Abstract
Real-time PCR with melting curve analysis of PCR products is a rapid procedure for detecting and genotyping herpes simplex virus (HSV). When testing mucocutaneous samples for HSV by a real-time PCR assay targeting the DNA polymerase gene, we found that some PCR products had atypical melting curves that did not conform to the expected melting temperatures for HSV type 1 or 2. Sequence analysis showed that these strains had base-pair mismatches over the probe binding sites. An alternative assay is required to type such atypical isolates.
The development of real-time PCR assays has provided a rapid alternative to traditional viral cell culture isolation for detection and typing of herpes simplex virus (HSV). (3-6, 8) These assays enable detection of HSV within a few hours in a closed system that reduces the risk of contamination by amplicon carryover. Furthermore, genotyping is possible through analysis of the melting curve of the PCR product, with each genotype being distinguished by a different melting temperature (Tm).
While evaluating real-time PCR for detection of HSV in clinical samples, we noticed that some HSV PCR products had atypical melting curves that did not conform to the expected Tm for HSV type 1 or HSV type 2. These samples and amplicons were further analyzed to determine the reasons for these findings.
For detection of HSV in mucocutaneous samples, we use a modification of the LightCycler PCR assay developed by Olfert Landt (TIB Molbiol, Berlin, Germany) and described by Burrows et al. (3) This assay targets the HSV DNA polymerase gene, and our modification involves the use of 4.0 mM MgCl2, a 0.5 μM concentration of forward primer, a 0.1 μM concentration of reverse primer (asymmetric primers to minimize the hook effect [2]), and a 0.2 μM concentration of each probe. All PCRs were carried out by using the LightCycler-FastStart-DNA Master Hybridization Probe kit (Roche Diagnostics).
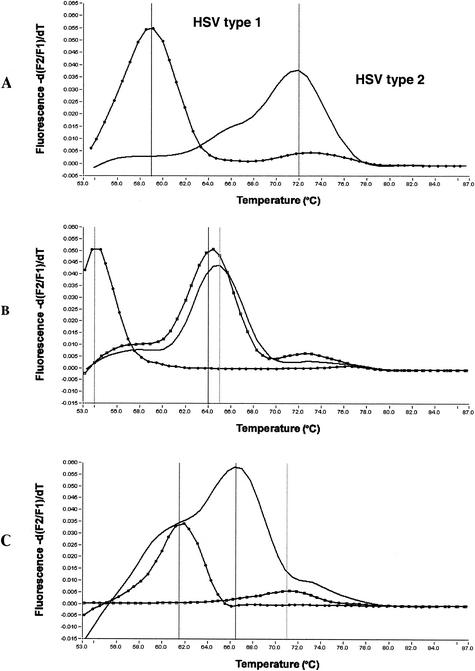
PCR products were analyzed by using the LightCycler melting curve analysis software and designated HSV type 1 or type 2 based on the Tm. The melting curve process involves the continuous acquisition of fluorescence as the temperature is steadily increased from 55 to 90°C. The presence of two mismatch base pairs over the LightCycler 640 labeled probe binding site for HSV type 1 causes the probe to separate at a lower temperature (59°C) than for HSV type 2 (72°C), which has a perfect match (Fig. 1A).
FIG. 1.
Melting curves for HSV PCR products. (A) Expected melting curves for HSV types 1 and 2; (B) atypical HSV type 1 melting curves; (C) atypical HSV type 2 melting curves.
Of the 745 genital and cutaneous samples that we tested, 266 (36%) were determined to be positive for HSV. Of the positive samples, 124 (46.6%) were found to be positive for HSV type 1, 127 (47.7%) were found to be positive for HSV type 2, and 15 (5.7%) failed to exhibit the expected melt curve for HSV type 1 or HSV type 2. The atypical melting curves from the 15 samples conformed to six different Tm patterns (Fig. 1).
We sequenced the PCR product from each representative pattern by using Megabase (Amersham Pharmacia Biotech) at the Waikato Sequencing Facility (Waikato University, Hamilton, New Zealand). CLUSTALX (v.1.81) alignment of sequences showed base pair changes over the probe binding sites (Fig. 2). We presume these changes are responsible for the atypical melt curves.
FIG. 2.
Alignment of the DNA polymerase gene sequences of six HSV strains with atypical melting curve patterns. The temperatures indicate the melting temperature for each strain. The reverse primer and probe binding sites are boxed.
The 15 samples with atypical melting curves were also tested for HSV by a PCR assay with a different target (glycoprotein D), and genotyping was performed by restriction enzyme analysis (1, 7). HSV was detected in all samples, and all could be classified as either HSV type 1 or HSV type 2 (Fig. 1). We were also able to retrospectively culture 9 of the 15 samples with atypical melting curves on A549 cells. Failure to isolate HSV from the remaining six samples may have been due to inadequate transport, prolonged storage under suboptimal conditions before culturing, or the reduced sensitivity of culture compared to the PCR analyses (3).
Our findings indicate that sequence variation in the HSV DNA polymerase gene may complicate melting curve analysis for genotyping HSV by producing Tm values that differ from expect values for HSV type 1 or HSV type 2. If genotyping is required for strains with atypical melting curves, an alternative method, such as restriction enzyme analysis, may be required.
Acknowledgments
We thank the staff of the Virology Unit, Canterbury Health Laboratories, for their support and enthusiasm.
REFERENCES
- 1.Anderson, N. E., K. F. Powell, and M. C. Croxson. 1993. A polymerase chain reaction assay of cerebrospinal fluid in patients with suspected herpes simplex encephalitis. J. Neurol. Neurosurg. Psychiatr. 56:520-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barratt, K., and J. F. Mackay. 2002. Improving real-time PCR genotyping assays by asymmetric amplification. J. Clin. Microbiol. 40:1571-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burrows, J., A. Nitsche, B. Bayly, E. Walker, G. Higgins, and T. Kok. 2002. Detection and subtyping of herpes simplex virus in clinical samples by LightCycler PCR, enzyme immunoassay, and cell culture. BMC Microbiol. 2:2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Espy, M. J., J. R. Uhl, P. S. Mitchell, J. N. Thorvilson, K. A. Svien, A. D. Wold, and T. F. Smith. 2000. Diagnosis of herpes simplex virus infections in the clinical laboratory by LightCycler PCR. J. Clin. Microbiol. 38:795-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kessler, H. H., G. Muhlbauer, B. Rinner, E. Stelzl, A. Berger, H. W. Dorr, B. Santner, E. Marth, and H. Rabenau. 2000. Detection of herpes simplex virus DNA by real-time PCR. J. Clin. Microbiol. 38:2638-2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nicoll, S., A. Brass, and H. A. Cubie. 2001. Detection of herpes viruses in clinical samples using real-time PCR. J. Virol. Methods 96:25-31. [DOI] [PubMed] [Google Scholar]
- 7.Powell, K. F., N. E. Anderson, R. W. Frith, and M. C. Croxson. 1990. Non-invasive diagnosis of herpes simplex encephalitis. Lancet 335:357-358. [DOI] [PubMed] [Google Scholar]
- 8.Schalasta, G., A. Arents, M. Schmid, R. W. Braun, and G. Enders. 2000. Fast and type-specific analysis of herpes simplex virus types 1 and 2 by rapid PCR and fluorescence melting-curve-analysis. Infection 28:85-91. [DOI] [PubMed] [Google Scholar]