Abstract
A new variant of Shiga toxin 1 (Stx1), designated Stx1d, which deviates considerably more than any other known variant from Stx1 encoded by phage 933J, was identified in an Escherichia coli strain, ONT:H19, isolated from bovine feces. The complete stx1 gene of this strain was amplified and sequenced. Nucleotide sequence homology with stx1 from phage 933J was only 91%, resulting in the substitution of 20 amino acids in the A subunit and 7 amino acids in the B subunit of the protein. Cell culture supernatant of this strain, which was negative for stx2 by PCR testing, was cytotoxic to Vero cells and gave positive results in two commercial enzyme-linked immunosorbent assays for Stx. PCR primers were constructed for the specific detection of the new variant. The findings of this study suggest that Stx1 is not as conserved as thought before and that there might be more variants which cannot be detected by commonly used PCR methods.
Shiga toxins (Stx) are highly potent cytotoxins and essential virulence factors of enterohemorrhagic Escherichia coli (EHEC). Based on antigenic, cytotoxic and genotypic differences, two major types of these toxins, Stx1 and Stx2, are known, both of which are phage encoded. Stx2 includes several subtypes, Stx2c (26), Stx2d (22), Stx2e (30), and Stx2f (25) being the most important ones. In contrast, Stx1 is rather homogenous and basically identical with Shiga toxin of Shigella dysenteriae, only one amino acid is replaced and three nucleotide changes were detected (28). However, during the last decade, several minor variants of Stx1 have been described as well. Paton et al. (20, 21) identified three variants which differed by two amino acids in the A subunit. Nucleotide sequence homology of these variants with stx1 of phage 933J was more than 99%. A more substantial deviation was observed only in strain OX3:H8 131/3, which differed from Stx1 of phage 933J by 9 amino acids within the A subunit and 3 amino acids within the B subunit (21). Recently it was shown that this variant, designated Stx1c and Stx1OX3, respectively, is widely distributed among isolates of human and ovine origin, but it could not be found in bovine and caprine strains (17, 33). None of the Stx1c-positive strains belonged to EHEC serogroup O26, O103, O111, O145, or O157, and all lacked eae (33). So far, there is very limited knowledge about the clinical and biological characteristics of these variants.
Although certain serotypes of Shiga toxin-producing E. coli (STEC)—foremost O157, but also, e.g., O26, O111, or O103—are more frequently identified as the cause of human illness, a large number of different somatic and flagellar antigens has been found in isolates from patients with gastrointestinal disease (2, 3, 8, 10). Besides the ability to produce Shiga toxin, the attaching and effacing factor intimin seems to be the most important virulence marker, especially with respect to the risk of extraintestinal complications (10, 14). On the other hand, there is evidence that STEC strains lacking the eae gene which encodes intimin, a genotype found frequently in less common serotypes, can also cause disease (1, 10, 31). Therefore, regardless of the serotype and the presence of eae, STEC should be considered as potential pathogens.
In the present study, we describe a new variant of Stx1 from an E. coli strain, ONT:H19, of bovine origin which has only 93% amino acid sequence homology with Stx1 from phage 933J within the A subunit and 92% sequence homology within the B subunit.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Strain E. coli MHI 813 (ONT:H19) was originally isolated by the Bavarian State Institute for Health and Food Safety, Oberschleißheim, Germany, from bovine feces. STEC (see Table 2) and E. coli lacking stx (12 strains) from the institute's strain collection and E. coli ATCC 43895 were used for optimization of PCRs and for reference in phenotypic testing. Serotyping of strain MHI 813 was done by the Federal Institute for Health Protection of Consumers and Veterinary Medicine, Berlin, Germany.
TABLE 2.
Results for genotypic and phenotypic testing of E. coli MHI 813 and further STEC from the institute's strain collection
| Method | Resulta for genotype and strain
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| stx1d MHI813 ONT:H19 |
stx1
|
stx1 + stx2
|
stx2
|
|||||||||
| MHI804 | MHI814 O26:H11 | ATCC 43895 O157:H7 | MHI802 O116:H21 | MHI803 O22:H8 | MHI807 O157:H7 | MHI809 O157:H7 | MHI810 O157:H7 | MHI811 O111:H− | MHI812 O157:H− | MHI805 O157:H7 | ||
| PCR | ||||||||||||
| MK1-MK2 | ± | + | + | + | + | + | + | + | + | + | + | + |
| LP30-LP31 | − | + | + | + | + | + | + | + | + | + | − | − |
| VT1FP-VT1RP | + | + | + | + | + | + | + | + | + | + | − | − |
| VT1AF-VT1AR | + | + | + | + | + | + | + | + | + | + | − | − |
| VT1AvarF-VT1AvarR | + | − | − | − | − | − | − | − | − | − | − | − |
| ELISA | ||||||||||||
| Premier EHEC | + | + | + | + | + | + | + | + | + | + | + | + |
| Verotoxin 1 + 2 | + | NTb | NT | NT | NT | NT | NT | NT | NT | NT | NT | NT |
| Sandwich MAb 13C4/MAb 2H3 | − | NT | + | + | NT | NT | NT | NT | NT | NT | NT | NT |
| Cytotoxicity | 3,300c | NT | >52,000 | >52,000 | NT | NT | NT | NT | NT | NT | NT | NT |
Symbols: +, positive; −, negative; ±, ambiguous.
NT, not tested.
Reciprocal dilution that caused 50% cell death.
For PCR testing and for commercial enzyme-linked immunosorbent assay (ELISA) test kits, strains were grown overnight with agitation in tryptic soy broth (TSB) (Oxoid) at 37°C. For the production of Stx1 and for in-house ELISA tests, mitomycin (200 ng/ml) was added to the culture medium to stimulate toxin production.
Primer design and PCR conditions.
E. coli strain MHI 813 was tested for Shiga toxin genes stx1 and stx2 using several published PCR methods and was further characterized using newly designed primers. Primer sequences and PCR conditions are summarized in Table 1. PCRs were performed in a total volume of 50 μl containing 5 μl of 10-fold PCR buffer, 5 μl of MgCl2 (25 mM), 2.5 μl of each primer (10 μM), 1 μl of deoxynucleoside triphosphate (10 mM each nucleotide), and 1 μl of Taq polymerase (2.5 U/μl; ABgene, Hamburg, Germany). Primers and TaqMan probes were synthesized by MWG Biotech (Ebersberg, Germany).
TABLE 1.
Oligonucleotide primers and PCR conditions used in this study
| Primer | Nucleotide sequencea | Target(s) | Product size (bp) | PCR conditions | Reference |
|---|---|---|---|---|---|
| MK1 | TTTACGATAGACTTCTCGAC | stxA | 230 | 94°C, 60 s | 13 |
| MK2 | CACATATAAATTATTTCGCTC | 44°C, 60 s | |||
| 72°C, 90 s | |||||
| LP30 | CAGTTAATGTGGTGGCGAAGG | stxA1 | 348 | 94°C, 90 s | 6 |
| LP31 | CACCAGACAATGTAACCGCTG | 64°C, 90 s | |||
| 72°C, 90 s | |||||
| LP43 | ATCCTATTCCCGGGAGTTTACG | stxA2 + variants | 584 | 94°C, 90 s | 6 |
| LP44 | GCGTCATCGTATACACAGGAGC | 64°C, 90 s | |||
| 72°C, 90 s | |||||
| hlyA1 | GGTGCAGCAGAAAAAGTTGTAG | EHEC | 1,551 | 94°C, 30 s | 24 |
| hlyA4 | TCTCGCCTGATAGTGTTTGGTA | hlyA | 57°C, 60 s | ||
| 72°C, 90 s | |||||
| SK1 | CCCGAATTCGGCACAAGCATAAGC | eaeA | 800 | 94°C, 30 s | 23 |
| SK2 | CCCGGATCCGTCTCGCCAGTATTCG | 52°C, 60 s | |||
| 72°C, 90 s | |||||
| VT1FP | TTATCGCTTTGCTGATTTTTCACA | stxA1 | 147 | 94°C, 60 s | 4 |
| VT1RP | GAAGTAGTCAACGAATGGCGATTT | 60°C, 60 s | |||
| VT1P | CCTTTCCAGGTACAACAGCGGTTACATTGTC | ||||
| VT2FP | CAACGGACAGCAGTTATACCACTCT | stxA2 + variants | 108 | 94°C, 60 s | 4 |
| VT2RP | ACTCCATTAACGCCAGATATGATGA | 60°C, 60 s | |||
| VT2P | TCCGGAATGCAAATCAGTCGTCACTCA | ||||
| VT1AF | TCGTATGGTGCTCAAGGAGTb | stxA1 | 966 | 94°C, 60 s | This study |
| VT1AR | AGTTCTGCGCATCAGAATTG | 52°C, 60 s | |||
| VT1BR2 | AGAACCGGCAACAACTGACTb | 1,309c | 72°C, 60 s | ||
| VT1BF | CGCCTGATTGTGTAACTGGA | stxB1 | 189 | 94°C, 60 s | This study |
| VT1BR | TGAATCCCCCTCCATTATGA | 52°C, 60 s | |||
| 72°C, 60 s | |||||
| VT1AvarF | CTTTTCAGTTAATGCGATTGCT | stxA1d | 192 | 94°C, 60 s; 62°C (5 cycles), 58°C (5 cycles), 54°C (20 cycles) (60 s each); 72°C, 60 s | This study |
| VT1AvarR | AACCCCATGATATCGACTGC |
Underlining, nucleotide mismatching stx1d; italics, degenerate position that allows simultaneous detection of stx1 and stx2.
Primer used for sequencing; no information about mismatches available.
Product size when using VT1BR2 as reverse primer.
For DNA extraction, an aliquot (1 ml) of an overnight culture was centrifuged and the cell pellet was washed three times with NaCl (0.9 M). The cell pellet was resuspended in double-distilled water (100 μl) and heated at 100°C for 10 min. Two μl of DNA preparation were used for each PCR. For the simultaneous detection of stx1 and stx2, degenerate primers MK1-MK2 (13) were used. Additionally, PCRs with primer pairs LP30-LP31 and LP43-LP44 (6), were performed. LP30-LP31 specifically detect the A subunit of stx1, whereas LP43-LP44 detect the A subunit of stx2 and its variants. Furthermore, two TaqMan PCR tests for stx1 and stx2, respectively, were performed (5). New primers were designed for the amplification of stx1a (VT1AF-VT1AR) and stx1b (VT1BF-VT1BR) and for the amplification of the complete coding sequence of stx1 (VT1AF-VT1BR2). The sequences of these primers, which were derived from GenBank accession no. M19473 (12), were selected with the aid of the Rozen and Skaletsky program Primer3 (http://www-genome.wi.mit.edu/genome_software/other/primer3.html). For the specific detection of the new variant gene stx1d, primers VT1AvarF-VT1AvarR were derived from the sequence obtained for strain MHI 813 with primers VT1AF-VT1AR. To obtain the desired specificity for this reaction, the annealing temperature was stepwise reduced from 62 to 54°C during the PCR, polymerase concentration was reduced to 0.625 U per reaction, and MgCl2 was reduced to 2 mM. Specificity of the primers was checked by testing of 12 STEC strains and 12 stx-lacking E. coli strains. Strain MHI 813 was also tested for the genes coding for EHEC hemolysin (hlyA [using primers hlyA1-hlyA4]) and for the attaching and effacing factor (eae [using primers SK1-SK2]).
Sequencing.
PCR products obtained with primer pairs VT1AF-VT1AR and VT1BF-VT1BR, respectively, were sequenced by MWG Biotech. PCR products obtained with primers VT1AF-VT1BR2 (complete coding sequence for stx1) were sequenced in both directions to publication quality by MWG Biotech by primer walking.
Antibodies.
Monoclonal antibody (MAb) 13C4 directed against the B subunit of Stx1 was purchased from the American Type Culture Collection (ATCC CRL 1794). MAb 2H3 was produced in our laboratory following conventional procedures. For the production of an immunogen, Stx1-positive E. coli O22:H8 (MHI 803) was enriched in TSB-mitomycin, and Stx1 was purified by using a combination of ion-exchange chromatography on DEAE-Sephacel (Amersham Pharmacia) and immunoaffinity chromatography utilizing MAb 13C4 as the immunosorbent (16). The affinity-purified Stx1 was neutralized through incubation with MAb 13C4 and injected to mice. Splenocytes of seropositive animals were fused with mouse myeloma cells. The screening of the antisera and the selection of the hybridoma cell lines was performed using an indirect ELISA as described below. MAb 2H3 was finally selected to establish a sandwich ELISA for the detection of Stx1. Further characterization of MAb 2H3 was performed using a mouse hybridoma subtyping kit (Roche Diagnostics) and by Western blotting of Stx1 subunits of the toxin preparation described above.
ELISA.
Culture supernatants of strain MHI 813 were repeatedly tested with two commercially available ELISA kits, Premier EHEC (no longer available; Hiss Diagnostics) and Novitec Verotoxin 1 and 2 (Hiss Diagnostics) according to the manufacturer's instructions, but without addition of mitomycin to the culture medium.
Mouse antisera and hybridoma cell lines were screened for antibodies against Stx1 using an indirect ELISA. Microtiter plates coated with affinity-purified Stx1 (2 μg/ml in carbonate-bicarbonate buffer, 0.05 M, pH 9.6; 100 μl per well) were incubated for 1 h with dilution series of the respective antisera or cell culture supernatants. The plates were washed, and rabbit anti-mouse peroxidase was added. After 1 h of incubation, the plates were washed again and enzyme substrate was added. The enzyme reaction was stopped after 20 min and the absorbance at 450 nm was measured.
A sandwich ELISA with MAb 2H3 as capture antibody and with MAb 13C4 (ATCC CRL 1794) as detector antibody was used to examine E. coli strain MHI 813 for production of Stx1. As positive controls, STEC ATCC 43895 and MHI 814 were tested simultaneously. Microtiter plates coated with MAb 2H3 (10 μg/ml) were incubated for 1 h with 100 μl of a twofold dilution series (in 0.5% Tween 20-phosphate-buffered saline) of culture supernatants. The plate was washed, and 100 μl of biotinylated MAb 13C4 (1 μg/ml in 1% casein-phosphate-buffered saline-0.5% Tween 20) was added. After 1 h of incubation, the plate was washed again and 100 μl of ExtrAvidin-horseradish peroxidase (Sigma-Aldrich) was added to each well and incubated for 1 h. Finally, 100 μl of substrate solution was added to the washed plate. After 20 min the enzyme reaction was stopped and the absorbance at 450 nm was measured.
Cytotoxicity testing.
For determination of cytotoxicity, E. coli MHI 813, STEC MHI 814 (Stx1 positive), and STEC ATCC 43895 (Stx1 and Stx2 positive) were grown overnight in TSB-mitomycin (200 ng/ml). Vero cell toxicity testing was essentially performed as described by Karmali et al. (15) with some modifications. Briefly, culture supernatant was filtered through a 0.22-μm-pore-size membrane and serial twofold dilutions of the filtrate were pipetted onto monolayers of Vero cell cultures. In parallel, heat-treated (10 min, 80°C) supernatants were tested as a control to verify heat inactivation of the heat-labile Stx. The cultures were incubated for 3 days at 37°C. Cell viability was measured spectrophotometrically as described by Dietrich et al. (9), using the cell proliferation agent WST-1 (Roche Diagnostics), a water-soluble tetrazolium salt. For this purpose, from each well 100 μl of supernatant was removed and 10 μl of WST-1 medium was added. After an incubation step of 60 min at 37°C, optical density was measured at 450 nm. Cytotoxicity titers were defined as the reciprocal dilution that caused 50% reduction of mitochondrial enzyme activity.
Nucleotide sequence accession number.
The nucleotide sequence of the stx1d variant gene was entered into the GenBank database under accession number AY170851.
RESULTS
Results of PCRs.
Strain E. coli MHI 813 had repeatedly given positive results in a commercial ELISA kit for Shiga toxins, which could not be verified by PCR with primers LP30-LP31. Therefore, this strain has been tested with several primer pairs specific for stx1 and stx2, and the results let one assume that the strain harbored an stx1 variant gene. Primers were designed to amplify the complete coding sequence of the gene, and from the sequence of the resulting amplicon, primers for the specific detection of this new variant were derived.
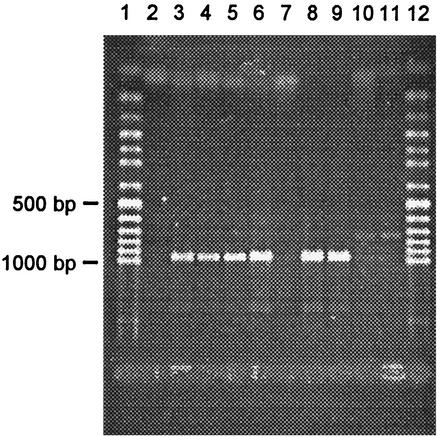
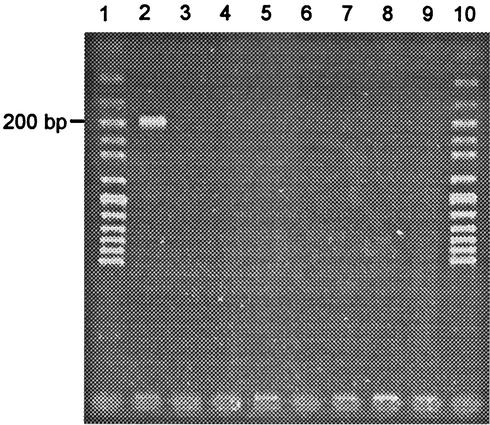
PCRs for the detection of stx2 were negative when using DNA extracted from E. coli MHI 813. Results for stx1 depended on the primers used. With the TaqMan PCR, positive signals were obtained regularly, while with conventional PCR using primers LP30-LP31 no band of the expected size appeared on the gel, and with primers MK1-MK2 occasionally a faint band was visible. Results for other STEC tested with these primers correlated with the genotype of the strains (Table 2). Using the newly designed primers for stx1 in the combinations VT1AF-VT1AR, VT1BF-VT1BR, and VT1AF-VT1BR2, strong bands were obtained with DNA extracts from E. coli MHI 813 and with DNA extracts from all other tested stx1-positive STEC (Fig. 1; Table 2). Optimized PCRs with the variant-derived primers VT1AvarF-VT1AvarR produced strong bands for E. coli MHI 813, and no amplification products for all other STEC and stx-lacking E. coli tested (Fig. 2). The genes eae and hlyA could not be detected in strain MHI 813.
FIG. 1.
Agarose gel electrophoresis of DNA fragments amplified by PCR with primers VT1AF-VT1AR from selected STEC strains. Lanes: 1 and 12, 50-bp ladder; 2, distilled water; 3, MHI 813 (stx1d); 4, MHI 814 (stx1); 5, ATCC 43895 (stx1 + stx2); 6, MHI 803 (stx1 + stx2); 7, MHI 806 (stx2); 8, MHI 802 (stx1 + stx2); 9, MHI 811 (stx1 + stx2); 10 MHI 812 (stx2); 11, MHI 805 (stx2).
FIG. 2.
Agarose gel electrophoresis of DNA fragments amplified by PCR with primers specific for stx1d (VT1AvarF-VT1AvarR) from selected STEC strains. Lanes: 1 and 10, 50-bp ladder; 2, MHI 813 (stx1d); 3, MHI 814 (stx1); 4, ATCC 43895 (stx1 + stx2); 5, MHI 803 (stx1 + stx2); 6, MHI 807 (stx1 + stx2); 7, MHI 802 (stx1 + stx2); 8, MHI 811 (stx1 + stx2); 9, MHI 812 (stx2).
Sequencing.
Sequencing of PCR products obtained with primer pairs VT1AF-VT1AR and VT1BF-VT1BR revealed that E. coli MHI 813 harbors a new variant of stx1. Consequently, the complete stx1 gene of this strain was sequenced. Analysis of the sequence data showed that there was only 91% nucleotide sequence homology with stx1 from phage 933J (12). The translated sequence deviated within the A subunit in 20 amino acids and within the B subunit in 7 amino acids from the sequence coded by phage 933J (Fig. 3). Multiple sequence alignment with CLUSTALW at EMBL-EBI (http://www.ebi.ac.uk/clustalw) revealed that the highest degree of nucleotide homology is obtained with stx1c (92%). Of 123 nucleotides that were not identical for the three genes stx1, stx1c, and stx1d, in 28 cases there was concordance between stx1c and stx1d, in 12 cases there was concordance between stx1 and stx1d, and in 83 cases the deviation found in stx1d had no parallel in any of the other sequences. Based on the proposed nomenclature for stx (5), the new variant was designated stx1d.
FIG. 3.
Alignment of the amino acid sequences of Stx1 (12), Stx1c (21), and the deduced amino acid sequence of the new variant Stx1d. Dots indicate amino acids identical to those of the reference sequence Stx1. Signal peptides are underlined, and amino acids that are putatively important for the function of the protein are printed in boldface type.
Cytotoxicity.
The 50% cytotoxicity titer determined for sterile filtered culture supernatant of strain MHI 813 was relatively low (3,300) in comparison to those determined for STEC ATCC 43895 (stx1, stx2) and MHI 814 (stx1), both of which were >52,000.
ELISA and antibodies.
ELISA testing of the culture supernatants of strain E. coli MHI 813 for Stx regularly gave positive results when using any one of the two commercial kits. However, the toxin was not detected by a sandwich ELISA with MAb 2H3 as capture antibody and MAb 13C4 as detector antibody, although the positive controls gave strong signals (absorbance > 4.0). As determined by Western blotting, both MAbs are specific for the B subunit of Stx1. MAb 2H3 was subtyped as IgG1κ. Additional experiments gave evidence that neither MAb 2H3 nor MAb 13C4 reacted with Stx1d (data not shown).
Serotyping.
Strain MHI 813 was serotyped as ONT:H19.
DISCUSSION
During the last decade, stx genes of many different STEC strains of various origin have been characterized. The results revealed that several subtypes of stx2 exist, while stx1 seemed to be highly conserved. An explanation for the low variability of stx1 could be that this gene entered E. coli significantly later than stx2, as a phylogenetic model of stx genes by Wittham (32) proposes. However, there is evidence that stx1 might be not as conserved as thought at first. Paton et al. (20, 21) described heterogeneity of stx1 operons in four strains. In all four variants, the single amino acid deviation between stx1 and stx from S. sonnei could be found in the A subunit. Three strains each had an additional amino acid substitution in the A subunit (G-132S, G-249E, and S-171L, respectively). stx of strain OX3:H8 131/3 contained nine amino acids deviating from the sequence encoded by the bacteriophage 933J within the A subunit. This strain was the only one that additionally contained three amino acid deviations within the B subunit, of which two were in the signal peptide. So far, this strain contained the only known substantially deviating variant of stx1. Recent investigations of Zhang et al. (33) demonstrated that this variant, designated stx1c, frequently occurs in human STEC isolates (36 out of 212 strains tested). Strains positive for stx1c originated from patients with watery, nonbloody diarrhea or from asymptomatic patients. Only one strain was associated with a case of hemolytic-uremic syndrome, but this strain additionally harbored the stx2 gene. A high proportion of the 36 stx1c positive strains belonged to the genotype carrying stx1c and stx2d. The eae gene, which codes for the attaching and effacing factor, an important virulence factor of STEC, could not be detected in any of these strains, and none belonged to enterohemorrhagic serogroups O157, O26, O103, O111, or O145. The sequencing of stx1c genes of 14 strains resulted in identical nucleotide sequences for all strains and 100% identity with stx1 of strain OX3:H8 131/3. The findings of Koch et al. (17) demonstrated that serotypes O146:H21 and O128:H2 were most frequently associated with stx1OX3 carrying STEC from sheep and humans. Sheep might be the main source of stx1OX3-carrying STEC which could be transmitted to humans, while the variant could not be found in isolates from cattle and goats. The authors could isolate a lysogenic phage carrying the stx1OX3 gene, thus proving that the variant gene is phage encoded like stx1 and stx2 but unlike stx of Shigella sonnei and certain stx variants. The variant seems to be associated with milder disease in humans, but the clinical relevance has to be further investigated.
Strain E. coli MHI 813 attracted attention as a consequence of unusual and supposedly false-positive reactions in several phenotypic and genotypic tests for STEC. The strain reacted repeatedly positively in Premier EHEC, a commercial ELISA for verotoxins, but these results could not be verified using PCR with primers LP30-LP31, and the results were ambiguous when primers MK1-MK2 were used. Further PCR testing with various primer pairs and subsequent sequencing of the PCR products revealed that this strain harbors a new variant of stx1 which deviates significantly more from stx1 than any other known variant of stx1. According to the systematic nomenclature for stx proposed by Calderwood et al. (5), the new variant was designated stx1d. Analysis of the nucleotide sequence of stx1d showed that the PCR results can be explained through the positions of the PCR primers with respect to nucleotides deviating from stx1 (Table 1). Primer pair LP30-LP31 has four and one mismatches, respectively, with the mismatch of primer LP31 positioned at the very 3′ end. For the degenerate primer pair MK1-MK2, only two nucleotides of the stx1d gene are deviating from stx1 within the sequence of MK1, but there are two degenerate positions in the sequence of MK1 and one in MK2. This could explain why with primers LP30-LP31 no PCR product was obtained for strain MHI 813 and with primers MK1-MK2 a weak but not well-reproducible signal was obtained. In contrast, the primers and the probe of the TaqMan PCR for stx1 have only a low number of nucleotides mismatching the sequence of stx1d. With the TaqMan PCR, a positive result was regularly observed. Although only a limited number of strains have been tested with the variant-specific primers VT1AvarF-VT1AvarR, this PCR seems to be suitable for the detection of stx1d, since no false-positive results were observed.
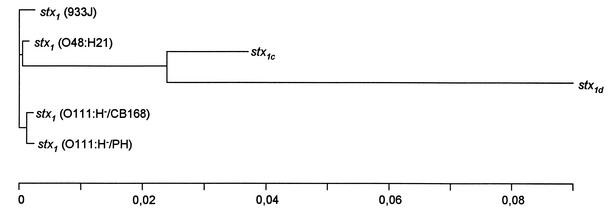
Interestingly, stx1d not only harbors additional nucleotide deviations from stx1 in comparison to stx1c but is in some positions homologous with stx1c and in others with stx1. These findings may be a hint that evolutionarily, this variant did not originate from stx1c but from a common ancestor of both variants. This hypothesis is supported by the calculation of phylogenetic distances of known stx1 genes by the program PHYLIP (version 3.6 [http://evolution.gs.washington.edu/phylip]) (Fig. 4). Deviations unique for stx1d were particularly found in the first 265 nucleotides of the sequence, with highly variable sections at nucleotides 2 to 62 and nucleotides 195 to 265. The section from nucleotide 266 to 670 is much more conserved, with only 19 nucleotides not identical for all three sequences. More mutations can be found again in the second half of the sequence, with another highly variable section towards the end of the sequence (nucleotides 1175 onwards).
FIG. 4.
Phylogenetic distances of stx1 genes as determined by PHYLIP 3.6 (http://evolution.gs.washington.edu/phylip). Sequences were analyzed using the neighbor-joining method. For information on stx1 (933J), see reference 12; for information on stx1 (O111:H−/CB 168), stx1 (O111:H−/PH), stx1 (O48:H21), and stx1c, see references 20 and 21); for information on stx1d, see this study.
The results of the phenotypic characterization of strain E. coli MHI 813 were ambiguous. Two commercial ELISA test for the simultaneous detection of Stx1 and Stx2 gave positive results, but ELISAs based on MAbs 13C4 and 2H3, which react specifically with different epitopes of the B subunit of Stx1 (16, 27), consistently gave negative results for this strain. The analysis of the deduced amino acid sequence of Stx1d showed that within the B subunit there are seven amino acids deviating from Stx1, three of those in the mature protein. The mutations were at positions T-1A, G-25A, and N-55T of the mature protein (Fig. 3). According to a computational model of the three-dimensional structure of Stx1B (19) deposited in the protein data bank (http://www.rcsb.org/pdb), amino acids T1 and N55 are located closely together on the surface of the protein. If the MAbs used for the ELISAs were directed against this protruding epitope, the mutations observed in Stx1d might reduce the affinity of the antibodies to the protein drastically, thus causing a negative ELISA result for strain MHI 813. As for the subunit specificity of the antibodies used in the commercial kits, no information could be obtained.
Whether the relatively low cytotoxicity titer for strain E. coli MHI 813 is attributable to a low rate of protein expression or to the functionality of the protein cannot be decided. As the positive results of the ELISA kits have shown, Stx1d is expressed, but there is no information on the quantity of the protein. On the other hand, the function of Stx1 has been intensively investigated, and for both subunits several amino acids that are presumably important for the functionality of the protein have been identified. Like Stx2, Stx1 consists of two subunits, with the A subunit representing the enzymatically active part and the B subunit forming a pentamer which is responsible for the specific binding to the receptor molecule globotriaosylceramide (Gb3). According to a computational model (19), each B monomer forms a β-barrel of six antiparallel β-sheets capped by an α-helix and has three binding sites for Gb3. The findings of this group suggest that amino acids D16, D17, F30, and probably W34 play essential roles in the receptor binding of Stx1B. These amino acids are all conserved in Stx1d. However, N55, which in the model participates in one of the three binding sites, in Stx1d is replaced by tryptophan. For Stx1A also several positions in the protein are discussed in the literature as functionally important. Deresiewicz et al. (7) identified five putative active sites: Y77, Y114, E167, R170, and W203. None of these amino acids is altered in Stx1d, and neither the disulfide bridge between C242 and C261 nor the trypsin-sensitive cleavage site R248-X-X-R251, which is cleaved following toxin entry into mammalian cells by the ubiquitous protease furin (11), is changed. But two deviating amino acids are directly adjacent to this cleavage site, and A253 and S254 are discussed as alternative cleavage sites (29). This could result in no or less effective cleavage and thus reduced toxicity, as proteolytic cleavage is essential for maximum toxicity (18). To summarize, functionally important sites of Stx1A and Stx1B are widely conserved in Stx1d, but the deviating amino acids could cause changes in the conformation of the protein that are responsible for the reduced cytotoxicity titers. As no information on the in vivo toxicity of Stx1d is available, further studies on the characteristics and on the prevalence of this toxin have to be done.
STEC of the serotype ONT and/or the flagellar type H19 have been found by many authors in isolates of human and bovine origin, including several cases of (bloody) diarrhea in humans (1, 8, 10, 31), and therefore have to be considered as potential pathogens.
This study also gives strong evidence that further variants of stx1 exist which possibly cannot be detected by commonly used PCR methods, and that not only sheep but also cattle are a reservoir of such variants.
Acknowledgments
We thank Marion Lorber and Brunhilde Minich for excellent technical assistance.
REFERENCES
- 1.Beutin, L., S. Aleksic, S. Zimmermann, and K. Gleier. 1994. Virulence factors and phenotypical traits of verotoxigenic strains of Escherichia coli isolated from human patients in Germany. Med. Microbiol. Immunol. 183:13-21. [DOI] [PubMed] [Google Scholar]
- 2.Beutin, L., S. Zimmermann, and K. Gleier. 1998. Human infections with Shiga toxin-producing Escherichia coli other than serogroup O157 in Germany. Emerg. Infect. Dis. 4:635-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boerlin, P., S. A. McEwen, F. Boerli-Petzold, J. B. Wilson, R. B. Johnson, and C. L. Gyles. 1999. Associations between virulence factors of Shiga toxin-producing Escherichia coli and disease in humans. J. Clin. Microbiol. 37:497-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bürk, C., I. G. B. Braumiller, H. Becker, and E. Märtlbauer. 2002. Nuclease fluorescence assay for the detection of verotoxin genes in raw milk. Lett. Appl. Microbiol. 35:153-156. [DOI] [PubMed] [Google Scholar]
- 5.Calderwood, S. B., D. W. K. Acheson, G. T. Keusch, T. J. Barrett, P. M. Griffin, N. A. Strockbine, B. Swaminathan, J. B. Kaper, M. M. Levine, B. S. Kaplan, H. Karch, A. D. O'Brien, T. G. Obrig, Y. Takeda, P. I. Tarr, and I. K. Wachsmuth. 1996. Proposed new nomenclature for SLT (VT) family. ASM News 62:118-119. [Google Scholar]
- 6.Cebula, T. A., W. L. Payne, and P. Feng. 1995. Simultaneous identification of strains of Escherichia coli serotype O157:H7 and their Shiga-like toxin type by mismatch amplification mutation assay-multiplex PCR. J. Clin. Microbiol. 33:248-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deresiewicz, R. L., S. B. Calderwood, J. D. Robertus, and R. J. Collier. 1992. Mutations affecting the activity of the Shiga-like toxin I A-chain. Biochemistry 31:3272-3280. [DOI] [PubMed] [Google Scholar]
- 8.DesRosiers, A., J. M. Fairbrother, R. P. Johnson, C. Desautels, A. Letellier, and S. Quessy. 2001. Phenotypic and genotypic characterization of Escherichia coli verotoxin-producing isolates from humans and pigs. J. Food Protect. 12:1904-1911. [DOI] [PubMed] [Google Scholar]
- 9.Dietrich, R., C. Fella, S. Strich, and E. Märtlbauer. 1999. Production and characterization of monoclonal antibodies against the hemolysin BL enterotoxin complex produced by Bacillus cereus. Appl. Environ. Microbiol. 65:4470-4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eklund, M., F. Scheutz, and A. Siitonen. 2001. Clinical isolates of non-O157 Shiga toxin-producing Escherichia coli: serotypes, virulence characteristics, and molecular profiles of strains of the same serotype. J. Clin. Microbiol. 39:2829-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garred, Ø., E. Dubinina, A. Polesskaya, S. Olsnes, J. Kozlov, and K. Sandvig. 1997. Role of the disulfide bond in Shiga toxin A-chain for toxin entry into cells. J. Biol. Chem. 17:11414-11419. [DOI] [PubMed] [Google Scholar]
- 12.Jackson, M. P., J. W. Newland, R. K. Holmes, and A. D. O'Brien. 1987. Nucleotide sequence analysis of the structural genes for shiga-like toxin I encoded by bacteriophage 933J from Escherichia coli. Microb. Pathog. 2:147-153. [DOI] [PubMed] [Google Scholar]
- 13.Karch, H., and T. Meyer. 1989. Single primer pair for amplifying segments of distinct Shiga-like-toxin genes by polymerase chain reaction. J. Clin. Microbiol. 27:2751-2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karch, H., H.-I. Huppertz, J. Bockemühl, H. Schmidt, A. Schwarzkopf, and R. Lissner. 1997. Shiga toxin-producing Escherichia coli infections in Germany. J. Food Protect. 60:1454-1457. [DOI] [PubMed] [Google Scholar]
- 15.Karmali, M. A., M. Petric, C. Lim, P. C. Fleming, G. S. Arbus, and H. Lior. 1985. The association between idiopathic hemolytic uramic syndrome and infection by verotoxin producing Escherichia coli. J. Infect. Dis. 151:775-782. [DOI] [PubMed] [Google Scholar]
- 16.Keil, C. 1999. Reinigung von Escherichia coli Shigatoxinen und Produktion von poly- und monoklonalen Antikörpern. Ph.D. thesis. University of Munich, Munich, Germany.
- 17.Koch, C., S. Hertwig, R. Lurz, B. Appel, and L. Beutin. 2001. Isolation of a lysogenic bacteriophage carrying the stx1OX3 gene, which is closely associated with Shiga toxin-producing Escherichia coli strains from sheep and humans. J. Clin. Microbiol. 39:3992-3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lea, N., M. Lord, and L. M. Roberts. 1999. Proteolytic cleavage of the A subunit is essential for maximal cytotoxicity of Escherichia coli O157:H7 Shiga-like toxin 1. Microbiology 145:999-1004. [DOI] [PubMed] [Google Scholar]
- 19.Ling, H., A. Boodhoo, B. Hazes, M. D. Cummings, G. D. Armstrong, J. L. Brunton, and R. J. Read. 1998. Structure of the Shiga-like toxin B-pentamer complexed with an analogue of the receptor Gb3. Biochemistry 37:1777-1788. [DOI] [PubMed] [Google Scholar]
- 20.Paton, A. W., J. C. Paton, P. N. Goldwater, M. W. Heuzenroeder, and P. A. Manning. 1993. Sequence of a variant shiga-like toxin type-I operon of Escherichia coli O111:H−. Gene 129:87-92. [DOI] [PubMed] [Google Scholar]
- 21.Paton, A. W., L. Beutin, and J. C. Paton. 1995. Heterogeneity of the amino acid sequences of Escherichia coli shiga-like toxin type-I operons. Gene 153:71-74. [DOI] [PubMed] [Google Scholar]
- 22.Piérard, D., G. Muyldermas, L. Moriau, D. Stewens, and S. Lauwers. 1998. Identification of new verotoxin type 2 variant B-subunit genes in human and animal Escherichia coli isolates. J. Clin. Microbiol. 36:3317-3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmidt, H., H. Rüssmann, A. Schwarzkopf, S. Aleksic, J Heesemann, and H. Karch. 1994. Prevalence of attaching and effacing Escherichia coli in stool samples from patients and controls. Zentbl. Bakteriol. 281:201-213. [DOI] [PubMed] [Google Scholar]
- 24.Schmidt, H., L. Beutin, and H. Karch. 1995. Molecular analysis of the plasmid-encoded hemolysin of Escherichia coli O157:H7 strain EDL 933. Infect. Immun. 63:1055-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmidt, H., J. Scheef, S. Morabito, A. Caprioli, L. H. Wieler, and H. Karch. 2000. A new Shiga toxin 2 variant (Stx2f) from Escherichia coli isolated from pigeons. Appl. Environ. Microbiol. 66:1205-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmitt, C. K., M. L. McKee, and A. D. O'Brien. 1991. Two copies of Shiga-like toxin II-related genes common in enterohemorrhagic Escherichia coli strains are responsible for the antigenic heterogeneity of the O157:H− strain E32511. Infect. Immun. 59:1065-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strockbine, N. A., L. R. M. Marques, R. K. Holmes, and A. D. O'Brien. 1985. Characterization of monoclonal antibodies against Shiga-like toxin from Escherichia coli. Infect. Immun. 50:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strockbine, N. A., M. P. Jackson, L. M. Sung, R. K. Holmes, and A. D. O'Brien. 1988. Cloning and sequencing of the genes for Shiga toxin from Shigella dysenteriae type 1. J. Bacteriol. 170:1116-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takao, T., T. Tanabe, Y.-M. Hong, Y. Shimonishi, H. Kurazono, T. Yutsudo, C. Sasakawa, M. Yoshikawa, and Y. Takeda. 1988. Identity of molecular structure of Shiga-like toxin I (VT1) from Escherichia coli O157:H7 with that of Shiga toxin. Microb. Pathog. 5:357-369. [DOI] [PubMed] [Google Scholar]
- 30.Weinstein, D. L., M. P. Jackson, J. E. Samuel, R. K. Holmes, and A. D. O'Brien. 1988. Cloning and sequencing of a Shiga-like toxin type II variant from an Escherichia coli strain responsible for edema disease of swine. J. Bacteriol. 170:4223-4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Welinder-Olsson, C., M. Badenfors, T. Cheasty, E. Kjellin, and B. Keijser. 2002. Genetic profiling of enterohemorrhagic Escherichia coli strains in relation to clonality and clinical signs of infection. J. Clin. Microbiol. 40:959-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whittam, T. S. 1998. Evolution of Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains, p. 195-209. In J. B. Kaper and A. D. O'Brien (ed.), Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. ASM Press, Washington, D.C.
- 33.Zhang, W., M. Bielaszewska, P. Kuczius, and H. Karch. 2002. Identification, characterization and distribution of a Shiga toxin 1 gene variant (stx1c) in Escherichia coli strains isolated from humans. J. Clin. Microbiol. 40:1441-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]