence, Florence, Italy,23 Laboratory of Molecular Microbiology, Saint Petersburg Pasteur Institute, Saint Petersburg, Russia,21 Les Centres Gheskio, INLR, Port au Prince, Haïti,24 Institut Pasteur de Madagascar, Tananarive, Madagascar,25 Institute of Medical Microbiology and Immunology, Göteborg University, Göteborg, Sweden,26 IVIC, Centro de Microbiología y Biología Celular, Laboratorio de Genética Molecular,30 Tuberculosis Laboratory, Instituto de Biomedicina, Caracas, Venezuela32
1,*