Abstract
We have analyzed the variability of minisatellite sequences (also called variable-number tandem repeats [VNTRs]) in the genome of Legionella pneumophila. Based upon the genome sequence of the Philadelphia-1 strain (serogroup 1), 25 minisatellites were selected and their polymorphisms were analyzed by PCR with the DNA of serogroup 1 to 14 reference strains. For 22 markers, a PCR product of the expected size was found with the DNA of the Philadelphia-1 strain. Most of these markers did not amplify the DNA of other Legionella species or other bacteria used as controls. A polymorphism was observed for seven markers among the L. pneumophila strains tested. To check whether these markers could be used to compare strains of L. pneumophila, we analyzed two groups of isolates from clinical and environmental samples which had been independently genotyped by other methods. The results showed that, for the isolates in these two sets of samples, VNTR typing is as informative as pulsed-field gel electrophoresis for comparison of strains. Sequencing of one minisatellite from 14 reference strains was performed. Comparison of the sequences allowed a classification and confirmed the existence of subspecies of L. pneumophila. We also tested the usefulness of one very polymorphic marker as a tool for the rapid screening of colonies grown from water samples. This allowed the rapid identification of the L. pneumophila colonies and gave a first hint as to the presence of several strains in a single sample.
Legionella pneumophila is the agent of Legionnaires' disease and Pontiac fever. This bacterium is present in aquatic environments, where it replicates within protozoan hosts (for reviews, see references 6 and 17). The bacteria infect alveolar macrophages following inhalation of contaminated aerosols. The organism is responsible for a large number of cases of nosocomial pneumonia in immunocompromised patients. The family Legionellaceae contains more than 40 species that have been isolated from either clinical or environmental sources. Three subspecies of the species L. pneumophila have been described to be the most frequently associated with disease: L. pneumophila subsp. pneumophila, L. pneumophila subsp. fraseri, and L. pneumophila subsp. pascullei (2). Up to 15 serogroups of L. pneumophila can be identified, and these are mostly in the subspecies L. pneumophila subsp. pneumophila and L. pneumophila subsp. fraseri. Different molecular techniques have been developed to characterize and analyze the different strains. Macrorestriction analysis and PCR-based methods such as amplified fragment length polymorphism (AFLP) analysis (19) and arbitrarily primed PCR (8) have been used to genotype L. pneumophila isolates. A comparative evaluation of these methods has been performed and showed that AFLP analysis was the most simple and reproducible (10). Sequencing of the rpoB and dotA genes (11) or the dotA and mip genes (3) was used to study the genetic population structures of L. pneumophila isolates. These studies confirmed the differences between the three subspecies and suggested that although a clonal population exists, horizontal transfer plays an important role in the evolution of L. pneumophila.
The use of variable-number tandem repeats (VNTRs) to genotype bacteria has been described for Haemophilus influenzae (20), Mycobacterium tuberculosis (16), and more recently, Bacillus anthracis and Yersinia pestis (12) and is known as multiple-locus VNTR analysis (MLVA). The availability of genome sequences and of appropriate algorithms for data analysis allows the search for new markers of this type.
We have performed a study with reference serogroup 1 to 14 strains and 27 strains isolated from the environment and patients, and we have selected several informative markers useful for epidemiological studies.
MATERIALS AND METHODS
Strains and growth conditions.
Fourteen reference strains of L. pneumophila (Table 1) and four strains of other Legionella species (Legionella dumoffii NY, Legionella micdadei TATLOCK, Legionella bozemanii WIGA, Legionella longbeachae 1) were obtained from the American Type Culture Collection (ATCC). The environment and clinical isolates are listed in Table 2. Some are described elsewhere (18).
TABLE 1.
L. pneumophila reference strains used in this study
| Strain | Serogroup |
|---|---|
| ATCC 33152 (Philadelphia-1) | 1 |
| ATCC 33154 (Togus 1) | 2 |
| ATCC 33155 (Bloomington-2) | 3 |
| ATCC 33156 (Los Angeles 1) | 4 |
| ATCC 33216 (Dallas 1E) | 5 |
| ATCC 33215 (Chicago 2) | 6 |
| ATCC 33823 (Chicago 8) | 7 |
| ATCC 35096 (Concord 3) | 8 |
| ATCC 35289 (International 23) | 9 |
| ATCC 43283 (Leiden 1) | 10 |
| ATCC 43130 (797-PA-H) | 11 |
| ATCC 43290 (570-CO-H) | 12 |
| ATCC 43736 (82A3105) | 13 |
| ATCC 43703 (1169-MN-H) | 14 |
TABLE 2.
L. pneumophila strains isolated from patients and water
| Source of isolate | Strain designation | Serogroup | Molecular typing methoda |
|---|---|---|---|
| Paris seriesb | |||
| Water, hospital N | NW1 | 3 | RFLP |
| NW2 | 3 | RFLP | |
| NW3 | 3 | RFLP | |
| NW4 | 3 | RFLP | |
| Patients, hospital N | NP1 | 3 | RFLP |
| NP2 | 3 | RFLP | |
| NP3 | 3 | RFLP | |
| NP4 | 3 | RFLP | |
| Patients, hospital P | PP1 | 3 | RFLP |
| PP2 | 3 | RFLP | |
| PP3 | 3 | RFLP | |
| PP4 | 3 | RFLP | |
| PP5 | 3 | RFLP | |
| PP6 | 3 | RFLP | |
| Greek series | |||
| Water hotel rooms | A1 | 1 | PFGE |
| A2 | 6 | PFGE | |
| B1 | 1 | PFGE | |
| B2 | 1 | PFGE | |
| H1 | 1 | PFGE | |
| H2 | 6 | PFGE | |
| Patient | P1 | 6 | PFGE |
| P2 | 6 | PFGE | |
| Control | ATCC 33152 | 1 | PFGE |
| ATCC 33215 | 6 | PFGE |
Other than VNTR typing.
Described by Tram et al. (18).
Isolation of Legionella from water samples was performed according to the recommendations of Association Française de Normalisation (report no. AFNOR-NFT 90-431). Samples of 500 ml of water were filtered on a 0.2-μm-pore-diameter polycarbonate membrane (Nuclepore, Corning, New York). After filtration, the bacteria on the filters were resuspended in 10 ml of distilled water and shaken. Then, 0.1 ml of the suspension was spread on a 90-mm petri dish containing solid buffered charcoal-yeast extract medium (BCYE; bioMérieux Sa, Marcy l'Etoile, France) supplemented with l-cysteine and ferric pyrophosphate, with or without antibiotics (Oxoid, Biolyon, Dardily, France). When observed under a microscope 3 days after primary culture, the characteristic Legionella colonies were round, convex, white with pink or green iridescent edges, and glistening with a ground-glass appearance. Up to 15 colonies were picked from each plate and subcultured onto BCYE and sheep blood agar. Organisms growing on BCYE but not on sheep blood agar were tested by direct immunofluorescence with antibodies against L. pneumophila antigens (MONOFLUO; Sanofi Diagnostics Pasteur, Marnes la Coquette, France) (4, 13). For direct immunofluorescence a colony was dispersed in sterile water and a drop of suspension (containing no more than 100 bacteria) was put on a glass slide, air dried for 10 to 15 min, and treated with acetone for 10 min. Then, the fluorescent monoclonal antibody was added and the slide was held for 30 min at 37°C in a humidifying chamber. Extensive washes with phosphate-buffered saline (PBS) were performed before the glass slide was mounted with 10% glycerol in PBS. The positive colonies were subcultured on solid medium, and pooled colonies were used for further studies.
Some primary colonies were also tested by PCR with minisatellite markers (this study).
The isolates from patients and water samples from Necker and Pitié Hospitals in Paris, France (Table 2), have been described by Tram et al. (18). Each clinical strain was from a single patient.
The L. pneumophila isolates from a Dutch patient and from a hotel in Greece (Heraklion, Crete) were obtained in 1989 from a lung specimen and from the hotel water, respectively (unpublished work) (Table 2). Hot water was collected from the showers of six different rooms (which had been unused for 24 h), including that of the Dutch patient, and bacteria were isolated as described above. About 600 colonies/liter had the phenotypic characteristics of Legionella, and 10 colonies from each water sample were cultured and further analyzed. Pulsed-field gel electrophoresis (PFGE) analysis was performed with one isolate from each room and two isolates from the patient.
Thermolysates and DNA extraction.
For thermolysate preparation, a large primary colony was picked and resuspended in 100 μl of distilled water, incubated for 10 min at 100°C, and then quickly chilled on ice.
For DNA purification, 10 to 20 colonies of a secondary culture were pooled, and the pool was resuspended in water. One volume of lysis buffer containing 20 mM Tris (pH 8), 2 mM EDTA, 20 mM NaCl, and 1% sodium dodecyl sulfate was added along with 100 μg of proteinase K per ml, and the mixture was incubated at 56°C for 2 h. DNA was extracted with phenol and phenol-chloroform (1/1) by using a Phase Lock gel (Eppendorf) and precipitated with 200 mM NaCl-70% ethanol.
For PFGE the DNA was prepared, digested with SfiI (Pharmacia), and analyzed as described previously (14). A bacteriophage lambda DNA ladder (FMC BioProducts, Vallensbaek Stand, Denmark) was used as a size marker.
PCR.
Thirty-five cycles of amplification were performed in a 96-well MJ Research PTC200 thermocycler. Annealing was for 30 s at 60°C, and elongation was for 45 s at 72°C. The PCR products were analyzed on 3% agarose gels by using the Gene Ruler 100-bp DNA ladder plus (MBI Fermentas) as a size marker. The oligonucleotide sequences are shown in Table 3.
TABLE 3.
Oligonucleotides
| Oligonucleotide namea | Directionb | Sequence (5′ → 3′) |
|---|---|---|
| Lpms1 | F | CAGGGAAATGCTCTAGCACAC |
| R | TCGCTTCGGACTGAATTTCT | |
| Lpms4 | F | CCAATTCCAGTTCGAGTTCC |
| R | GACCACTCCCGCTGTTAGAA | |
| Lpms4b | F | AGTCCGAGTTCGAGCAGTACTA |
| R | GGCGTACTTGTATTTGTATTCG | |
| Lpms5 | F | CCATGCATTGGCTCAAAAA |
| R | TTGATTTGGATTAATATTCGTTGG | |
| Lpms11 | F | TGTGAAAAAGCACCACAACC |
| R | ACCCAGAAACAGGAAGCAGA | |
| Lpms13 | F | CAATAGCATCGGACTGAGCA |
| R | TGCCTGTGTATCTGGAAAAGC | |
| Lpms17 | F | CAGCTCACCCCGTATCACTT |
| R | TAACATCAATGACCGCGAAA | |
| Lpms19 | F | AGGGAGGCATTGAGGTTTTT |
| R | CTCAGGCAACTCGGGATAAC | |
| Lpms25 | F | GTTAGCGCGTCTGCGATTAG |
| R | TCCTGCTTCCCTTTCCTTTT |
Each oligonucleotide is given the name of the minisatellite locus.
F, forward; R, reverse.
For cloning, PCR products were purified with the QIAquick PCR purification kit (Qiagen) and were ligated into the pGEM-T Easy Vector system (Promega, Charbonniéres, France). The inserts were sequenced by using M13 universal and reverse primers (MWG-Biotech, Ebersborg, Germany). The clustering analyses were performed with BioNumerics software (Applied Maths, Kortrijk, Belgium). The categorical coefficient and unweighted pair group method with arithmetic means clustering method were used.
RESULTS
Characterization of polymorphic markers.
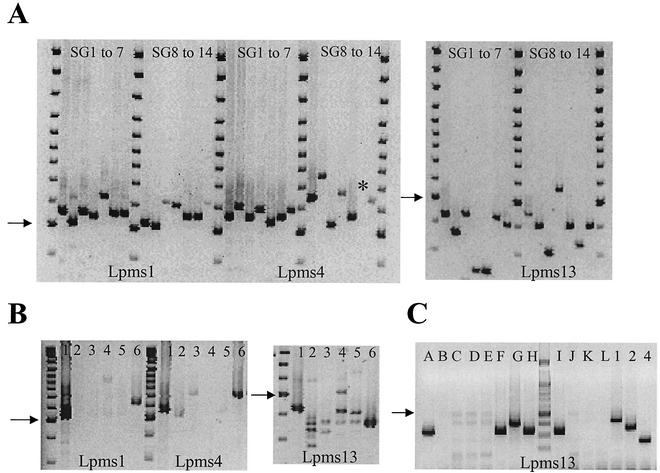
Using the software developed by G. Benson (1) and the derived database described by Vergnaud and Denoeud (21), we selected 25 sequences containing a tandem repeat in the sequence of L. pneumophila Philadelphia-1, provided by the Columbia Genome Center Legionella Project (http://genome3.cpmc.columbia.edu/∼legion/). We chose oligonucleotides from both sides of the nucleotide sequence and performed PCRs with DNA from reference strains of serogroups 1 to 14. We did not get amplification for three markers, and polymorphisms were observed for seven markers. Markers Lpms1, Lpms4, and Lpms13 are highly informative, as they possess three, seven, and eight alleles, respectively, among the 14 strains tested (Fig. 1A). These markers did not significantly amplify the DNA from the four other Legionella species included in this study (Fig. 1B), although in some cases a faint band was visible.
FIG. 1.
(A) Polymorphisms of minisatellites Lpms1, Lpms4, and Lpms13 in strains of serogroups (SG) 1 to 14. The PCR products were analyzed by agarose gel electrophoresis. The asterisk indicates a very faint band. (B) Analysis of different Legionella species. Lanes: 1 and 6, L. pneumophila; 2, L. dumoffii; 3, L. micdadei; 4, L. bozemanii; 5, L. longbeachae 1. (C) Analysis with Lpms13 of 12 colonies isolated from a water sample and of reference strains of serogroups 1, 2, and 4. The arrows point to the size marker of 500 bp.
Markers Lpms5, Lpms11, Lpms17, and Lpms19 are polymorphic, but they did not amplify the DNA of all the isolates. Lpms25 is not polymorphic but gave a very good amplification with all of the L. pneumophila isolates tested. The reference DNA of serogroups 1, 3, and 6 had the same allele sizes with all markers except Lpms5 and Lpms19.
We tested whether Lpms13 could be used to identify an L. pneumophila isolate freshly isolated from the environment. Figure 1C shows the result of an analysis performed with thermolysates (samples A to L) prepared from one colony of bacteria isolated from a water sample after 72 h of growth on solid medium. As a control, the DNA of serogroup 1, 2, and 4 reference strains was amplified at the same time. The results suggest that five colonies were L. pneumophila and that at least two different strains were present in the same water sample. These results were confirmed by immunofluorescence with a monoclonal antibody that detects L. pneumophila and by differential growth on medium with or without l-cysteine and iron.
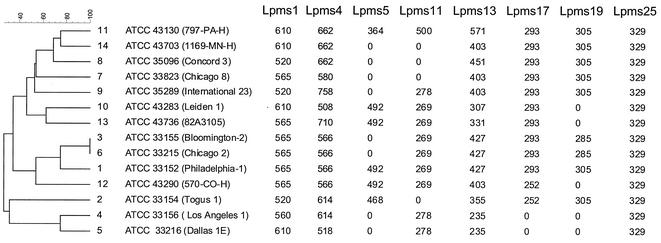
A dendrogram was produced by clustering analysis by using the genotyping data, as shown in Fig. 2. It confirms that strains Los Angeles 1 and Dallas 1E (L. pneumophila subsp. fraseri) are dissociated from the other strains and allows the grouping of the strains representing serogroups 1, 3, 6, and 12.
FIG. 2.
Dendrogram produced by categorical clustering of genotyping data by use of BioNumerics software. The numbers to the right of the dendrogram are serogroup numbers. Repeated amplification failure was considered a reflection of primer mispriming and was used in the analysis (null values in the table). The allele size estimates (in base pairs), deduced from gel image analysis with BioNumerics software, were adjusted to correspond to the expected allele sizes by taking into account the repeat unit length and the sequence data for reference strain L. pneumophila Philadelphia-1.
Comparison of MLVA with RFLP analysis by use of serogroup 3 strains.
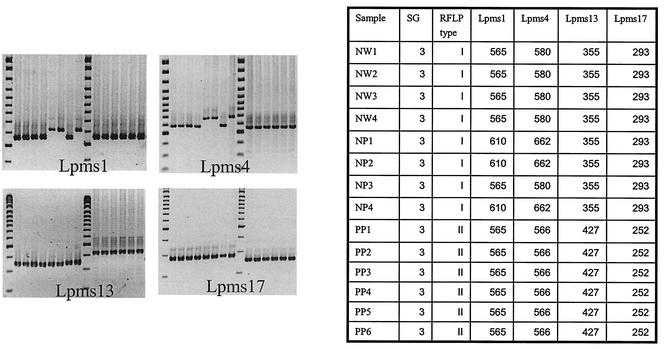
In order to test the usefulness of our markers for comparison of clinical and environmental strains of L. pneumophila, we genotyped a group of strains described by Tram et al. (18). It consisted of 14 strains of serogroup 3 isolated from four water samples (strains NW1 to NW4), 4 strains from patients (strains NP1 to NP4) hospitalized in Necker Hospital during the same time period, and 6 strains from patients at Pitié Hospital (strains PP1 to PP6). Restriction fragment length polymorphism (RFLP) analysis was performed as described by Tram et al. (18) and identified two profiles, the Necker profile (water and patient isolates) and the Pitié profile. We performed a PCR with DNA prepared from all these strains, using primers for Lpms1, Lpms4, Lpms5, Lpms11, Lpms13, LPms17, and Lpms19. Figure 3 shows the results of part of that analysis. Markers Lpms5 and Lpms19 were not informative. The strains with the Pitié profile were identical and different from all the others. Two profiles were identified among the strains with the Necker profile, with the strains from the four water samples providing a single profile similar to that of the strain from one of the patients (strain NP3). Another profile was observed with the strains from three other patients. That profile differed from that for the strain from the water sample by use of markers Lpms1 and Lpms4.
FIG. 3.
Genotyping of 14 serogroup (SG) 3 isolates from Necker and Pitié Hospitals (described in Table 2) with markers Lpms1, Lpms4, Lpms13, and Lpms17. The sizes of the PCR products (in base pairs) are shown in the accompanying table.
Comparison of MLVA and PFGE by use of serogroup 1 and 6 strains.
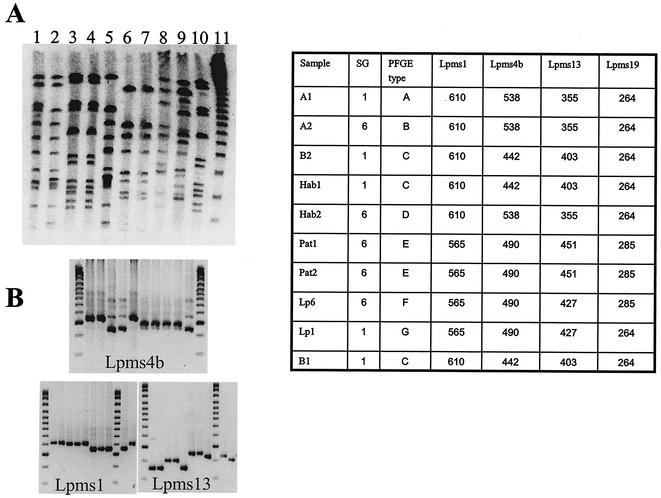
We performed a second analysis with a group of serogroup 1 and 6 strains isolated in 1989 from a patient and from environmental samples and for which a PFGE analysis had been performed (unpublished work). A Dutch patient died of pneumonia shortly after a stay in a Greek hotel. At that time, L. pneumophila strains were isolated from the shower water in four rooms of the hotel (strains A1, A2, B1, and B2), in another building of the hotel (strains Hab1 and Hab2), and from the patient (strains Pat1 and Pat2). A molecular study was performed following digestion of total genomic DNA by use of the restriction enzyme SfiI and PFGE. The profiles obtained for the different strains are shown in Fig. 4A and clearly indicate that the strains from the patient were not related to the environmental strains isolated at the hotel. Lanes 1 to 5 and lane 10 correspond to the strains recovered from the water in the hotel. Lanes 1, 2, and 5 show similar but not completely identical patterns (types A, B, and D, respectively), whereas lanes 3, 4, and 10 show the same pattern (type C). The two strains from the patient (lanes 6 and 7) showed similar patterns (type E) different from the patterns for the environmental isolates and those for the reference serogroup 6 and 1 strains (lanes 8 and 9, types F and G, respectively). We analyzed the different strains with the polymorphic minisatellite markers using reference serogroup 1 strain L. pneumophila Philadelphia-1 and serogroup 6 strain L. pneumophila Chicago 2 as controls. The Lpms4 marker-specific primers did not give a product for the strains in lanes 3, 4, and 10 (data not shown), so we made use of sequencing data for the different serogroups (see below) to select new primers, the Lpms4b primers (Table 3). The results for markers Lpms1, Lpms4b, and Lpms13 are shown in Fig. 4B. Three different profiles were observed, one for the strains in lanes 1, 2, and 5 (A1, A2, and Hab2, respectively), one for the strains in lanes 3, 4, and 10 (B2, Hab2, and B1, respectively), and one for the patient strains. The data match the results of the RFLP analysis concerning the different nature of the patient and environmental strains.
FIG. 4.
Macrorestriction analysis and minisatellite genotyping of the Greek isolate series (described in Table 2). (A) SfiI-cleaved genomic DNA of strains A1 (lane 1), A2 (lane 2), B2 (lane 3), Hab1 (lane 4), Hab2 (lane 5), Pat1 (lane 6), and Pat2 (lane 7), control serogroup 6 strain (lane 8), control serogroup 1 strain (lane 9), and strain B1 (lane 10). Bacteriophage lambda concatemers (lane 11) were used as size markers. (B) Genotyping of the strain in lanes 1 to 10 of panel A with markers Lpms4b, Lpms1, and Lpms13. The PFGE type and the sizes of the PCR products (in base pairs) are shown in the accompanying table. SG, serogroup.
Comparison of Lpms4 sequences in 14 strains.
The PCR product of Lpms4 was frequently of low abundance and of an unexpected size with respect to the tandem repeat unit. Thus, we decided to sequence the minisatellites from the reference strains of serogroups 1 to 14. We cloned the amplification products in a plasmid vector and sequenced the inserts from one or two clones from both ends.
All the sequences showed a repeat motif, usually of 48 bp, like that in the Philadelphia-1 strain, but of 54 bp in two strains, reference strains of serogroups 10 and 14, due to the insertion of 6 bases in the 48-bp motifs (Table 4). The number of repeats varied, explaining the size polymorphism. The sequence identity was high between the different serogroups except for serogroups 4 and 5. These two serogroups clearly belong to a separate group, with the sequence of serogroup 4 having an additional motif compared to the sequence of serogroup 5. The primer pair for Lpms4b was selected to match the sequences of serogroups 4 and 5. For the other strains, point mutations were found inside the consensus motif, and their number and repartition allowed the tentative distribution of the different strains into groups. The sequences of serogroup 1, 3, and 6 strains, on the one hand, and the sequence of the serogroup 12 strain, on the other, differed at 1 of 363 bases. The sequence of serogroup 2 is very similar to those of serogroups 1, 3, 6, and 12, but it possesses one additional motif and a 6-bp deletion. It was not possible to group strains of serogroups 7, 8, 9, 11, and 13 on the basis of their nucleotide sequences. The results were different when the protein sequences were examined. The translation in the six reading frames produced only one open reading frame (ORF), which was identical in the 14 alleles analyzed. Figure 5 shows a multiple-sequence alignment of the ORFs, in which the additional motifs have all been arbitrarily localized at a single position for clarity. A BLAST search gave no hit either for microbial genomes or for the complete nucleotide database.
TABLE 4.
Alignment of Lpms4 consensus motifs
| Group | Serogroup | Sequencea | Repeat motif (bp) | No. of motifs |
|---|---|---|---|---|
| I | 1 | GCAGCGACAGCCAGTCTTCATCACCGCCTACAGACAGTAGCAGCAGTG | 48 | 7.8 |
| 3 | ................................................ | 48 | 7.8 | |
| 6 | ................................................ | 48 | 7.8 | |
| 12 | ................................................ | 48 | 7.8 | |
| 2 | ................................................ | 48 | 8.8 | |
| 9 | ................................................ | 48 | 10.9 | |
| 11 | ................................................ | 48 | 9.1 | |
| 13 | ................C............................... | 48 | 9.8 | |
| 7 | ..........T.....C............................... | 48 | 7.9 | |
| 8 | .T........T.....C............................... | 48 | 8.9 | |
| II | 10 | AT....G.........C.......................TAACAG........ | 54 | 5.7 |
| 14 | AT....G.........C.......................TAACAG........ | 54 | 7.7 | |
| III | 4 | .............A..C...........A..G.....C.....T..C. | 48 | 8.1 |
| 5 | .............A..C...........A..G.....C.....T..C. | 48 | 7.1 |
Boldface indicates nucleotide changes, and dots indicate nucleotides in all motifs.
FIG. 5.
Multiple-peptide-sequence alignment. The putative ORFs derived from the Lpms4 minisatellite sequences from 14 reference strains were aligned manually. SG, serogroup.
DISCUSSION
We have characterized several polymorphic minisatellites in L. pneumophila which can be used for genotyping. Three of these PCR markers have been able to amplify the DNA of the strains from all samples tested, although in some cases the PCR product was not abundant, suggesting that the primer sequences were not perfectly homologous. A large proportion of the markers tested did not amplify the DNA from all strains, showing that the level of diversity is high within the species L. pneumophila. For this reason, we have not investigated all the minisatellite loci identified with the Tandem Repeat Finder software. The availability of sequence data for another strain (possibly L. pneumophila subsp. fraseri) would greatly facilitate the selection of primers that are specific for most if not all the L. pneumophila strains and the identification of additional polymorphic tandem repeat loci. Indeed, using sequencing data for the Lpms4 locus, we were able to design new primers that amplified the DNA of all strains tested.
Strains with identical serotypes can have different genotypes, confirming the data obtained by other techniques, including protein polymorphism analysis (15), ribotyping and RFLP analysis with PFGE (5, 14) repetitive-element PCR (7), or gene sequencing (3, 11).
Interestingly, in the study performed with serogroup 3 strains isolated from Necker Hospital, the profiles obtained suggested that one patient was infected with a strain present in the hospital water, whereas the other three patients were infected with another strain which was not recovered from the water samples. The latter strain differed from the strain from hospital water only at markers Lpms1 and Lpms4, which could mean that the strain is of a different origin or that it is a variant of the strain isolated from water. This is reminiscent of the results of Harrison et al. (9), who suggested that phenotypic variation of a single parent organism could account for the phenotypically different strains isolated from a single cooling tower. A larger number of colonies isolated from water could have been analyzed by tandem repeat typing at the time of the infection (as shown in Fig. 1C), possibly allowing the isolation of additional environmental strains.
We performed a second retrospective study with Legionella isolates from a Dutch patient and from water samples in a Greek hotel. The comparison of the RFLP profiles performed in 1989 (unpublished work) had concluded that the patient strain was not present in the water of the hotel. The water samples were taken from the showers in hotel rooms that had not been used for 24 h. Isolates of two different serogroups were found, and their genotypes were different from the genotype of the patient isolate, as determined by using four minisatellite markers and by RFLP analysis and PFGE. It might have been interesting to analyze a larger number of colonies from different environmental sources by a simple and rapid technique such as MLVA. However, when the first study was conducted in 1989, it was not easy to genotype large numbers of isolates.
Because we noticed that the primers whose sequences were selected from that of a sequenced strain, Philadelphia-1, did not systematically work with all the strains that we tested, we decided to sequence the PCR products of the Lpms4 markers of the 14 reference strains. The data confirm the existence of important nucleotide variations inside the minisatellite and in the flanking sequences from which the sequences of the primers used for PCR were selected. We tentatively chose degenerated primers that amplified all strains tested, although they did not always give a single product (data not shown). Concerning the minisatellite structure and internal variations, the sequencing data are very instructive and complementary to the minisatellite length polymorphism data. There were more variations at the nucleotide level than at the protein level, and these variations allow some clustering of the different strains that were not clustered by VNTR analysis. Reference strains Los Angeles 1 (serogroup 4) and Dallas 1E (serogroup 5), which belong to L. pneumophila subsp. fraseri, are the most divergent. Reference strains of serogroups 1, 3, 6, and 12 were distinguished by only one to three base changes in the complete minisatellite sequence. The reference strains of serogroups 10 and 14 have a particular motif of 54 bp instead of the motif of 48 bp found for all other strains tested, and the consensus motif sequence contains four base changes compared to the consensus motif of strain Philadelphia-1. Our data are in agreement with those of Ko et al. (11), who analyzed the rpoB sequence, and they suggest that sequencing of the Lpms4 loci of a large number of strains could improve phylogenetic studies with L. pneumophila. We are analyzing the sequence polymorphisms at the Lpms1 and Lpms13 loci, and this may eventually lead to proposed two-step assay: MLVA followed, if necessary, by allele sequencing for greater resolution.
Acknowledgments
We thank France Denoeud for help in recovering and analyzing the genomic sequences and Guy Baranton for constant support. We thank the Columbia Genome Center Legionella Project for access to the unpublished genome sequence data.
MLVA typing of the bacterial pathogens was supported by grants from the Délégation Générale de l'Armement (grants DGA/DSA/STTC and DGA/DSA/SP-Nuc). C. Pourcel is a member of the Pasteur Institute.
REFERENCES
- 1.Benson, G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27:573-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brenner, D. J., A. G. Steigerwalt, P. Epple, W. F. Bibb, R. M. McKinney, R. W. Starnes, J. M. Colville, R. K. Selander, P. H. Edelstein, and C. W. Moss. 1988. Legionella pneumophila serogroup Lansing 3 isolated from a patient with fatal pneumonia, and descriptions of L. pneumophila subsp. pneumophila subsp. nov., L. pneumophila subsp. fraseri subsp. nov., and L. pneumophila subsp. pascullei subsp. nov. J. Clin. Microbiol. 26:1695-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bumbaugh, A. C., E. A. McGraw, K. L. Page, R. K. Selander, and T. S. Whittam. 2002. Sequence polymorphism of dotA and mip alleles mediating invasion and intracellular replication of Legionella pneumophila. Curr. Microbiol. 44:314-322. [DOI] [PubMed] [Google Scholar]
- 4.Cherry, W. B., B. Pittman, P. P. Harris, G. A. Hebert, B. M. Thomason, L. Thacker, and R. E. Weaver. 1978. Detection of Legionnaires' disease bacteria by direct immunofluorescent staining. J. Clin. Microbiol. 8:329-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Zoysa, A. S., and T. G. Harrison. 1999. Molecular typing of Legionella pneumophila serogroup 1 by pulsed-field gel electrophoresis with SfiI and comparison of this method with restriction fragment-length polymorphism analysis. J. Med. Microbiol. 48:269-278. [DOI] [PubMed] [Google Scholar]
- 6.Fields, B. S., R. F. Benson, and R. E. Besser. 2002. Legionella and Legionnaires' disease: 25 years of investigation. Clin. Microbiol Rev. 15:506-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Georghiou, P. R., A. M. Doggett, M. A. Kielhofner, J. E. Stout, D. A. Watson, J. R. Lupski, and R. J. Hamill. 1994. Molecular fingerprinting of Legionella species by repetitive element PCR. J. Clin. Microbiol. 32:2989-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gomez-Lus, P., B. S. Fields, R. F. Benson, W. T. Martin, S. P. O'Connor, and C. M. Black. 1993. Comparison of arbitrarily primed polymerase chain reaction, ribotyping, and monoclonal antibody analysis for subtyping Legionella pneumophila serogroup 1. J. Clin. Microbiol. 31:1940-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harrison, T. G., N. A. Saunders, A. Haththotuwa, G. Hallas, R. J. Birtles, and A. G. Taylor. 1990. Phenotypic variation amongst genotypically homogeneous Legionella pneumophila serogroup 1 isolates: implications for the investigation of outbreaks of Legionnaires' disease. Epidemiol. Infect. 104:171-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jonas, D., H. G. Meyer, P. Matthes, D. Hartung, B. Jahn, F. D. Daschner, and B. Jansen. 2000. Comparative evaluation of three different genotyping methods for investigation of nosocomial outbreaks of Legionnaires' disease in hospitals. J. Clin. Microbiol. 38:2284-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ko, K. S., H. K. Lee, M. Y. Park, M. S. Park, K. H. Lee, S. Y. Woo, Y. J. Yun, and Y. H. Kook. 2002. Population genetic structure of Legionella pneumophila inferred from RNA polymerase gene (rpoB) and DotA gene (dotA) sequences. J. Bacteriol. 184:2123-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Le Flèche, P., Y. Hauck, L. Onteniente, A. Prieur, F. Denoeud, V. Ramisse, P. Sylvestre, G. Benson, F. Ramisse, and G. Vergnaud. 2001. A tandem repeats database for bacterial genomes: application to the genotyping of Yersinia pestis and Bacillus anthracis. BioMed Central Microbiol. 1:2.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McKinney, R. M., L. Thacker, P. P. Harris, K. R. Lewallen, G. A. Hebert, P. H. Edelstein, and B. M. Thomason. 1979. Four serogroups of Legionnaires' disease bacteria defined by direct immunofluorescence. Ann. Intern. Med. 90:621-624. [DOI] [PubMed] [Google Scholar]
- 14.Schoonmaker, D., T. Heimberger, and G. Birkhead. 1992. Comparison of ribotyping and restriction enzyme analysis using pulsed-field gel electrophoresis for distinguishing Legionella pneumophila isolates obtained during a nosocomial outbreak. J. Clin. Microbiol. 30:1491-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Selander, R. K., R. M. McKinney, T. S. Whittam, W. F. Bibb, D. J. Brenner, F. S. Nolte, and P. E. Pattison. 1985. Genetic structure of populations of Legionella pneumophila. J. Bacteriol. 163:1021-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Supply, P., E. Mazars, S. Lesjean, V. Vincent, B. Gicquel, and C. Locht. 2000. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Mol. Microbiol. 36:762-771. [DOI] [PubMed] [Google Scholar]
- 17.Swanson, M. S., and B. K. Hammer. 2002. Legionella pneumophila pathogenesis: a fateful journey from amoebae to macrophages. Annu. Rev. Microbiol. 54:567-613. [DOI] [PubMed] [Google Scholar]
- 18.Tram, C., M. Simonet, M. H. Nicolas, C. Offredo, F. Grimont, M. Lefevre, E. Ageron, A. Debure, and P. A. Grimont. 1990. Molecular typing of nosocomial isolates of Legionella pneumophila serogroup 3. J. Clin. Microbiol. 28:242-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Valsangiacomo, C., F. Baggi, V. Gaia, T. Balmelli, R. Peduzzi, and J. C. Piffaretti. 1995. Use of amplified fragment length polymorphism in molecular typing of Legionella pneumophila and application to epidemiological studies. J. Clin. Microbiol. 33:1716-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Belkum, A., S. Scherer, W. van Leeuwen, D. Willemse, L. van Alphen, and H. Verbrugh. 1997. Variable number of tandem repeats in clinical strains of Haemophilus influenzae. Infect. Immun. 65:5017-5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vergnaud, G., and F. Denoeud. 2000. Minisatellites: mutability and genome architecture. Genome Res. 10:899-907. [DOI] [PubMed] [Google Scholar]