Abstract
The objective of this study was to evaluate the advantages of cytomegalovirus (CMV) real-time PCR in blood plasma to monitor CMV infection in a population of adult and pediatric bone marrow recipients in comparison with the pp65 antigenemia method. Fifty allogeneic bone marrow transplant recipients from our center, including 23 adults and 27 children, were enrolled. A CMV real-time PCR designed to amplify a well-conserved region of the UL123 gene was evaluated for its results with whole blood and blood plasma. The CMV real-time PCR assay and the CMV antigenemia method were performed in parallel with 558 blood samples. The results obtained by the two techniques were significantly correlated (r = 0.732; P < 0.0001). Twenty patients developed at least one episode of CMV replication, with a total of 24 episodes detected by CMV PCR; antigenemia assays were positive in 17 of these 24 episodes. The first positive PCR test preceded the first positive antigenemia by a median of 8 days. The median time interval necessary to obtain a negative CMV PCR test after implementation of preemptive treatment was 28 days. CMV PCR of plasma was positive in two children with CMV disease (one with early CMV pneumonia and one with CMV gastroenteritis), while CMV antigenemia remained negative. The use of CMV PCR with plasma to guide both implementation and discontinuation of CMV preemptive therapy might reduce the risk of occurrence of CMV disease since patients would be treated earlier, and it might also help to reduce the duration of treatment, which could attenuate the side effects of antiviral drugs.
Human cytomegalovirus (CMV) infection is frequent after bone marrow transplantation. Viral reactivation occurs in 30 to 50% of allogeneic bone marrow recipients and may lead to the development of CMV disease. Although the outcome of CMV disease and particularly CMV pneumonia has been improved by the use of antiviral therapy, it remains a life-threatening disease. Two strategies are currently used to prevent the development of CMV disease in bone marrow transplant recipients. Universal prophylaxis consists of effective viral therapy given to all recipients at risk of CMV reactivation. Preemptive therapy consists of viral therapy given only to patients with proven CMV reactivation. In the latter strategy, sensitive and specific virologic markers are necessary to ensure early detection of CMV infection. Preemptive therapy based on pp65 antigen detection in peripheral blood leukocytes has been widely used to monitor CMV infection in bone marrow transplant recipients, with subsequent reduction in the incidence of CMV disease in this population (2). However, this antigen-based diagnostic method has some disadvantages. It has a low sensitivity for detecting early CMV infection or disease that may occur before engraftment due to the lack of leukocytes readily examinable at this period. It has also a low predictive value for the occurrence of CMV-related gastroenteritis (3, 14). Moreover, this technique requires immediate processing, is time-consuming, and cannot be automated.
Molecular techniques based on quantitative PCR have been used to monitor CMV infection in bone marrow recipients. Some groups are routinely using in-house quantitative PCR tests based on either serial dilutions or competition with an internal positive control (10). However, these methods are fastidious and time-consuming. A commercial quantitative PCR assay is now available, but this assay does not detect CMV infection significantly earlier than the pp65 antigen detection method (5, 7, 19) and is more expensive. Real-time PCR technology is simple, reliable, and cost-effective (12). Different groups have developed real-time PCR assays for the diagnosis of CMV infection (9, 11, 16, 17, 18, 20, 21, 22, 25). The results of these assays in CMV DNA quantification in blood demonstrated a good correlation with those of pp65 antigenemia assay (9, 11, 24). Evaluation of quantitative real-time CMV PCR in monitoring bone marrow recipients gave encouraging results; it was able to detect virus reactivation earlier than was the CMV antigenemia assay in patients with CMV infection and disease (15, 17, 24).
We have developed an in-house quantitative real-time CMV PCR targeted to the major immediate-early gene. To make the technique simpler, we chose to detect CMV DNA in blood plasma rather than in blood leukocytes. Both adult and pediatric allogeneic bone marrow recipients were prospectively monitored during the posttransplantation period by both the real-time CMV PCR test and the antigenemia assay. The pediatric bone marrow recipients in our center are mainly infants affected with congenital immunodeficiency syndromes. CMV infection and disease are frequent and severe in these children, and the advantages of real-time CMV PCR to monitor viral replication in this population has not been previously described. Our study aimed to evaluate the feasibility and advantages of real-time PCR to monitor CMV infection in allogeneic bone marrow recipients in comparison with the pp65 antigenemia reference method.
MATERIALS AND METHODS
Patients.
Adults and children who underwent allogeneic bone marrow transplantation in our center between December 2000 and December 2001 were enrolled in this study. All patients or children's parents gave their informed consent to participate to the study.
Monitoring of CMV infection.
Monitoring of CMV infection was routinely performed with the help of CMV culture and CMV pp 65 antigenemia.
(i) CMV culture.
Peripheral blood leukocytes isolated from blood, nasopharyngeal aspirates, and bronchoalveolar lavage (BAL) fluid were inoculated into conventional flask culture or rapid shell cell culture seeded with MRC-5 cells as described elsewhere (13).
(ii) CMV pp65 antigenemia assay.
CMV antigenemia was measured weekly from the initiation of the conditioning regimen. A 5-ml volume of EDTA-treated blood was collected from each patient and 2 ml of whole blood was used for the CMV antigenemia assay. The CMV antigenemia assay was performed by indirect immunofluorescence detection of pp65 internal matrix phosphoprotein in peripheral blood leukocytes as specified by the manufacturer (CINA kit; Argene Biosoft, Varilhes, France). The results were reported as the number of positively stained cells per 100,000 leukocytes.
CMV real-time PCR test. (i) Samples
From the 5 ml of EDTA-treated blood drawn for routine antigenemia tests, 1 ml of total blood and 1 ml of plasma were recovered and stored at −20°C. CMV PCR assays were performed twice a week, but the CMV PCR results were not communicated to the clinicians who took care of the patients before the end of the study.
(ii) DNA extraction from blood plasma or whole blood.
DNA was extracted from 200 μl of total blood or blood plasma with the QIAamp DNA blood minikit as recommended in the protocol supplied by the manufacturer (Qiagen, Courtaboeuf, France). DNA was eluted from the columns with 50 μl of provided elution buffer. A 10-μl volume of extracted DNA was used for PCR.
(iii) Real-time PCR probe and primers.
We chose the primers and probe in the UL 123 exon 4 gene because nucleotide sequences of this region from different strains had been published in GenBank (1, 8). These sequences were aligned with Clustal software to choose an interstrain-conserved region of this gene for primers and probe, which were designed with the help of Primer Express 1.0 software (Applera, Courtaboeuf, France). The sequences of the forward and reverse primers were 5′-2791AGCGCC GCA TTG AGG A2806-3′ and 5′-2833CAG ACT CTC AGA GGA TCG GCC2853-3′, respectively. The sequence of the probe was 5′-2808ATC TGC ATG AAG GTC TTT GCC CAG TAC ATT2827-3′. The TaqMan probe was labeled at the 5′ end with 6-carboxyfluorescin and at the 3′ end with 6-carboxytetramethylrhodamine (Applera). The PCR amplification was performed in a total volume of 25 μl in the presence of TaqMan Universal PCR master mix 2X (Applera), with each primer present at 300 nM and the probe present at 200 nM.
PCR was performed with the ABI prism 7700 instrument (Applera) under the following conditions: 2 min at 50°C, 10 min at 95°C, and 45 cycles of 95°C for 15 s and 60°C for 1 min.
A plasmid containing the amplified sequence of AD 169 strain was constructed with the pADV TA cloning kit (Clontech, Saint Quentin en Yvelines, France). This plasmid was used as an external standard. For each batch, serial plasmid dilutions (range, 105 to 10 copies) were amplified; this allowed the construction of a standard curve and the quantification of CMV in clinical samples. The results were reported as the number of CMV genome copies/ml of whole blood or plasma.
The detection rate of the CMV PCR was 100% when the copy number was ≥10 copies per well, and therefore the threshold value was estimated at 250 copies/ml of plasma or whole blood. The specificity of the CMV PCR was assessed by testing strains of others herpesviruses (herpes simplex virus types 1 and 2 and varicella-zoster virus) in cell culture and plasma of patients with known Epstein-Barr virus primary or chronic infection. The CMV PCR assay was negative for all those samples.
Definition of CMV infection and CMV disease.
CMV reactivation or primary infection was defined as the presence of at least one antigen-positive cell per 100,000 cells. Symptomatic CMV infection was described as the association of pyrexia (≥37.5°C) with either thrombocytopenia, neutropenia (less than 0.5 × 109 neutrophils/liter) or elevated liver aminotransferase levels immediately preceding or coinciding with the diagnosis of CMV infection. CMV disease was diagnosed by the association of clinical symptoms and histologic or virologic confirmation of CMV infection of the organ. Definitions of organs involvement were as follows: (i) pneumonia, as evidenced by clinical symptoms of pneumonia (radiographic and/or hypoxemia plus CMV in BAL fluid or lung biopsy specimen); (ii) gastrointestinal disease, as evidenced by gastrointestinal symptoms plus histopathological evidence of CMV from gut biopsy specimens; and (iii) hepatitis, as evidenced by a combination of abnormal liver aminotransferase levels and histopathological changes consistent with viral hepatitis together with CMV in liver biopsy specimens by culture or PCR.
Preemptive therapy for prevention of CMV disease.
The decision to use preemptive therapy was based on a positive antigenemia test (≥1 antigen-positive cell/100,000 cells) or diagnosis of CMV disease- or CMV infection-related symptoms. In this study, PCR results were not used to make decisions about preemptive therapy. Preemptive treatment involved intravenous infusion of either ganciclovir at 10 mg/kg/day or foscavir at 180 mg/kg/day for 2 to 3 weeks followed by either ganciclovir at 5 mg/kg/day or foscavir at 90/mg/kg/day for 2 to 3 weeks.
Statistical analysis.
Nonparametric Spearman correlation coefficients were used to assess the association between continuous variables (number of antigen-positive cells and level of CMV DNA in whole blood and in plasma). When analyzing CMV DNA loads as a continuous variable, values below the limit of detection were replaced by half the threshold value.
The Wilcoxon test was used for matching samples to compare value over time (days of first positive antigenemia and first positive PCR as well as days of first negative antigenemia and negative PCR after treatment).
A P value of <0.05 was accepted as statistically significant.
RESULTS
Patients.
Fifty consecutive allogeneic bone marrow transplants recipients were enrolled. Twenty-three were adults requiring bone marrow transplantation for the following underlying diseases: 9 for acute myeloblastic leukemia, 4 for chronic myelogenous leukemia, 3 for acute lymphoblastic leukemia, 2 for non-Hodgkin's lymphoma, 2 for multiple myeloma, 1 for chronic lymphocytic leukemia, 1 for primary amyloidosis, and 1 for severe aplastic anemia. Twenty-seven of the patients were children. One child required bone marrow transplantation for acute lymphoblastic leukemia. The other 26 children had congenital diseases: 7 with combined immunodeficiency syndrome (SCID), 4 with Wiskott-Aldrich syndrome, 3 with Hurler syndrome, 2 with congenital aplasia, 2 with CD40 ligand deficiency, 2 with familial lymphohistiocytosis, 1 with FAS ligand deficiency, 1 with gamma interferon deficiency, 1 with major histocompatibility complex class II deficiency, 1 with DiGeorge syndrome, 1 with osteopetrosis, and 1 with chronic granulomatous disease.
The serostatus for CMV of donors and recipients before transplantation was as follows: donor positive/recipient positive in 18 cases, donor negative/recipient negative in 13 cases, donor positive/recipient negative in 8 cases, and donor negative/recipient positive in 11 cases. The 50 enrolled patients were followed up for a median of 107 days (range, 37 to 328), and a median of 11.5 (range, 5 to 22) samples per patient were analyzed.
Comparison of CMV real-time PCR results obtained from blood plasma and whole blood.
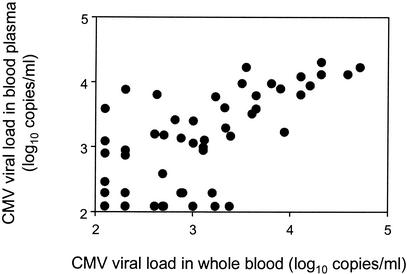
A total of 202 samples were tested simultaneously in blood plasma and in whole blood; 140 samples were positive in blood plasma, while 145 were positive in whole blood. The correlation between CMV loads obtained in these two types of samples were highly significant (r = 0.873 and P < 0.0001 by the Spearman test) (Fig. 1). On the basis of these results obtained with the first 202 samples, we decided to use blood plasma as PCR templates for the rest of the study.
FIG. 1.
Correlation between CMV DNA loads in blood plasma and CMV DNA loads in whole blood for 202 samples (Spearman test: r = 0.873, P < 0.0001). CMV DNA loads below the limit of detection were replaced by half the threshold value (2.09 log10 copies/ml).
Comparison of the pp65 antigenemia assay with the CMV real-time PCR in blood plasma.
Real-time PCR in plasma and the pp65 antigenemia assay were assessed on 558 samples (Table 1). A total of 371 samples were negative for both assays and 69 were positive in the two tests. The sensitivity of the CMV antigenemia assay was much lower: 108 samples were CMV PCR positive and CMV antigenemia negative. Ten samples were negative in the CMV PCR test but tested positive with the CMV antigenemia assay. For these 10 samples, only one positive cell/100,000 cells was reported. These iterative low positive antigenemias associated with a negative CMV PCR were obtained from four different patients during their follow-up after discontinuation of ganciclovir or foscavir treatment for CMV infection.
TABLE 1.
Results of CMV pp65 antigenemia and real-time CMV PCR of blood plasma for 558 blood samples
| CMV PCR in blood plasma | No. of samples with following CMV pp65 antigenemia result:
|
Total no. of samples | |
|---|---|---|---|
| Positive | Negative | ||
| Positive | 69 | 108 | 177 |
| Negative | 10 | 371 | 381 |
| Total | 79 | 479 | 558 |
The results obtained for the two diagnostic methods with the 558 samples were significantly correlated (R = 0.732, P < 0.0001 by the Spearman test). In the 177 PCR-positive samples, the number of CMV DNA copies was correlated with the number of pp65-positive cells (r = 0.609, P < 0.0001 by the Spearman test), as shown in Fig. 2. CMV PCR-positive samples were classified into three groups according to the results of pp65 antigenemia (Fig. 3a). By definition, samples were negative for pp65 antigenemia in group 1 (n = 108), positive but with ≤10 positive cells in group 2 (n = 45), and positive with >10 positive cells in group 3 (n = 24). The median CMV DNA level in samples from group 1 was 700 (2.9 log10) copies/ml (range, 250 to 53,900). The median CMV DNA level in group 2 was 3,000 (3.5 log10) copies/ml (range, 250 to 130,000) and was significantly higher than in group 1 (P = 0.007 by the Wilcoxon test). In group 3, the CMV DNA median was 17,500 (4.2 log10) copies/ml (range, 2,000 to 335,000) and was therefore significantly higher than in group 2 (P = 0.0009 by the Wilcoxon test).
FIG. 2.
Correlation between CMV DNA loads in blood plasma and CMV antigen-positive cells in the 177 PCR-positive samples (Spearman test: r = 0.609, P < 0.0001).
FIG. 3.
(a) CMV DNA loads in plasma based on the number of antigen-positive cells. PCR-positive samples were classified into three groups according to the results of the pp65 antigenemia assay. Samples in group 1 (n = 108) were negative for antigenemia, samples in group 2 (n = 45) were positive with ≤10 antigen-positive cells, and samples in group 3 (n = 24) were positive with >10 antigen-positive cells. Median CMV DNA loads were significantly higher in group 2 than in group 1 (Wilcoxon test: P = 0.007) and in group 3 than in goup 2 (Wilcoxon test: P = 0.0009). (b) Comparison of peak CMV DNA viral loads in patients with CMV infection-related symptoms (n = 13) and in asymptomatic patients (n = 9). The median peak CMV DNA load was significantly higher in the symptomatic group than in the asymptomatic group (Wilcoxon test: P = 0.0109). Horizontal bars indicate the medians.
Comparison of the day of first positive CMV PCR and the day of first positive CMV antigenemia.
Twenty patients developed a positive CMV PCR result, and altogether there were 24 episodes of CMV reactivation detected by PCR. In 17 of these episodes, both the CMV PCR and CMV antigenemia assays were positive. For these 17 episodes, the CMV PCR test was positive before the antigenemia assay in 15 cases and on the same day in 2 cases. CMV antigenemia was positive before CMV PCR in none of these episodes. When both CMV antigenemia and CMV PCR assays were positive, the median day for the first positive PCR result and the first positive antigenemia result after transplantation were 28 days (range, 0 to 112) and 45 days (range, 0 to 119), respectively. The CMV PCR test was therefore positive significantly earlier than the CMV antigenemia test (P = 0.001 by the Wilcoxon test). The first positive CMV PCR result preceded the first positive antigenemia result by a median of 8 days (range, 0 to 46).
Comparison of treatment follow-up for CMV PCR and the pp65 antigenemia assay.
Treatment induced a rapid decrease of both the number of viral copies in plasma and of the number of CMV antigen-positive cells. All but one of the treated patients achieved a negative CMV antigenemia result earlier than a negative CMV PCR result. The median time interval necessary to obtain a negative antigenemia result and a negative PCR result was 16 days (range, 3 to 37) and 28 days (range, 7 to 80), respectively. The first negative antigenemia result preceded the first negative PCR result by a median of 10 days (range, 0 to 50). CMV antigenemia was therefore negative significantly earlier than was CMV PCR (P = 0.005 by the Wilcoxon test).
Comparison of plasma CMV viral loads in patients with symptoms of CMV infection and in asymptomatic patients.
Twenty patients developed a positive CMV PCR result in plasma, with a total of 24 episodes of positive CMV PCR. Thirteen episodes of CMV replication were associated with CMV-related symptoms, nine patients were asymptomatic, and for one patient (patient 12) it was impossible to determine if the clinical symptoms observed were linked to CMV infection since this patient concomitantly had a severe lymphoproliferative disease. The peak CMV viral load in plasma was significantly higher in the symptomatic group, with a median of 15,000 (4.2 log10) copies/ml (range, 1,000 to 450,000) than in the asymptomatic group, with a median CMV load of 3,000 (3,5 log10) copies/ml (range, 250 to 12,600) (P = 0.0109 by the Wilcoxon test) (Fig. 3b).
Incidence of CMV replication, CMV infection, and CMV disease in the adult population.
Thirteen adults (2 donor positive/recipient positive [D+/R+], 1 D+/R−, 1 D−/R+, 9 D−/R−) did not develop CMV reactivation or infection: all of their samples tested negative for both CMV PCR and antigenemia tests. A positive CMV PCR result was observed in 10 (43%) of the 23 adult patients (Table 2). For these 10 patients, it was a reactivation of CMV replication. A positive CMV PCR result was followed by a positive antigenemia result for 8 patients. Two patients (patients 9 and 10) developed a positive PCR result with a negative antigenemia result a few days before their death, and there was no follow up. Among the 10 patients with a positive CMV PCR result, 5 (50%) had CMV infection-related symptoms but none developed CMV disease. Of the 10 patients with a positive CMV PCR result, 8 had been given ganciclovir or foscavir preemptive therapy because of a positive antigenemia and 2 were not treated because they died the day after the first positive test.
TABLE 2.
Characteristics of the 20 patients who developed a positive CMV PCR result
| Patient no. | Age (yr) | Sexa | CMV serostatus (D/R) | Day of first positive PCR result | Day of first positive antigenemia resulta | Peak CMV level (copies/ml) by:
|
CMV infection symptomsa | CMV disease | Treatment drug/ first daya | Outcome | Cause of deatha | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PCR | Antigene- mia assaya | |||||||||||
| Adults | ||||||||||||
| 1 | 33 | F | −/+ | 28 | 46 | 21,000 | 88 | Yes | No | GCV/D56 | Alive | |
| 2 | 40 | F | −/+ | −1 | 45 | 15,000 | 43 | Yes | No | FOS/D45 | Alive | |
| 3 | 28 | M | +/+ | 28 | 57 | 1,400 | 6 | Yes | No | GCV/D57 | Alive | |
| 4 | 51 | F | −/+ | 25 | 42 | 12,000 | 5 | Yes | No | GCV/D42 | Dead (D87) | Fungus |
| 5 | 31 | F | +/+ | 42 | 42 | 105,000 | 100 | Yes | No | GCV/D42 | Dead (D142) | GVH |
| 6 | 39 | M | −/+ | 34 | 44 | 300 | 2 | No | No | No | Dead (D45) | Relapse |
| 7 | 38 | F | −/+ | 44 | 51 | 2,000 | 2 | No | No | GCV/D51 | Alive | |
| 8 | 50 | M | −/+ | 41 | 49 | 4,200 | 5 | No | No | GCV/D49 | Alive | |
| 9 | 56 | M | +/+ | 38 | NF | 250 | NF | No | No | No | Dead (D39) | Relapse |
| 10 | 34 | M | −/+ | 79 | NF | 250 | NF | No | No | No | Dead (D80) | Sepsis |
| Children | ||||||||||||
| 11 | 2 | F | −/+ | 13 | 30 | 8,000 | 10 | No | No | GCV + FOS/D35 | ||
| 69 | 76 | 9,000 | 12 | No | No | GCV/D76 | Alive | |||||
| 12 | 4 | F | +/+ | −8 | −8 | 450,000 | 50 | Yes | Hepatitis | GCV + FOS/D-7 | ||
| 57 | 64 | 4,000 | 26 | Int | No | GCV + FOS/D64 | ||||||
| 97 | 104 | 3,000 | 9 | Int | No | GCV + FOS/D104 | Dead (D125) | PTLD | ||||
| 13 | 3 | F | +/+ | 30 | Neg | 1,700 | Neg | Yes | No | FOS + GCV/D30 | ||
| 112 | 119 | 24,000 | 17 | Yes | No | FOS/D119 | Dead (D145) | Pneumonia | ||||
| 14 | 8 | M | +/+ | −11 | −1 | 125,000 | 12 | Yes | No | FOS/D0 + GCV/D24 | Alive | |
| 15 | 0.3 | M | +/+ | −10 | −3 | 130,000 | 2 | Yes | No | GCV/D8 | Alive | |
| 16 | 2 | M | +/+ | −10 | 30 | 12,600 | 3 | No | No | FOS/D15 | Alive | |
| 17 | 1 | F | +/+ | 35 | Neg | 2,400 | Neg | Yes | No | GCV/D30 | Alive | |
| 18 | 0.5 | M | +/+ | 84 | Neg | 3,000 | Neg | No | No | No | Alive | |
| 19 | 0.9 | M | +/+ | 0 | Neg | 4,300 | Neg | Yes | Colitis | CID/D-40 | Alive | |
| 20 | 1 | F | +/− | 31 | Neg | 1,000 | Neg | Yes | Pneumonia | GCV/D30 | Dead (D42) | CMV pneumonia |
F, female; M, male; GCV, ganciclovir; FOS, foscavir; CID, cidofovir; PTLD, posttransplantation lymphoproliferative disease; NF, no follow-up; Neg, negative; Int, indeterminate.
Incidence of CMV replication, CMV infection, and CMV disease in the pediatric population.
Of the 27 children, 17 (5 D+/R+, 6 D+/R−, 2 D−/R+, 4 D−/R−) did not develop CMV replication: antigenemia and CMV PCR were both negative during the follow-up of these 17 children.
Ten children (37%) developed a positive CMV PCR result with a total of 14 positive CMV PCR episodes (Table 2). Of these 10 children, 9 (patients 11 to 19) had been treated for severe symptomatic CMV infection in the pretransplantation period. The last one (patient 20) had a primary CMV infection posttransplantation, linked to a seropositive donor. Two children (patients 11 and 13) had two episodes of CMV reactivation, and one (patient 12) had three episodes. CMV reactivation was symptomatic in eight episodes of replication, asymptomatic in four, and indeterminate in two. Three children developed CMV disease: one developed CMV pneumonia, one developed CMV enterocolitis, and one developed CMV hepatitis (Table 2).
A positive CMV PCR result was followed by a positive CMV antigenemia result in nine episodes. Four children had repeatable positive CMV PCR results in plasma with negative antigenemia tests. Of these four children, two developed CMV disease and one developed CMV infection-related symptoms. The first child (patient 17) developed fever and showed a rise of liver aminotransferase levels associated with the recovery of CMV in urine and nasopharyngeal aspirates. The second patient (patient 18) developed a positive blood plasma PCR result on day 84, associated with the recovery of CMV in the urine and nasopharyngeal aspirates without any related symptoms. The third child (patient 19) developed symptoms of gastrointestinal disease around day 110, with histological signs of viral infection and a positive CMV PCR result in gut biopsy specimens. The fourth child (patient 20) developed a very severe interstitial pneumonia on day 30, with a positive CMV PCR result for BAL fluid and CMV recovery by culture in blood.
In 13 of the 14 positive CMV PCR episodes, preemptive treatment was implemented either because of a positive antigenemia or because of CMV-related symptoms. Only one episode of positive CMV PCR was not treated, because antigenemia was negative and there were no associated symptoms. In this case, the positive CMV PCR result occurred late after transplantation (day 84), the CMV viral load was not high (3,000 copies/ml), and after 3 weeks the CMV PCR result became spontaneously negative and the child did not develop CMV disease.
DISCUSSION
This work aimed to evaluate the sensitivity of a real-time PCR assay of blood plasma to detect CMV replication in adult and pediatric bone marrow transplant recipients and its clinical relevance. This evaluation was done by comparing the CMV PCR assay of blood plasma with the CMV antigenemia assay used as a reference method.
In this study, the real-time CMV PCR assay of plasma was highly sensitive and the results of CMV DNA detection in whole blood and in blood plasma were highly correlated with identical sensitivity of this assay in the two types of blood samples. In previous studies, both in-house and commercial CMV PCR assays of blood plasma had demonstrated a lower sensitivity than the antigenemia assay or than PCR of blood leukocytes (3, 5, 19, 23) and comparison of CMV detection with a real-time PCR method in blood plasma, whole blood, and blood leukocytes showed a trend for a more sensitive CMV DNA detection in blood leukocytes and whole blood than in blood plasma (16). In a recent study, CMV DNA of plasma was found to be highly fragmented (6). In this study, because of this fragmentation, quantitative PCR targeting a small amplicon consistently gave higher quantitative results than did assays amplifying a larger amplicon. In our study, the PCR target (74 bp) was smaller than those used in other studies (3, 5, 19, 23). This could explain why the sensitivity of our PCR test was equivalent for plasma and whole blood.
We found a significant correlation between the number of CMV antigen-positive cells and the CMV DNA load in plasma, as already reported (22, 24). However, in this study there were samples which had negative or low antigenemia counts along with large numbers of CMV DNA copies in plasma. Others also reported such discrepancies when comparing the antigenemia assay results and viral loads obtained with the real-time PCR assay of blood leukocytes (9, 11). Quantitative CMV PCR performed on a blood sample evaluates the number of CMV genome copies present in this sample. This quantification of genome copies is a good reflection of the number of viral particles present in the blood (since one particle contains only one copy of the genome) and therefore is a good reflection of CMV replication in the blood compartment. Conversely, CMV antigenemia detects the tegument phosphoprotein (pp65) present in blood polynuclear cells; this protein is synthesized in excess during viral multiplication and accumulates in blood cells. Therefore, quantification of CMV antigen-positive cells roughly reflects the level of CMV multiplication, explaining its correct predictive value for CMV disease, but probably gives a less accurate reflection of CMV replication than does quantification of viral particles by PCR.
Real-time CMV PCR in blood plasma allowed an early diagnosis of CMV replication after transplantation, with a positive CMV PCR result occurring before a positive CMV antigenemia result by a median of 8 days. Griscelli et al. (11) also found that CMV replication in blood leukocytes was detectable by PCR a mean of 15 days prior to antigenemia. If the decision to use preemptive therapy on our population had been based on the first positive PCR result, most patients would have been treated between 1 and 2 weeks earlier. Furthermore, monitoring of CMV infection treatment showed that a negative CMV PCR result was achieved after a median of 28 days (10 days after a negative antigenemia result). Since real-time CMV PCR of plasma is very sensitive for detection of CMV replication, a negative PCR might be an indicator of a completely successful treatment. Therefore, discontinuation of therapy based on a negative PCR result is probably safe. By contrast, discontinuation based on the first negative antigenemia result has led to a significant number of CMV pneumonia (2). For this reason in our center, patients with CMV replication are empirically treated with intravenous ganciclovir at 10 mg/kg/day for 2 to 3 weeks and then at 5 mg/kg/day for 2 to 3 weeks; they therefore receive antiviral treatment for at least 5 weeks. Monitoring of treatment efficacy by CMV PCR might help to adapt the duration of treatment to each individual. Therefore, the use of real-time CMV PCR to guide preemptive treatment might have the advantages of reducing the occurrence of CMV disease, since patients would be treated earlier, and of allowing a shorter duration of treatment, with potential attenuation of antiviral side effects. Indeed, a shorter course of antiviral treatment might allow patients to overcome the increase in invasive fungal infection described in studies using either antigenemia-guided early treatment at low level of antigenemia or ganciclovir prophylaxis (2, 4).
In our center, the majority of pediatric bone marrow transplant recipients have congenital immunodeficiency syndromes. These children frequently either have severe CMV primary infection in the pretransplantation period or are at high risk for developing posttransplantation primary infection. They often experience two or more episodes of CMV replication and receive long courses of antiviral treatment. Monitoring CMV infection with a sensitive quantitative CMV PCR assay in this population appears to be particularly useful. For example, in our study, CMV PCR of blood plasma gave positive results for one child who developed early CMV pneumonia posttransplantation and for another child with CMV gastroenteritis, while CMV antigenemia assays were repeatedly negative. In both cases, the CMV viral load in plasma was not high, reflecting a low level of CMV replication in the blood compartment, in contrast to the a strongly positive detection in the organs involved. These results confirm the lack of sensitivity of the antigenemia assay and the advantage of CMV PCR in cases of early CMV disease or CMV infection of the gut (3, 14, 17).
We were concerned that a policy of real-time plasma PCR-guided preemptive therapy would increase the total number of bone marrow recipients treated for CMV infection, since this technique is more sensitive than the antigenemia assay. However, in our study, all early positive CMV PCR results (<80 days) had been associated with a positive antigenemia and/or CMV related infection or disease, and we therefore think that an initial positive CMV PCR result with in blood plasma within the first 80 days of transplantation should lead to implementation of preemptive treatment. Alternatively, an initial positive CMV PCR result occurring in the late posttransplantation period, when bone marrow recipients have recovered part of their immune defenses, should be carefully interpreted: either the CMV viral load in plasma is high and the preemptive treatment should not be delayed or the viral load is low and only a further increase of CMV load should lead to preemptive treatment.
In conclusion, CMV real-time PCR with blood plasma demonstrated a better sensitivity than the CMV antigenemia assay and also showed good clinical relevance. Moreover, this CMV PCR test has the advantage of being easily performed with a small amount of blood plasma and with stored specimens, while the CMV antigenemia test requires larger samples and immediate processing. We have implemented routine CMV real-time PCR of plasma to monitor CMV infection in bone marrow transplant recipients in our center. All patients are monitored weekly by a CMV PCR test, and preemptive treatment is started at the first positive PCR result when it occurs within the first 80 days posttransplantation. After 80 days posttransplantation, preemptive treatment is implemented only if the CMV viral load is high or increasing. After a minimum of 14 days, preemptive treatment is discontinued on obtaining a negative CMV PCR. The impact of this new protocol on the occurrence of early and late CMV disease, early invasive fungal infection, and mortality will be evaluated in our clinical settings.
REFERENCES
- 1.Akrigg, A., G. W. Wilkinson, and J. D. Oram. 1985. The structure of the major immediate early gene of human cytomegalovirus strain AD169. Virus Res. 2:107-121. [DOI] [PubMed] [Google Scholar]
- 2.Boeckh, M., T. A. Gooley, D. Myerson, T. Cunningham, G. Schoch, and R. A. Bowden. 1996. Cytomegalovirus pp65 antigenemia-guided early treatment with ganciclovir versus ganciclovir at engraftment after allogeneic marrow transplantation: a randomized double-blind study. Blood 88:4063-4071. [PubMed] [Google Scholar]
- 3.Boeckh, M., G. M. Gallez-Hawkins, D. Myerson, J. A. Zaia, and R. A. Bowden. 1997. Plasma polymerase chain reaction for cytomegalovirus DNA after allogeneic marrow transplantation. Transplantation 64:108-113. [DOI] [PubMed] [Google Scholar]
- 4.Boeckh, M., R. A. Bowden, T. Gooley, D. Myerson, and L. Corey. 1999. Successful modification of a pp65 antigenemia-based early treatment strategy for prevention of cytomégalovirus disease in allogeneic marrow transplant recipients. Blood 93:1781-1782. [PubMed] [Google Scholar]
- 5.Boivin, G., R. Belanger, R. Delage, C. Beliveau, C. Demers, N. Goyette, and J. Roy. 2000. Quantitative analysis of cytomegalovirus (CMV) viremia using the pp65 antigenemia assay and the COBAS AMPLICOR CMV MONITOR PCR test after blood and marrow allogeneic transplantation. J. Clin. Microbiol. 38:4356-4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boom, R., C. J. A. Sol, T. Schuurman, A. van Breda, J. F. L. Weel, M. Beld I. J. M. ten Berge, P. M. E. Wertheim-van Dillen, and M. D. de Jong. 2002. Human cytomegalovirus DNA in plasma and serum specimens of renal transplant recipients is highly fragmented. J. Clin. Microbiol. 40:4105-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caliendo, A. M., K. St George, S. Y. Kao, J. Allega, B. H. Tan, R. LaFontaine, L. Bui, and C. R. Rinaldo. 2000. Comparison of quantitative cytomegalovirus (CMV) PCR in plasma and CMV antigenemia assay: clinical utility of the prototype AMPLICOR CMV MONITOR test in transplant recipients. J. Clin. Microbiol. 38:2122-2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chou, S. 1992. Effect of interstrain variation on diagnostic DNA amplification of the cytomegalovirus major immediate-early gene region. J. Clin. Microbiol. 30:2307-2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gault, E., Y. Michel, A. Dehée, C. Belabani, J. C. Nicolas, and A. Garbarg-Chenon. 2001. Quantification of human cytomegalovirus DNA by real-time PCR. J. Clin. Microbiol. 39:772-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gor, D., C. Sabin, H. G. Prentice, N. Vyas, S. Man, P. D. Griffiths, and V. C. Emery. 1998. Longitudinal fluctuations in cytomegalovirus load in bone marrow transplant patients: relationship between peak load, donor/recipient serostatus, acute GVHD and CMV disease. Bone Marrow Transplant. 21:597-605. [DOI] [PubMed] [Google Scholar]
- 11.Griscelli, F., M. Barrois, S. Chauvin, S. Lastere, D. Bellet, and J. H. Bourhis. 2001. Quantification of human cytomegalovirus DNA in bone marrow transplant recipients by real-time PCR. J. Clin. Microbiol. 39:4362-4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heid, C. A., J. Stevens, K. J. Livak and P. M. Williams. 1996. Real-time quantitative PCR. Genome Res. 6:986-994. [DOI] [PubMed] [Google Scholar]
- 13.Legendre, C., C. Rouzioux, D. Ducloux, C. Delamarre, N. Chkoff, E. Thervet, B. Page, and H. Kreis. 1994. Strategy of cytomegalovirus infection prevention in renal transplantation. Adv. Nephrol. Necker Hosp. 23:331-345. [PubMed] [Google Scholar]
- 14.Limaye, A. P., R. A. Bowden, D. Myerson, and M. Boeckh. 1997. Cytomegalovirus disease occuring before engraftment in marrow transplant recipients. Clin. Infect. Dis. 24:830-835. [DOI] [PubMed] [Google Scholar]
- 15.Limaye, A. P., M. L. Huang, W. Leisenring, L. Stensland, L. Corey, and M. Boeckh. 2001. Cytomegalovirus (CMV) DNA load in plasma for the diagnosis of CMV disease before engraftment in hematopoietic stem-cell transplant recipients. J. Infect. Dis. 183:377-382. [DOI] [PubMed] [Google Scholar]
- 16.Machida, U., M. Kami, T. Fukui, Y. Kazuyama, M. Kinoshita, Y. Tanaka, Y. Kanda, S. Ogawa, H. Honda, S. Chiba, K. Mitani, Y. Muto, K. Osumi, S. Kimura, and H. Hirai. 2000. Real-time automated PCR for early diagnosis and monitoring of cytomegalovirus infection after bone marrow transplantation. J. Clin. Microbiol. 38:2536-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mori, T., S. Okamoto, S. Matsuoka, T. Yajima, M. Wakui, R. Watanabe, A. Ishida, Y. Iwao, M. Mukai, T. Hibi, and Y. Ikeda. 2000. Risk-adapted pre-emptive therapy for cytomegalovirus disease in patients undergoing allogeneic bone marrow transplantation. Bone Marrow Transplant. 25:765-769. [DOI] [PubMed] [Google Scholar]
- 18.Nitsche, A., N. Steuer, C. A. Schmidt, O. Landt, H. Ellerbrok, G. Pauli, and W. Siegert. 2000. Detection of human cytomegalovirus DNA by real-time quantitative PCR. J. Clin. Microbiol. 38:2734-2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piiparinen, H., K. Hockerstedt, C. Gronhagen-Riska, M. Lappalainen, J. Suni, and I. Lautenschlager. 2001. Comparison of plasma polymerase chain reaction and pp65-antigenemia assay in the quantification of cytomegalovirus in liver and kidney transplant patients. J. Clin. Virol. 22:111-116. [DOI] [PubMed] [Google Scholar]
- 20.Razonable, R. R., R. A. Brown, M. J. Espy, A. Rivero, W. Kremers, J. Wilson, C. Groettum, T. F. Smith, and C. V. Paya. 2001. Comparative quantification of cytomegalovirus (CMV) DNA in solid-organ transplant recipients with CMV infection by using two high-throughput automated systems. J. Clin. Microbiol. 39:4472-4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schaade, L., P. Kockelkorn, K. Ritter, and M. Kleines. 2000. Detection of cytomegalovirus DNA in human specimens by lightcycler PCR. J. Clin. Microbiol. 38:4006-4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanaka, N., H. Kimura, K. Iida, Y. Saito, I. Tsuge, A. Yoshimi, T. Matsuyama, and T. Morishima. 2000. Quantitative analysis of cytomegalovirus load using a real-time PCR assay. J. Med. Virol. 60:455-462. [DOI] [PubMed] [Google Scholar]
- 23.Woo, P. C., S. K. Lo, K. Y. Yuen, J. S. Peiris, H. Siau, E. K. Chiu, R. H. Liang, and T. K. Chan. 1997. Detection of CMV DNA in bone marrow transplant recipients: plasma versus leucocyte polymerase chain reaction. J. Clin. Pathol. 50:231-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yakushiji, K., H. Gondo, K. Kamezaki, K. Shigematsu, S. Hayashi, M. Kuroiwa, S. Taniguchi, Y. Ohno, K. Takase, A. Numata, K. Aoki, K. Kato, K. Nagafuji, K. Shimoda, T. Okamura, N. Kinukawa, N. Kasuga, M. Sata, and H. Harada. 2002. Monitoring of cytomegalovirus reactivation after allogeneic stem cell transplantation: comparison of an antigenemia assay and quantitative real-time polymerase chain reaction. Bone Marrow Transplant. 29:599-606. [DOI] [PubMed] [Google Scholar]
- 25.Yun, Z., I. Lewensohn-Fuchs, P. Ljungman, and A. Vahlne. 2000. Real-time monitoring of cytomegalovirus infections after stem cell transplantation using the TaqMan polymerase chain reaction assays. Transplantation 69:1733-1736. [DOI] [PubMed] [Google Scholar]