Abstract
Nonchlamydial, nongonococcal urethritis (NCNGU) is suggested to be a sexually transmitted disease in men. NCNGU patients were compared to control subjects with regard to the presence of potentially infectious bacteria in the first void urine. Patients' pre- and post-antibiotic-treatment urine samples and two samples obtained 2 weeks apart from healthy volunteers, who did not receive antibiotic therapy, were analyzed with broad-spectrum PCR tests aiming at eubacterial small subunit rRNA genes. Restriction fragment length polymorphism analysis of the amplicons cloned from the mixtures of PCR products revealed that many different species of microorganisms were found to be colonizing the male urethra. We document here clear differences in the composition of the resident urethral flora between samples obtained from various individuals and between samples obtained at various points in time for a single individual. No major changes in population complexity were found upon antimicrobial treatment. In two of five patients a previously suggested pathogen (Mycoplasma genitalium or Haemophilus parainfluenzae) was accurately identified on the basis of DNA sequencing. No ubiquitous, azithromycin-sensitive organism was identified as a common pathogen in all patients, but up to 40% of all clones represented as-yet-unclassified bacterial species. Relatively often Pseudomonas spp. or Pseudomonas-like organisms were identified in the bacterial flora of patients. Interestingly, an as-yet-uncharacterized microbial species was identified as a negative predictor of NCNGU. This species was identified in all control subjects and was absent from all of the patient' samples (5 of 5 versus 0 of 5, P = 0.0079). This suggests that NCNGU might also be diagnosed by assessing the absence rather than the presence of certain bacterial species.
Chlamydia trachomatis and Neisseria gonorrhoeae are common causes of urethritis in males (7, 14, 16). Nongonococcal urethritis is diagnosed in over two million cases per year in the United States. A significant fraction of the urethritis patients (up to 50%), however, are not infected by either one of these pathogens (7). In these cases, the clinical syndrome is referred to as nonchlamydial nongonococcal urethritis (NCNGU). NCNGU is a common condition, frequently diagnosed in sexually transmitted disease (STD) clinics around the world. In the pathogenesis of acute NCNGU, microorganisms other than C. trachomatis or N. gonorrhoeae seem to play a role as well, since a significant fraction of the patients involved respond well to antibiotic treatment (1). Furthermore, data showing that condoms are protective against NCNGU support the hypothesis that NCNGU is an infectious disease (8, 25). Despite extensive microbiological studies, no single causative microbial species has been identified as the main cause of NCNGU. Although Mycoplasma genitalium seems to be an important candidate (4, 14, 17, 20, 28), with an incidence that may be as high as 36% (4), many clinically overt cases still remain microbiologically unexplained. Previous studies were limited to selective searches for one or more traditional pathogens that are putatively involved in inflammation of the lower genital tract in males. These pathogens included M. genitalium, Ureaplasma urealyticum, or Trichomonas vaginalis, and searches were performed by culture, PCR, or a combination of both methods (14, 20, 28). Efforts to really identify novel pathogens have employed culture methods mainly (10, 26). The downside of this approach is that culture-dependent techniques may not adequately elucidate the microbial diversity in the genitourinary tract. Molecular techniques have suggested that the number of pathogenic microorganisms that have been cultured to date probably equals only a fraction of the total (23). Consequently, culture-based approaches have probably prohibited the discovery of novel uropathogens, an omission that underscores the need for additional molecular-microbiological searches.
A potential solution to the inadequacy of microbiological culture appears to be the use of diagnostic broad-range ribosomal DNA (rDNA) amplification in combination with phylogenetic studies (21). Eubacterial, domain-specific PCR primers are particularly useful for the identification of putative human pathogens. Amplified rDNA from bacteria can be sequenced, and these sequences can be used in computerized database searches to identify the bacterial species involved (31). This approach turned out to be particularly successful in microbial-etiological studies in chronic idiopathic prostatitis. Bacterial rRNA genes could be detected in 77% of all patients, and certain Vibrio species were identified as putative agents of infection (19). In addition, the same strategy helped to identify novel bacterial species, both in natural environments and clinical syndromes (3, 11, 15, 22, 29).
The objective of the current research was to define the microbial communities present in the urethra of healthy male volunteers and NCNGU patients. We searched for putative pathogens and/or markers for a healthy microbial flora by ribosomal PCR, which should allow for a detailed comparison of the flora of control individuals and the spectrum of bacterial species present in pre- and post-antibiotic-treatment samples of NCNGU patients.
MATERIALS AND METHODS
Participants and procedures.
Men attending the STD Outpatient Clinic of the Department of Dermatology and Venereology of the Erasmus MC (University Medical Centre Rotterdam, Rotterdam, The Netherlands) for a sexual health assessment were eligible for the present study. Selected individuals provided informed consent (Erasmus MC Medical Ethical Committee, protocol 00-859). Major exclusion criteria were the use of antibiotics within the previous month and a history of urethritis within the previous 3 months. Personal interviews revealed that all five enrolled patients had clear symptoms of urethritis (penile irritation in combination with discharge and/or dysuria [see Table 1 ]). Urethritis was confirmed microscopically, on the basis of >6 polymorphonuclear leukocytes (PMNs) per μl in the sediment of 12 ml of first-pass urine (FPU). The numbers of PMNs were determined by using the standardized KOVA system (Hycor, Garden Grove, Calif.) in full accordance with the manufacturer's instructions. Control subjects were five asymptomatic volunteers with no signs of urethritis (≤6 PMNs/μl). After the urethral meatus of each patient was washed with a sterile gauze and tap water, ∼30 ml of FPU was collected into sterile tubes. In addition, urethral swabs were obtained for confirmatory purposes. Infection by N. gonorrhoeae was excluded microscopically (classical Gram stain) and by culture performed on a GC-LECT agar (Becton Dickinson, Alphen aan den Rijn, The Netherlands). C. trachomatis infection was excluded by PCR analysis of FPU by using the Cobas Amplicor Detection Reagent Kit and the Cobas Amplicor machine (Roche Diagnostics, Mannheim, Germany) according to instructions of the manufacturer. A total of 12 ml of FPU was used for the microscopic and diagnostic evaluations described above, whereas 500 μl was used for the Amplicor tests. In preparation of the broad-spectrum ribosomal PCRs, 10 ml of FPU was centrifuged for 10 min at 3,000 rpm. The sediment (ca. 75 to 300 μl) was kept at −80°C prior to processing. Patients with microscopically diagnosed urethritis and a negative Gram stain for N. gonorrhoeae were treated with a single oral dose of 1 g of azithromycin. Patients and control subjects were advised to abstain from any form of sexual intercourse (vaginal, anal, and oral) and were asked to return for reexamination after 2 weeks. At the second visit they delivered ca. 30 ml of FPU, which was collected at home in the early morning, according to the collection procedure described above. Again, 12 ml of FPU was microbiologically evaluated for the presence of PMNs. A 10-ml portion was centrifuged, and the sediment was kept at −80°C prior to PCR processing. The healthy volunteers were not treated with azithromycin between the two samplings.
TABLE 1.
Age, STD history, and clinical findings of patients and controls
| Patient or control no. | Age (yr) | STD historya | First visit
|
Second visit
|
Sexual orientation | ||
|---|---|---|---|---|---|---|---|
| Symptoms | PMNs/μl | Symptoms | PMNs/μl | ||||
| Patient | |||||||
| 1 | 37 | NG | Discharge, dysuria | 21 | None | 5 | Heterosexual |
| 2 | 29 | CT, PB | Discharge, dysuria | 31 | None | 5 | Homosexual |
| 3 | 25 | CT, GW | Dysuria | 107 | None | 5 | Heterosexual |
| 4 | 32 | NG, CT | Dysuria | 21 | None | 2 | Heterosexual |
| 5 | 44 | NG, NCNGU | Dysuria | 61 | None | 1 | Homosexual |
| Control | |||||||
| 1 | 36 | NCNGU | None | 1 | None | 1 | Homosexual |
| 2 | 30 | PB | None | 0 | None | 3 | Homosexual |
| 3 | 50 | None | None | 2 | None | 1 | Heterosexual |
| 4 | 29 | None | None | 2 | None | 2 | Heterosexual |
| 5 | 40 | None | None | 4 | None | 1 | Heterosexual |
NG, N. gonorrhoeae; CT, C. trachomatis; PB, pubic pediculosis; GW, genital warts. All patients suffered from penile irritation.
Tap water.
To determine the possible microbiological contamination factor of tap water used for washing the urethral meatus, 50 ml of first-run tap water was collected in a sterile tube. This was centrifuged in 10-ml parts for 10 min at 3,000 rpm. The sediment was kept at −80°C prior to processing.
DNA purification.
Part of the collected water and urine sediments (150 μl) was used for DNA extraction and purification. To the samples with a volume of <150 μl, a compensating amount of 50 mM Tris-HCl (pH 7.5)-0.1 mM EDTA-50 mM glucose buffer was added to a total volume of 150 μl. First, 75 μl of lysostaphin solution (10 mg/ml; Sigma, St Louis, Mo.) was added, and the mixture was heated to 37°C for 30 min. Thereafter, 1 ml of guanidinium lysis buffer (4 mM guanidinium isothiocyanate, 0.1 M Tris-HCl [pH 6.4], 0.2 M EDTA, 0.1% Triton X-100) was added, and the mixture was kept at room temperature for 1 h, after which 50 μl of Celite suspension was added. The samples were kept at room temperature and mixed at regular intervals for 10 min (5). After a vortexing step and centrifugation (15 s at 14,000 rpm in an Eppendorf centrifuge), the supernatant was discarded and the pellet was washed twice with a second chaotropic lysis buffer (4 M guanidinium isothiocyanate, 0.1 M Tris-HCl; pH 6.4), twice with ethanol (70%) and, finally, once with acetone. The pellet was vacuum dried and emulsified in 100 μl of 10 mM Tris-HCl (pH 8.0). The sample was heated to 56°C for 10 min and centrifuged (10 min at 14,000 rpm in an Eppendorf centrifuge). The resulting supernatant was used as a template for PCR.
PCR tests.
All PCRs were performed in GeneAmp 9600 or 9700 machines (PE Applied Biosystems, Foster City, Calif.). The primers used for the 16S rDNA PCR were EUB-L (5′-CTTTACGCCCATTTAATCCG-3′) and EUB-R (5′-AGA-GTTTGATCCTGGTTCAG-3′). These generate an ∼500-bp fragment deriving from the 3′-terminal end of the small-subunit (ssu) rRNA gene (30). A total of 45 μl of PCR mix was added to 5 μl of the purified DNA solution. The PCR mix consisted of 10 μl of a 20 mM desoxynucleotide triphosphate stock solution (Amersham Life Science, Cleveland, Ohio), 5 μl of a 10-fold-concentrated SuperTaq PCR buffer (HT Biotechnology, Cambridge, United Kingdom), 0.5 μl of both primers, 28.92 μl of distilled water, and 0.08 μl of SuperTaq polymerase (15 U/μl; HT Biotechnology, Cambridge, United Kingdom). The PCR consisted of 40 cycles of denaturation at 94°C (45 s), annealing at 55°C (45 s), and extension at 72°C (45 s). A precycling denaturation step at 94°C was applied for 5 min. As control sample, 50 μl of PCR mix without additional DNA samples was run in parallel. Then, 10-μl portions of the PCR products were analyzed on a 1% agarose gel containing ethidium bromide. Electrophoresis was performed in 0.5× TBE (50 mM Tris, 50 mM borate, 1 mM EDTA); gels were then stained in aqueous ethidium bromide (10 ng/ml) and photographed under UV illumination.
Cloning of amplification products.
The PCR amplification products (3 μl of a PCR mix) were used for ligation in pCR2.1 and transformed into competent Escherichia coli TOP10 cells by using the Original TA Cloning Kit (Invitrogen, San Diego Calif.). Clones were grown overnight at 37°C on 2YT agars (Yeast-Trypton; Gibco-BRL, Breda, The Netherlands) containing ampicillin (100 μg/ml) and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; 40 μg/ml). Possible transformants were identified by blue-white colony screening.
Screening for full-length inserts.
DNA was liberated from possible recombinants by suspending part of the colony in 100 μl of distilled water. The suspension was boiled for 10 min and centrifuged for 10 min at 14,000 rpm in an Eppendorf centrifuge. Next, 5 μl of the supernatant was used as a template for PCR. The primers used for amplification of the putative inserts in pCR2.1 were M13 and T7 sequence specific (AACAGCTATGACCATG and TAATACGACTCACTATAGGG, respectively). then, 45 μl of PCR mix, identical to the mix used for the ssu rDNA PCR, was added. The PCR consisted of 30 cycles of denaturation at 94°C (45 s), annealing at 56°C (45 s), and extension at 72°C (45 s). A precycling denaturation at 94°C was applied for 5 min. The PCR products were visualized as described above. Only the samples with a full-length insert (∼700 bp) were analyzed by restriction enzyme digestion. Per clinical sample, ca. 50 clones with full-length inserts were selected for further analysis.
RFLP analysis.
We digested 15 μl of the PCR product solution (M13/T7 PCR) by using the restriction endonuclease AluI (New England Biolabs, Beverly, Mass.). The restriction digests were analyzed in a 3% Metaphor agarose gel containing ethidium bromide. The electrophoresis was performed in 0.5× TBE. The gels were stained, examined, and photographed under UV illumination. Analysis of the different restriction fragment length polymorphism (RFLP) patterns was initially performed visually. If it was not possible to discriminate between certain types, the software program GelCompar version 4.0 was also used. When GelCompar was used, the position of DNA fragments shorter than 100 bp was ignored because these were not resolved well enough. For the remaining DNA fragments, bands were analyzed according to Dice with the tolerance set at 1.0% (optimization = 0.50%, minimal area = 0.1%). Two RFLP patterns were regarded as the same if they matched for 100%. Different RFLP types were given separate capital letter codes.
DNA sequencing.
For bidirectional sequencing of the insert, the ssu rDNA PCR (described above) was repeated. The nucleic acid sequence of the PCR product was analyzed by Sanger's method (BaseClear, Leiden, The Netherlands) by using the Big Dye terminator sequencing kits 373, 377, and 3100 (PE Applied Biosystems). The assembled ssu rDNA sequences were subjected to basic local alignment search tool (BLAST) analysis (http://www.-ncbi.nlm.nih.gov/blast/; version 1 June 2002). This analysis was used to determine which sequence in the GenBank depository was most similar to the partial 3′-terminal ssu rDNA sequence of the isolate. For the construction of phylogenetic trees, the sequence data were compared by using multiple sequence alignment software as available at www.genebee.msu (A. N. Belozersky Institute, Russian EMB Net Node) and were expressed as phylograms (6).
RESULTS
Participants.
The five patients had a mean age of 33 years; the mean age of the controls was 37 years (Table 1). Although patients were asked to return 2 weeks after the initial visit, logistical difficulties caused the interval time to vary between 13 and 22 days. The control subjects all presented with an interval time of precisely 14 days. All patients had a history of some sort of urethritis (chlamydial, gonococcal, or NCNGU) in the years prior to the study. In comparison, only one of the control subjects suffered from NCNGU in the past. Patients and control subjects refrained from sexual intercourse between visits, had negative tests for N. gonorrhoeae and C. trachomatis, and did not have signs of herpes genitalis or other overt sexually transmissible diseases. Table 1 presents the urine PMN cell counts, thereby illustrating that for all of the patients the NCNGU episode cleared between the two visits, probably due to the azithromycin therapy. Clinical complaints resolved during the 2-week period as well. The cell counts for the controls were always in the normal range (≤6).
RFLP analysis of ribosomal clones derived from tap water sediments.
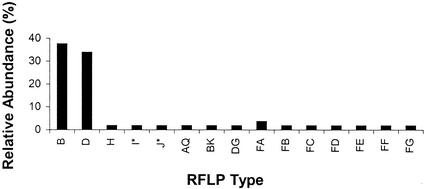
Many bacterial ribosomal clones were obtained by cloning the PCR products of the sediment of 50 ml of tap water. Upon analysis of 55 clones, 15 clearly different RFLP types were documented (see Table 2). Figure 1 shows the distribution of these clones across the RFLP types, indicating their relative frequencies of occurrence. There were seven RFLP types (FA to FG) present that were never obtained from controls and patients. Types B and D appeared to be most common. Sequencing of some of the clones revealed that these two types represented Pseudomonas spp., which are known to be associated with water-rich environments. The amounts of bacteria present in tap water probably mask the detection of rDNA contaminants in the reagents used for PCR.
TABLE 2.
Clones and RFLP types recovered from water, control, and patient samples
| Control no. sample type, or patient no. | Visit | No. of clones analyzed | No. of RFLP types |
|---|---|---|---|
| Controls | |||
| 1 | First | 55 | 22 |
| Second | 50 | 15 | |
| 2 | First | 46 | 19 |
| Second | 62 | 22 | |
| 3 | First | 49 | 15 |
| Second | 50 | 22 | |
| 4 | First | 49 | 14 |
| Second | 43 | 18 | |
| 5 | First | 48 | 15 |
| Second | 36 | 11 | |
| Watera | Singlea | 55 | 15 |
| Patients | |||
| 1 | First | 44 | 10 |
| Second | 49 | 15 | |
| 2 | First | 50 | 11 |
| Second | 49 | 14 | |
| 3 | First | 46 | 28 |
| Second | 37 | 15 | |
| 4 | First | 48 | 8 |
| Second | 49 | 11 | |
| 5 | First | 50 | 18 |
| Second | 50 | 21 |
The water sample was obtained only once.
FIG. 1.
Distribution of ssu rDNA PCR RFLP types derived from tap water. Indicated are the AluI types identified by uppercase letters versus their frequency of occurrence among the 55 clones studied. Note that types B and D, representing Pseudomonas spp., are by far the most prevalent. The relative abundance represents the proportion the number of representatives of a given RFLP type divided by the cumulative number of all clones analyzed. The latter is set at 100%.
RFLP analysis of ribosomal clones derived from urine sediments.
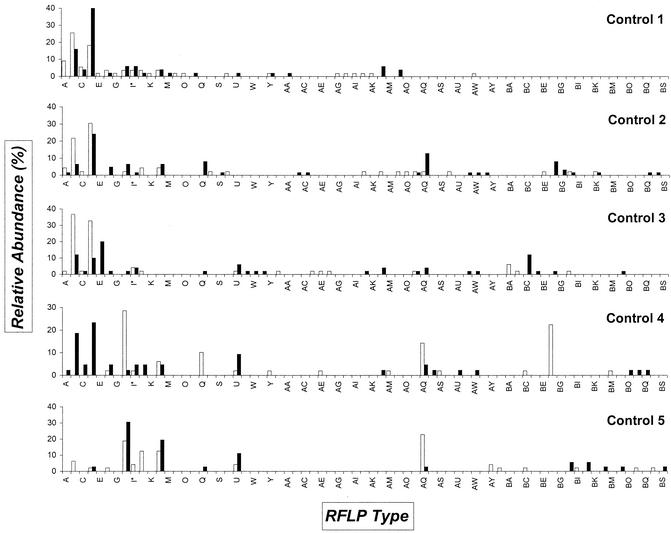
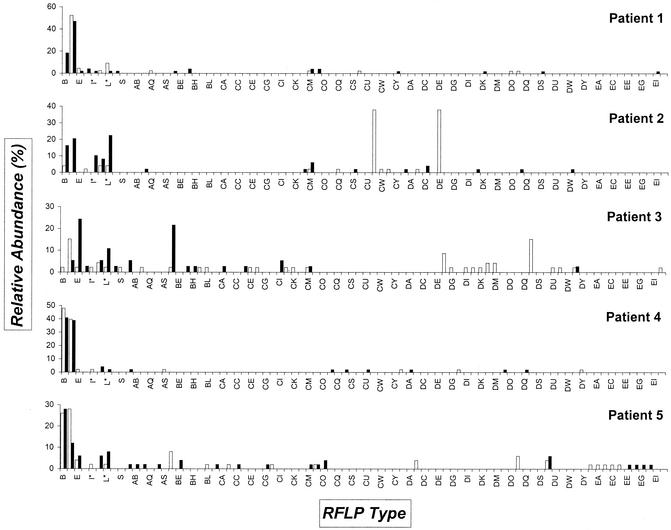
Table 2 indicates the number of clones that were analyzed for patients and volunteers by the RFLP approach. In all, RFLP patterns were recorded for 472 clones derived from patient samples, and 488 were documented for the healthy controls. Ultimately, the RFLP database is comprised of 960 ribosomal fingerprints. AluI digestion of full-length inserts generally resulted in two to five conveniently resolved bands (Fig. 2). Different RFLP types represent the different types of ssu rRNA genes that are present in microbes concentrated in the urine sediment. It has to be emphasized, since we only employed a single restriction enzyme, that clones with identical RFLP patterns do not necessarily represent identical bacterial species. The distribution of clones (both in RFLP type and abundance) in control subjects and in patients is plotted in Fig. 3 and 4. Overall, 71 and 84 different RFLP types were obtained in control subjects and patients, respectively. We found 62 different RFLP types present in patient samples that were absent in control samples (CA-EJ). All clone libraries had a relative dominance of a few types (e.g., the “watertypes” B and D in most samples or CV and DE in the sample obtained during the first visit of patient 2). It can be noted that some types were only present during one visit (e.g., in control 4, types Q and BF). Also, most of the types were not found in all of the persons (e.g., type E was only present in control 1 and 3 and all patients except number 2). Especially noteworthy is the fact that RFLP type H was encountered in 9 of 10 samples derived from control individuals. During NCNGU, this type was never detected. This difference was highly significant from the statistical viewpoint (9 of 10 versus 0 of 5; P = 0.0020). Even when calculated for the two groups of individuals, this significance was maintained (5 of 5 versus 0 of 5; P = 0.0079). For RFLP type AQ, a similar trend was observed; in this case statistical relevance was not reached.
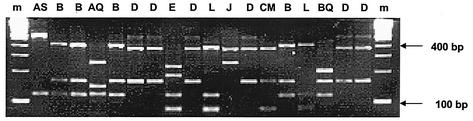
FIG. 2.
PCR RFLP analysis of a randomly selected subset of the ssu rDNA clones obtained from the urine sediments of patient 1. Both on the right and on the left, a molecular size marker is included (100-bp ladder; Bio-Rad, Veenendaal, The Netherlands); above the lanes, the uppercase letter code for the RFLP patterns is indicated.
FIG. 3.
Distribution of ssu rDNA PCR RFLP types derived from control individuals. Open bars indicate the clones identified from the urine sediment obtained during the first visit; solid bars indicate clones from the sediment collected at the second visit. On the horizontal axis the RFLP types are identified; on the vertical axis the relative abundance of the types is shown. For a definition of relative abundance, see the legend to Fig. 1. Codes indicated by an asterisk were hard to classify definitely and may represent heterogeneous types probably consisting of more than one sequence motif.
FIG. 4.
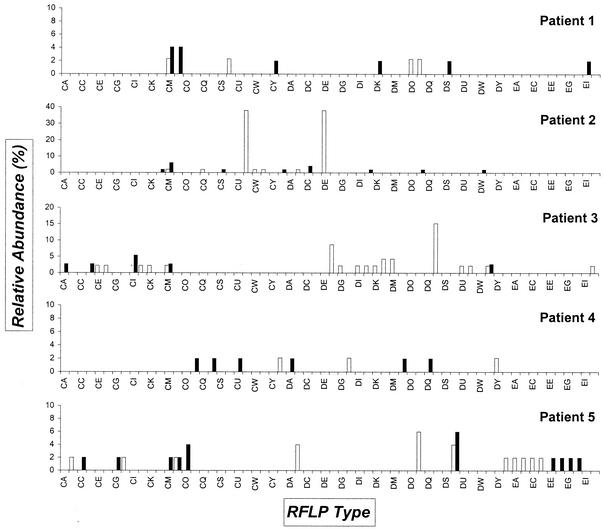
Distribution of ssu rDNA PCR RFLP types derived from the five patients. Open bars indicate the clones identified from the urine sediment obtained during the first visit; solid bars indicate clones from the sediment collected at the second visit. On the horizontal axis, the RFLP types are identified; on the vertical axis, the relative abundance of the types is shown. For a definition of relative abundance, see the legend to Fig. 1. Codes indicated by an asterisk were hard to classify definitely and may represent heterogeneous types probably consisting of more than one sequence motif.
On average, the fraction of variant clones identified at any sampling moment was comparable. The index of variation, i.e., the ratio between the number of RFLP types and the overall number of clones studied, was between 0.32 and 0.36. These results indicate that the urethral flora shows extensive intra- and interpersonal variation. It is interesting that in the volunteers, 47 of 138 cumulatively identified types disappeared during the 2-week monitoring period. In the patients, who were treated with antibiotics, 50 of 127 identified types seemed to disappear. The difference in microbial dynamics between the two groups is not statistically significant (two-sided Fisher exact test, P = 0.37). This implies that the azithromycin treatment does not seem to drastically induce microbial species extinction in the urethra: in both controls and patients, similar elimination rates are encountered. On the other hand, species also appear during the monitoring period. Again, no statistically relevant difference is noted between the groups (52 of 138 versus 51 of 127; P = 0.71). In conclusion, azithromycin treatment does not seem to severely affect the composition and dynamics of the urethral flora. Interestingly, however, there still is a relatively large group of ssu rRNA PCR RFLP types that only occur in patients and not in controls (n = 62). A number of these disappear during the 2-week posttreatment period (n = 34). We assume that these represent antibiotic-susceptible pathogenic bacteria and, therefore, these clones will be addressed specifically below (see Fig. 5).
FIG. 5.
Overview of the 16S rDNA RFLP types that are present in the patients and absent from the controls. Open bars indicate the clones identified from the urine sediment obtained during the first visit (t = 0); solid bars indicate clones from the sediment collected at the second visit (t = 1). On the horizontal axis, the RFLP types are identified; on the vertical axis, the relative abundance of the types is shown. For a definition of relative abundance, see legend to Fig. 1. Table 4 shows the sequence-based identification of a selection of these clones.
DNA sequencing.
Due to the large number of different RFLP types (140 in all), sequence analysis of all 16S rDNA types was impractical. Some of the RFLP types found more frequently (relative abundance of >10%) were sequenced to assess the nature of the constant factors in the bacterial urethral flora in more detail. Table 3 surveys the resemblance of some of the more dominant clones with known bacterial species based on BLAST searches in GenBank. All sequencing analyses involved two independent clones per RFLP type. The fact that in eight of nine cases identical or highly similar sequences were obtained corroborates the reliability of the RFLP analysis. It should be noted that clones with different RFLP types (e.g., B and D, CV and DE, etc.) sometimes revealed a close relatedness to the same bacterial genus. In conclusion, Table 3 highlights that many water-borne bacterial species, including Pseudomonas, Ralstonia, and Sphingomonas spp., were identified as possible members of the healthy male urethral flora, some of which are also identified as water contaminants. The latter observation raises the question of whether these species are tap water contaminants or genuine members of the male urethral flora.
TABLE 3.
Sequencing results of RFLP clones dominating one or more of the samples obtained from healthy control subjects
| RFLP type | Sequencing result | Score |
|---|---|---|
| B | Pseudomonas gessardii | 1092 |
| Pseudomonas libaniensis | 1092 | |
| Pseudomonas synxantha | 1092 | |
| B | Pseudomonas libaniensis | 1067 |
| Pseudomonas gessardii | 1067 | |
| Pseudomonas synxantha | 1065 | |
| D | Pseudomonas fluorescens ATCC 49642 | 852 |
| Pseudomonas fluorescens ATCC 17574 | 852 | |
| Uncultured manure pit bacterium P320 | 852 | |
| Unidentified γ-proteobacterium OM93 | 852 | |
| Pseudomonas sp. clone NBO.1H | 852 | |
| D | Pseudomonas veronii | 1074 |
| Unidentified γ-proteobacterium | 1061 | |
| E | Streptococcus sp. oral strain H6 | 1043 |
| Streptococcus sp. oral strain B5SC | 1041 | |
| Streptococcus sp. oral clone BW009 | 1037 | |
| E | Streptococcus mitis | 1065 |
| H | Uncultured bacterium GKS2-124 | 979 |
| Unidentified gamma proteobacterium | 965 | |
| Sphingomonas echinoides | 955 | |
| H | Uncultured bacterium GKS2-124 | 981 |
| Unidentified γ-proteobacterium | 973 | |
| Sphingomonas echinoides | 963 | |
| L | Ralstonia pickettii | 850 |
| Ralstonia sp. strain APF11 | 850 | |
| Burkholderia pickettii ATCC 27511 | 835 | |
| L | Ralstonia pickettii | 850 |
| Ralstonia sp. strain APF11 | 850 | |
| Burkholderia pickettii ATCC 27511 | 835 | |
| U | Pseudomonas fluorescens bv. | 771 |
| Pseudomonas gessardii | 771 | |
| Pseudomonas libaniensis | 771 | |
| Uncultured bacterium GR-296.II.35 | 771 | |
| Pseudomonas sp. strain IC038 | 771 | |
| Unidentified bacteria | 771 | |
| U | Ralstonia sp. strain APF11 | 1086 |
| Uncultured bacterium OSs75 | 1053 | |
| Pseudomonas pickettii | 1045 | |
| AQ | Sphingomonas echinoides | 765 |
| Sphingomonas echinoides | 759 | |
| Uncultured bacterium GKS2-124 | 757 | |
| Unidentified α-proteobacterium | 757 | |
| AQ | Unidentified α-proteobacterium | 997 |
| Uncultured bacterium GKS2-124 | 989 | |
| Star-like microcolonies | 977 | |
| Sphingomonas echinoides | 971 | |
| BC | Streptococcus mitis | 1019 |
| BC | Streptococcus mitis | 1080 |
| BF | Streptococcus agalactiae | 807 |
| Streptococcus agalactiae | 805 | |
| BF | Streptococcus agalactiae | 1082 |
| Streptococcus agalactiae | 1074 | |
| Streptococcus agalactiae | 1005 |
To determine which ssu rDNA types could possibly be associated with NCNGU, clones that appeared to be differentially distributed among patients and controls were subjected to sequencing. Type H, which, significantly, more often occurs in the flora of healthy men, gave rise to ribosomal sequence motifs that were most homologous to that of an uncultured bacterial species encoded GKS2-124. GKS2-124 belongs to the group of the α-proteobacteria and was initially isolated from a German freshwater lake (12). No obvious homology with known pathogenic bacterial species was observed, the sequence showed a somewhat more distant homology to water-thriving organisms such as Sphingomonas spp. A similar, though not statistically significant, trend was also observed for type AQ, occurring in four of five healthy controls and just one patient.
Sequence analysis was also performed for one or two clones with RFLP types that were present in the first samples provided by the patients but absent from the water sample, the patient's follow up samples, and both samples from the controls (Fig. 5). Of the 32 different types of clones, 24 were successfully sequenced (Table 4). Interestingly, patient 2 represents the only individual for whom a well-known putative uropathogen was identified. Four of the different RFLP clones were very similar to the rRNA sequence of M. genitalium, thereby providing evidence for the fact that this person was cured from an M. genitalium infection. This fact again illustrates that the experimental approach employed was solid and that the outcome is relevant. However, the diversity observed among the RFLP patterns is somewhat enigmatic since M. genitalium only harbors a single copy of the 16S rRNA gene. Whether this variation is due to mixed infections or PCR and cloning induced errors is subject of current investigation. Patient 3 appeared to be infected by Haemophilus parainfluenzae (clone DF/DL), a bacterial species that has been mentioned before, but less convincingly as M. genitalium, in relation to NCNGU (27). For the other patients, various previously identified bacterial species, but also some species that currently lack a detailed description, matched entries in the GenBank database.
TABLE 4.
Sequencing results of RFLP clones only present in patients’ t = 0 samples or randomly selected from t = 0 samples of controls
| Patient or control no. | RFLP | Sequencing result | Score |
|---|---|---|---|
| Patients | |||
| 1 | CT | Uncultured Veillonella spp. | 1096 |
| DO | Uncultured bacterium clone DJAT-434 | 797 | |
| Uncultured γ-proteobacterium | 797 | ||
| 2 | CQ | Streptococcus gordonii | 1068 |
| CV | Mycoplasma genitalium | 1019 | |
| CW | Mycoplasma genitalium | 1080 | |
| CX | Mycoplasma genitalium | 1096 | |
| DB | Uncultured bacterium clone DJAT-434 | 797 | |
| Uncultured γ-proteobacterium | 797 | ||
| DE | Mycoplasma genitalium | 1088 | |
| 3 | CJ | Unidentified bacterium 6C | 624 |
| Methylobacterium spp. | 595 | ||
| CK | Corynebacterium thomssenii | 1007 | |
| DF | Haemophilus parainfluenzae | 969 | |
| Haemophilus paraphrophilus | 946 | ||
| DI | Unidentified oral bacterium AP60-15 | 676 | |
| Gemella haemolysans | 670 | ||
| DL | Haemophilus parainfluenzae | 763 | |
| Pseudomonas spp. | 728 | ||
| DM | Actinomyces turicensis | 648 | |
| Bacterial spp. | 634 | ||
| DR | Haemophilus paraphrophilus | 959 | |
| Haemophilus parainfluenzae | 959 | ||
| DU | Corynebacterium genitalium | 946 | |
| Uncultured Corynebacterium sp. | 473 | ||
| DV | Pseudomonas fluorescens | 1051 | |
| 4 | DH | Uncultured bacterium clone DJAT-434 | 735 |
| Uncultured γ-proteobacterium | 735 | ||
| DY | Pseudomonas fluorescens | 1082 | |
| 5 | CB | Uncultured bacterium clone TFBME10 | 613 |
| Pseudomonas sp. G2 | 613 | ||
| CH | Variovorax sp. strain HAB-30 | 920 | |
| DB | Uncultured bacterium clone DJAT-434 | 706 | |
| Pseudomonas sp. | 706 | ||
| DP | Uncultured bacterium clone DJAT-434 | 706 | |
| Uncultured γ-proteobacterium | 706 | ||
| EB | Uncultured bacterium D29A | 1063 | |
| Streptococcus salivarius | 1061 | ||
| Controls | |||
| 1 | K | Mesorhizobium loti | 932 |
| Rhizobium sp. strain CJ5 | 924 | ||
| Rhizobium loti | 924 | ||
| M | Pseudomonas marginalis | 1072 | |
| Pseudomonas spp. | 1072 | ||
| Pseudomonas reactans | 1072 | ||
| Pseudomonas veronii | 1072 | ||
| H | Unidentified bacterium ox-SCC-25/5 | 946 | |
| Unidentified bacterium ox-SCC-36/29 | 930 | ||
| O | Uncultured Comamonas spp. | 876 | |
| Pseudomonas testosteroni | 876 | ||
| Comamonas testosteroni | 876 | ||
| P | Peptostreptococcus genospecies | 500 | |
| Peptostreptococcus spp. | 456 | ||
| T | Uncultured Comamonas spp. | 891 | |
| Pseudomonas testosteroni | 891 | ||
| Comamonas testosteroni | 891 | ||
| AJ | Uncultured eubacterium WD293 | 1017 | |
| Pseudomonas mephitica | 1005 | ||
| AK | Uncultured eubacterium WD293 | 1036 | |
| Agricultural soil bacterium | 1027 | ||
| AN | Pseudomonas spp. | 1052 | |
| Pseudomonas gessardii | 1046 | ||
| Pseudomonas libaniensis | 1046 | ||
| G | Uncultured bacterium clone DJAT-434 | 783 | |
| Uncultured γ-proteobacterium | 783 | ||
| 2 | BG | Sphingomonas spp. | 787 |
| Uncultured bacterium GKS2-124 | 785 | ||
| AO | Ralstonia detusculanense | 628 | |
| Ralstonia spp. | 628 | ||
| Uncultured bacterium OSs7 | 628 | ||
| 3 | V | Unidentified bacterium rM7 | 648 |
| Comamonas testosteroni | 640 | ||
| AE | Ralstonia detusculanense | 713 | |
| Ralstonia spp. | 713 | ||
| Burkholderia spp. | 682 | ||
| AF | Pseudomonas jessenii | 653 | |
| Pseudomonas spp. | 653 | ||
| Uncultured eubacterium | 647 | ||
| AG | Pseudomonas sp. strain G2 | 1036 | |
| Pseudomonas sp. strain NZ66/64/124/122/113/108/106/65 | 1028 | ||
| Pseudomonas synxantha | 1028 | ||
| 4 | AM | Streptococcus agalactiae | 624 |
| Streptococcus agalactiae | 600 | ||
| BQ | Actinomyces viscosus | 997 | |
| Actinomyces viscosus | 908 |
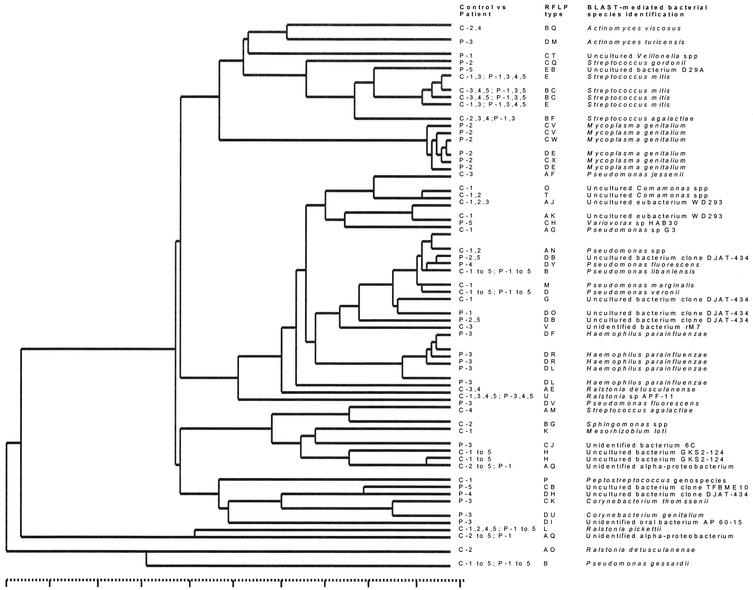
To strengthen the pathogen identification efforts, 18 minor clones from the controls were also sequenced (Table 4). Interestingly, 10 of 24 clones from the patients and 7 of 18 clones derived from the healthy controls were matched to microbial species that have never been cultured in vitro or have not yet been precisely classified as a species. Since the homology scores of the clone sequences with, for instance, the uncultured bacterial clone DJAT-434, occurring in four of five patients versus one of five control subjects, are variable, it is quite possible that we detected here novel bacterial species (Table 4). Figure 6 illustrates the gross interrelatedness of the individual bacterial species that were identified in Tables 3 and 4. Besides two heterogeneous groups of various bacterial species (Actinomyces, Veillonella, Corynebacterium, and Ralstonia spp., etc.), three major clusters can be discerned. Cluster A represents the streptococci, whereas cluster B derives from multiple clones analyzed for the patient suffering from an M. genitalium infection. Cluster C is the most interesting one since it is built from four subclusters, one of which (C3) gathers the various Haemophilus spp. The larger clusters C1 and C2 primarily contain Pseudomonas spp. Interestingly, the DJAT-434 homologues are clustered in the C2 group (except for the DJAT-434 homologue sequence derived from RFLP type DH from patient 4), a group consisting mainly of Pseudomonas-like organisms. Apparently, the DJAT-434 homologues closely resemble Pseudomonas spp. This leads to the conclusion that as-yet-unidentified Pseudomonas species may play a significant role, either as inducers of NCNGU or as opportunists occupying the environmental niche created during the disease process.
FIG. 6.
Phylogram based on comparative analysis of the ribosomal sequences obtained for the clones derived from urine sediments of patients and controls. The tree was constructed from a weighted residue weight table. Clones B and D are the types that are most prevalent among both patients and controls (present in P1 to P5 and C1 to C5). The entries derived from the patients' clones are identified by patient numbers (see also Table 4). The clones are identified to a putative species level on the basis of BLAST searching. Clusters harboring streptococci, the M. genitalium sequences, or the Haemophilus and Pseudomonas spp.-like sequences are obvious. The scale at the bottom runs from 0 to 100% homology.
DISCUSSION
With regard to the etiology of NCNGU in males, it is appropriate to distinguish between acute and “chronic” disease. Although no precise definition is available, NCNGU etiology is probably multifactorial (2, 14). In order to gain more insight in the pathogenesis of NCNGU in males, novel diagnostic approaches are mandatory (7). We focused here on acute cases of urethritis, i.e., those not caused by N. gonorrhoeae or C. trachomatis. To our knowledge, ours is the first study to approach the long-standing enigma of the etiology of acute NCNGU in men, employing broad-spectrum molecular-biological techniques, such as ssu rDNA PCR and subsequent RFLP and sequencing techniques. A downside of this approach is that eukaryotic pathogens, such as T. vaginalis, are not detected. These organisms are certainly important in a subset of NCNGU cases (7, 18). Second, the data could be influenced by the fact that some microorganisms are present in larger numbers or contain more ribosomal operons per genome than others, thereby facilitating their detection. Consequently, the spectrum of bacteria identified may not completely mimic the urethral flora in situ. Third, definite proof for the association of the bacteria identified with the urethral epithelium is not available, i.e., the bacteria could also originate from the bladder or other anatomical locations.
Despite these pitfalls, we successfully documented the cure of a M. genitalium infection in one of the subjects, which serves as an excellent technological process control. Recent data by Bjornelius et al. (4) revealed that M. genitalium could be detected in more than 36% of all patients. It was also shown that in 30% of all cases of chronic nongonococcal urethritis, U. urealyticum could be detected (14), although the general literature is inconclusive with respect to the importance of this bacterial species (14, 24, 27). We did not find evidence for the presence of infectious U. urealyticum among our acute NCNGU patients. In addition, various species of Haemophilus have been implicated in NCNGU (27). Interestingly, patient 3 of the present study seemed to be harboring various species of this particular genus as well. Haemophilus spp. were not encountered among the healthy controls. Finally, the presence of various species of oral streptococci (Streptococcus gordoni, an unidentified oral bacterium AP60-15, and Streptococcus salivarius in patients 2, 3, and 5, respectively) may corroborate an earlier suggestion concerning the involvement of oral sex in the pathogenesis of NCNGU (13).
For some species, involvement in NCNGU was denounced on the basis of previous studies performed by others (9, 32). On the other hand, many other species have been previously indicated as possibly involved in the establishment of NCNGU. Whether these candidates, including, for instance, U. urealyticum and Gardnerella vaginalis (9, 19), should be excluded from future studies cannot be decided on the basis of the present study, since the number of patients was kept low because of the experimental complexity of our in vitro work. We did not identify obvious and novel, putatively pathogenic bacterial species that are 100% associated with acute NCNGU. Although the RFLP analyses identified numerous types that were confined to pretreatment patient samples, DNA sequencing revealed that not a single bacterial species was exclusively present in all five disease-related, pretreatment samples and absent in all control samples. However, several uncharacterized bacterial species were identified more often (for instance, the DJAT 434 clone was detected in four of five patients versus one of five control subjects). In addition, Fig. 6 highlights the fact that, among the patients, Pseudomonas-like bacterial species were identified relatively frequently. The prevalence in NCNGU and the pathogenic potential of these bacterial species need to be defined more precisely. This requires the development of diagnostic tests for these elusive microorganisms that allow larger groups of patients to be screened. The reverse situation was documented once: RFLP type H occurred in all of the controls and in none of the patients. This difference, even with the limited number of individuals included in the current study, is statistically significant. The bacterial species most homologous to the RFLP type H DNA sequence appeared to have been detected in fresh lake water (12). Whether this bacterial species acts as a putative probiotic or whether it is simply outcompeted by the pathogens involved remains to be elucidated, just as diagnostic tests for this species need to be developed in order to more accurately determine its prevalence in healthy and diseased male urethras.
The major findings presented here are, first, the significant inter- and intrapersonal variability of the urethral flora, both in healthy and infected individuals, although this conclusion may be biased by the fact that only 50 clones were analyzed per urine sample. Second, azithromycin treatment seems to have little effect on the variability, complexity, and dynamics of the resident flora: many species not only disappear or appear during antibiotic treatment but also in the healthy, untreated situation. It is reassuring to see that in two of five patients, previously suggested pathogens were encountered that disappeared upon antibiotic treatment. The detection of a diversity of uncharacterized Pseudomonas-like bacterial species suggests that there is much more to explore in the bacterial flora of the male urethra, whereas the detection of bacterial species that disappear upon disease development may have important future implications to the therapy of NCNGU.
Acknowledgments
We thank the medical, nursing, and laboratory staff of the STD Outpatient Clinic of the Department of Dermatology and Venereology of the Erasmus MC (Rotterdam, The Netherlands) for their help and support during this study.
REFERENCES
- 1.Arya, O. P., D. Hobson, C. A. Hart, C. Bartzokas, and B. C. Pratt. 1986. Evaluation of ciprofloxacin 500 mg twice daily for one week in treating uncomplicated gonococcal chlamydial and nonspecific urethritis in men. Genitourin. Med. 62:170-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Association of Genitourinary Medicine and the Medical Society for the Study of Venereal Diseases. 1999. National guidelines for the management of non-gonococcal urethritis. Sex Transm. Infect. 75:S9-S12. [PubMed] [Google Scholar]
- 3.Becker, M. R., B. J. Paster, E. J. Leys, M. L. Moeschberger, S. G. Kenyon, J. L. Galvin, S. K. Boches, F. E. Dewhirst, and A. L. Griffen. 2002. Molecular analyses of bacterial species associated with childhood caries. J. Clin. Microbiol. 40:1001-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bjornelius, E., P. Lidbrink, and J. S. Jensen. 2000. Mycoplasma genitalium in non-gonococcal urethritis: a study in Swedish male STD patients. Int. J. STD AIDS 11:292-296. [DOI] [PubMed] [Google Scholar]
- 5.Boom, R., C. J. A. Sol, M. M. M. Salimans, C. L. Jansen, P. M. E. Wertheim van Dillen, and J. van der Noordaa. 1990. Rapid and simple method for the purfication of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brodsky, L. I., V. V. Ivanov, Y. L. Kalaidzidis, A. M. Leontovich, V. K. Nikolaev, S. I. Feranchuk, and V. A. Drachev. 1995. Genebee-NET: internet-based server for analyzing biopolymers structure. Biochemistry 60:923-928. [PubMed] [Google Scholar]
- 7.Burstein, G. R., and J. M. Zenilman. 1999. Nongonococcal urethritis: a new paradigm. Clin. Infect. Dis. 28:S66-S73. [DOI] [PubMed] [Google Scholar]
- 8.Cates, W. C., and K. K. Holmes. 1996. Condom efficacy against gonorrhoeae and nongonococcal urethritis. Am. J. Epidemiol. 143:843-844. [DOI] [PubMed] [Google Scholar]
- 9.Elsner, P., A. A. Hartmann, and I. Wecker. 1998. Gardnerella vaginalis is associated with other sexually transmittable microorganisms in the male urethra. Zentbl. Bakteriol. Mikrobiol. Hyg. 269:56-63. [DOI] [PubMed] [Google Scholar]
- 10.Gall, H., H. Beckert, H. Meier-Ewert, U. Tummers, R. A. Pust, and R. U. Peter. 1999. Pathogen spectrum of urethritis in the man. Hautartzt 50:186-193. [DOI] [PubMed] [Google Scholar]
- 11.Giovannoni, S. J., T. B. Britschgi, C. L. Moyer, and K. G. Field. 1990. Genetic diversity in Sargasso Sea bacterioplankton. Nature 345:60-63. [DOI] [PubMed] [Google Scholar]
- 12.Glockner, F. O., E. Zaichikov, N. Belkova, L. Denissova, J. Pernthaler, A. Pernthaler, and R. Amann. 2000. Comparative 16S rRNA analysis of lake bacterioplankton reveals globally distributed phylogenetic clusters including an abundant group of actinobacteria. Appl. Environ. Microbiol. 66:5053-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hernandes Aguado, I., C. Alvarez Dardet, M. Gili, et al. 1988. Oral sex as a risk factor for chlamydia-negative, ureaplasma-negative non-gonococcal urthritis. Sex Transm. Dis. 15:100-102. [DOI] [PubMed] [Google Scholar]
- 14.Horner, P., B. Thomas, B. C. Gilroy, M. Egger, and D. Taylor-Robinson. 2001. Role of Mycoplasma genitalium and Ureaplasma urealyticum in acute and chronic nongonococcal urethritis. Clin. Infect. Dis. 32:995-1003. [DOI] [PubMed] [Google Scholar]
- 15.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janier, M., F. Lassau, I. Casin, P. Grillot, C. Scieux, A. Zavaro, C. Chastang, A. Bianchi, and P. Morel. 1995. Male urethritis with or without discharge: a clinical and microbiological study. Sex Transm. Dis. 22:244-252. [DOI] [PubMed] [Google Scholar]
- 17.Keane, F. E., B. J. Thomas, C. B. Gilroy, A. Renton, and D. Taylor-Robinson. 2000. The association of Chlamydia trachomatis and Mycoplasma genitalium with non-gonococcal urethritis: observations on heterosexual men and their female partners. Int. J. STD AIDS 11:435-439. [DOI] [PubMed] [Google Scholar]
- 18.Krieger, J. N., C. Jenny, M. Verdon, et al. 1993. Clinical manifestations of trichomoniasis in men. Ann. Intern. Med. 118:844-849. [DOI] [PubMed] [Google Scholar]
- 19.Krieger, J. N., D. E. Riley, M. C. Roberts, and R. E. Berger. 1996. Prokaryotic DNA sequences in patients with chronic idiopathic prostatitis. J. Clin. Microbiol. 34:3120-3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morency, P., M. J. Dubois, G. Gresenguet, et al. 2001. Aetiology of urethral discharge in Bangui, Central African Republic. Sex Transm. Infect. 77:125-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Relman, D. A. 1993. The identification of uncultured microbial pathogens. J. Infect. Dis. 168:1-8. [DOI] [PubMed] [Google Scholar]
- 22.Rolph, H. J., A. Lennon, M. P. Riggio, W. P. Saunders, D. MacKenzie, L. Coldero, and J. Bagg. 2001. Molecular identification of microorganisms from endodontic infections. J. Clin. Microbiol. 39:3282-3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmidt, T. M., and D. Relman. 1994. Phylogenetic identification of uncultured pathogens using ribosomal RNA sequences. Methods Enzymol. 235:205-222. [DOI] [PubMed] [Google Scholar]
- 24.Schwartz, M. A., and T. M. Hooton. 1998. Etiology of nongonococcal nonchlamydial urethritis. Sex Transm. Dis. 4:727-733. [DOI] [PubMed] [Google Scholar]
- 25.Schwartz, M. A., W. E. Lafferty, J. P. Hughes, and H. A. Handsfield. 1997. Risk factors for urethritis in heterosexual men: the role of fellatio and other sexual practices. Sex Transm. Dis. 24:449-455. [DOI] [PubMed] [Google Scholar]
- 26.Stefanik, M., K. Rychna, and A. Valkoun. 1992. Microbial causative agents of male urethritis. J. Hyg. Epidemiol. Microbiol. Immunol. 36:111-118. [PubMed] [Google Scholar]
- 27.Sturm, A. W. 1986. Haemophilus influenzae and Haemophilus parainfluenzae in non-gonococcal urethritis. J. Infect. Dis. 153:165-167. [DOI] [PubMed] [Google Scholar]
- 28.Totten, P. A., M. A. Schwartz, K. Sjostrom, et al. 2001. Association of Mycoplasma genitalium with non-gonococcal urethritis in men. J. Infect. Dis. 183:269-276. [DOI] [PubMed] [Google Scholar]
- 29.Ward, D. M., R. Weller, and M. M. Bateson. 1990. 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature 345:63-65. [DOI] [PubMed] [Google Scholar]
- 30.Wilson, K. H., R. B. Blitchington, and R. C. Greene. 1990. Amplification of bacterial 16S rRNA with polymerase chain reaction. J. Clin. Microbiol. 28:1942-1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson, K. H. 1994. Detection of culture resistant bacterial pathogens by amplification and sequencing of ribosomal DNA. Clin. Infect. Dis. 18:958-962. [DOI] [PubMed] [Google Scholar]
- 32.Woolley, P. D., G. R. Kinghorn, M. D. Talbot, and B. I. Duerden. 1990. Microbiological flora in men with nongonococcal urethritis with particular reference to anaerobic bacteria. Int. J. STD AIDS 1:122-125. [DOI] [PubMed] [Google Scholar]