Abstract
The objectives of this study were to understand the molecular diversity of animal and human strains of Mycobacterium avium subsp. paratuberculosis isolated in the United States and to identify M. avium subsp. paratuberculosis-specific diagnostic molecular markers to aid in disease detection, prevention, and control. Multiplex PCR of IS900 integration loci (MPIL) and amplified fragment length polymorphism (AFLP) analyses were used to fingerprint M. avium subsp. paratuberculosis isolates recovered from animals (n = 203) and patients with Crohn's disease (n = 7) from diverse geographic localities. Six hundred bacterial cultures, including M. avium subsp. paratuberculosis (n = 303), non-M. avium subsp. paratuberculosis mycobacteria (n = 129), and other nonmycobacterial species (n = 168), were analyzed to evaluate the specificity of two IS900 integration loci and a newly described M. avium subsp. paratuberculosis-specific sequence (locus 251) as potential targets for the diagnosis of M. avium subsp. paratuberculosis. MPIL fingerprint analysis revealed that 78% of bovine origin M. avium subsp. paratuberculosis isolates clustered together into a major node, whereas isolates from human and ovine sources showed greater genetic diversity. MPIL analysis also showed that the M. avium subsp. paratuberculosis isolates from ovine and bovine sources from the same state were more closely associated than were isolates from different geographic regions, suggesting that some of the strains are shared between these ruminant species. AFLP fingerprinting revealed a similar pattern, with most isolates from bovine sources clustering into two major nodes, while those recovered from sheep or humans were clustered on distinct branches. Overall, this study identified a high degree of genetic similarity between M. avium subsp. paratuberculosis strains recovered from cows regardless of geographic origin. Further, the results of our analyses reveal a relatively higher degree of genetic heterogeneity among M. avium subsp. paratuberculosis isolates recovered from human and ovine sources.
Paratuberculosis or Johne's disease (JD) is a chronic granulomatous enteritis of ruminants caused by Mycobacterium avium subsp. paratuberculosis (48). It is estimated that 35% of the U.S. herds are infected with M. avium subsp. paratuberculosis, resulting in annual losses of $200 million (7). Crohn's disease (CD) is also a chronic inflammation of distal intestines exhibiting a pathology similar to that of JD in ruminants. The prevalence of CD is estimated to be 0.15% among the U.S. population resulting in substantial morbidity and medical costs (1). M. avium subsp. paratuberculosis has been implicated as a cause of CD in humans (13, 21, 36). Currently, the evidence for a link remains inconclusive since strain sharing or a causal role of M. avium subsp. paratuberculosis has not been demonstrated.
Comprehensive analysis of the molecular diversity and comparative molecular pathology of M. avium subsp. paratuberculosis will help establish the degree of heterogeneity in strains isolated from a variety of host species. The extent of strain sharing across a variety of hosts will reflect the degree of interspecies transmission. It will also identify any strain similarities between JD and CD. Until recently, an IS900-based restriction fragment length polymorphism (RFLP) fingerprinting has been applied to study molecular epidemiology of M. avium subsp. paratuberculosis isolates. Extensive analyses of the IS900-RFLP patterns have identified that JD in cattle and other species such as goats and rabbits (3, 20, 33) is caused by indistinguishable strains. Two M. avium subsp. paratuberculosis isolates from a CD patient were also shown to carry a bovine-like IS900-RFLP fingerprint (47). JD in sheep appears to be caused by a different M. avium subsp. paratuberculosis strain (3, 47). However, the occurrence of a “sheep strain” in cattle with JD has been reported, indicating that interspecies transmission cannot be ruled out (49). Use of other independent fingerprinting and molecular diversity analysis tools to verify strain sharing is essential to provide substantial evidence of interspecies transmission.
The control and eradication of JD is severely hindered by prolonged incubation time, the presence of undetected subclinical cases, the absence of M. avium subsp. paratuberculosis-specific diagnostic tools, and the lack of knowledge of strain diversity. Current diagnostic methods include isolation from fecal and tissue specimens, enzyme-linked immunosorbent assay (ELISA), and IS900-based PCR. Although culturing the organisms is considered a “gold standard,” this method is fraught with difficulties. For example, the organism takes at least 12 to 16 weeks to grow to detectable levels, and even the most sensitive culture methods have only 50% sensitivity (43). Serologic tests such as agar gel immunodiffusion (18), complement fixation (25), and ELISA are limited in their use because of low specificity and sensitivity since antibodies may not be detectable either due to anergy or until their late appearance in the pathogenesis of JD (32, 42).
The conventional wisdom is that M. avium subsp. paratuberculosis differs from other members of the M. avium complex (MAC) in having 14 to 18 copies of IS900 inserted into conserved loci in its genome. Insertion element IS900 is a 1,451-bp segment that lacks inverted terminal repeats and has features characteristic of the IS900 family such as homologous transposases and particular insertion sites (19). IS900-based PCR identification techniques have been routinely used for M. avium subsp. paratuberculosis diagnosis (22, 50), and a number of studies have used DNA probes based on IS900 for M. avium subsp. paratuberculosis detection in tissues (29, 50), fecal specimens (17, 43), and milk samples (35). There is growing concern about the zoonotic potential of M. avium subsp. paratuberculosis, and several studies have attempted to show a causal relationship between M. avium subsp. paratuberculosis and CD by demonstration of IS900 in tissue (22, 37) and milk (9, 31, 35) samples of CD patients. Additionally, based on the presence of IS900, several studies have implied that bovine milk may serve as a mode of transmission of M. avium subsp. paratuberculosis (9, 35). However, IS900-like elements have been found in MAC strains (30) and in some atypical mycobacteria (10, 14, 28). This implies that diagnosis of M. avium subsp. paratuberculosis based on detection of IS900 alone needs to be evaluated with caution. Hence, there is a need for identification and large-scale analysis of additional M. avium subsp. paratuberculosis-specific molecular targets other than IS900 to confirm diagnosis. We report here a molecular test that exploits the fact that IS900 is integrated into conserved loci in the genome and that these integration sites are unique to M. avium subsp. paratuberculosis. Microtiter plate hybridization to integration site-specific probes was used to confirm specificity of PCR amplification (40). A recently identified M. avium subsp. paratuberculosis unique sequence 251 (2) was also evaluated as a target sequence for use in molecular diagnosis. The presence of hsp65 polymorphism was used to confirm the identification of M. avium subsp. paratuberculosis (15, 26). Two different molecular typing methods, multiplex PCR of 28 IS900 integration loci (MPIL) (5) and amplified fragment length polymorphism (AFLP) analysis were used to fingerprint M. avium subsp. paratuberculosis isolates obtained from a variety of geographical localities and host species. An IS1311 PCR and restriction endonuclease analysis (PCR-REA) was used to differentiate between bovine and ovine M. avium subsp. paratuberculosis isolates (46) within MPIL and AFLP clusters.
MATERIALS AND METHODS
Bacteria.
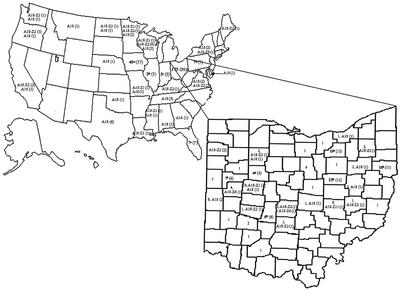
Mycobacterial strains were obtained from a variety of hosts and geographical localities (Table 1). A total of 173 field isolates were obtained from Ohio farms and represented 33 counties (Fig. 1). About 43% of the isolates were from Columbiana (n = 16), Holmes (n = 22), Logan (n = 18), and Medina (n = 19) counties in the state of Ohio. Nationally, 27 states were represented. A bovine isolate each from Argentina and Nova Scotia were also analyzed. Some human M. avium subsp. paratuberculosis strains analyzed in the present study have been described elsewhere (31). Most isolates were provided as distinct colonies on Herrold's egg yolk medium (HEYM) slants containing mycobactin J. Some mycobacterial strains that were received as bacterial suspensions in water or broth were subcultured on HEYM with mycobactin J. DNA from M. tuberculosis isolates, representing the extent of diversity within the species based on IS6110 fingerprinting and classification into the three major groups (41), were a kind gift from the collection of B. N. Kreiswirth (Public Health Research Institute, New York, N.Y.). DNA extracts from M. bovis and BCG strains were a kind gift of B. C. Cooksey (Centers for Disease Control, Atlanta, Ga.).
TABLE 1.
Bacterial isolates used for analysis
| Strain and source | No. of isolates used in each analysis
|
|||||
|---|---|---|---|---|---|---|
| L1-L9 PCRa | Locus 251 PCR | hsp65 PCR-REA | IS1311 PCR-REA | MPIL | AFLP | |
| Mycobacterium avium subsp. paratuberculosisa | ||||||
| ATCC (700535, 19698, and 19851) | 3 | 3 | 3 | 2 | 3 | |
| Cattle | 254 | 71 | 105 | 78 | 165 | 72 |
| Sheep | 18 | 11 | 18 | 10 | 16 | 4 |
| Goat | 19 | 19 | 8 | 10 | 17 | 7 |
| CD patients | 7 | 7 | 7 | 7 | 7 | 2 |
| Deer | 1 | 1 | 1 | 1 | 1 | |
| Mouse | 1 | 1 | 1 | 1 | 1 | 1 |
| K-10 | 1 | |||||
| MAC strains | ||||||
| TIGR 104 | 1 | 1 | 1 | 1 | ||
| CD patients | 31 | 15 | 30 | 24 | 20 | 10 |
| Unknown host | 53 | 45 | 53 | 30 | 17 | 6 |
| Mycobacterium tuberculosis (human) | 25 | 21 | 5 | 6 | ||
| Mycobacterium bovis (cattle and deer) | 9 | 3 | 2 | 2 | ||
| Mycobacterium bovis BCG | 4 | 3 | 3 | 3 | ||
| Atypical mycobacteriab (ATCC) | 6 | 6 | 6 | 6 | ||
| Staphylococci | 10 | 10 | 4 | 2 | ||
| Salmonella enterica subsp. (cattle) | 3 | 1 | 1 | |||
| Escherichia coli (cattle) | 20 | 20 | 4 | 4 | ||
| Mannheimia hemolytica (cattle) | 2 | 2 | 2 | 2 | ||
| Uncharacterized nonmycobacteria (human and animal) | 133 | 89 | 19 | |||
| Total | 600 | 328c | 272c | 190c | 247c | 104c |
PCR amplification and hybridization for IS900 integration loci L1 (left-side integration site of IS900 into an unknown ORF) and L9 (left-side integration of IS900 into alkA).
Atypical mycobacteria include M. chelonae, M. xenopi, M. celatum, M. marinum, M. scrofulaceum, and M. smegmatis.
The numbers represent a convenient subset of isolates analyzed for L1-L9 integration loci.
FIG. 1.
Distribution of MPIL and AFLP fingerprints of M. avium subsp. paratuberculosis isolates by U.S. states and Ohio counties. The numbers in parentheses indicate the numbers of isolates with each specific fingerprint. Isolates analyzed by both fingerprinting techniques are hyphenated. On the Ohio county map, the first number indicates the M. avium subsp. paratuberculosis isolates available from those regions, a subset of which was fingerprinted. Five of the states—Iowa, Illinois, Indiana, Pennsylvania, and Florida—had more than one fingerprint. The superscript letters a to e indicate fingerprint combinations as follows: a, A1, A2, A5, A10, A15-Z2 (3), A18-Z1 (3), A18-Z2 (8), A18-Z21, A18 (17), and B1; b, A18-Z2 and A18 (2); c, A18-Z1 (2) and A18-Z2; d, A1, A12, A18-Z2, and A18; (4); and e, A3, A13-Z15, A14, A18-Z10, and A18 (3). Similarly, Ohio counties with more than one fingerprint are indicated by superscript letters as follows: f (Auglaize), A1 (2), A2 (2), A6, and A18-Z1; g (Hardin), A3, A18-Z2, and B3; h (Holmes), A1 (4), A2, A3, A6-Z2, A18-Z1, A18-Z2, A18 (2), and B4; i (Medina), A12, A18 (8), B1 (2), B2-Z2, and B4; and j (Columbiana), A2 (2), A6, A8, A18-Z1, A18-Z2 (3), A18-Z21, and A18 (2). Isolates not included in the map are strains from Nova Scotia, Argentina, and areas for which the source was not known, including ATCC 19851.
DNA extraction.
Bacteria were harvested into 500 μl of sterile deionized water for DNA extraction. QIAamp DNA Blood Mini kit (Qiagen, Inc., Valencia, Calif.) was used with a few modifications. Briefly, the cell suspension was pelleted by centrifugation, and 180 μl of 20 mg of lysozyme (Sigma Aldrich, St. Louis, Mo.)/ml was added, followed by vigorous vortex mixing to obtain a homogeneous mixture. After incubation at 37°C for 30 min, 20 μl of protease and 200 μl of lysis buffer were added; the combination was vortex mixed thoroughly and then incubated at 70°C for 30 min. Protease was inactivated by incubating at 95°C for 15 min. Subsequently, the DNA was bound to columns, washed, and eluted in 200 μl of preheated sterile deionized water as suggested by the manufacturer. An extraction blank was included in each batch and used as negative control for molecular analyses.
Genetic fingerprinting by MPIL.
Genetic fingerprints of a subset of the M. avium subsp. paratuberculosis and MAC isolates were generated by using a method described previously (5) with several modifications. Briefly, six sets of multiplex PCRs referred as 9R1, 5R2, 4L1, 4L2, 3L3, and 3L4 were carried out as described earlier. The modifications to the published technique included the use of primer Loc14L in set 5R2 (instead of primer Loc14R) and primer Loc14R (6) in set 3L3 (instead of primer Loc14L). All primers were used at a concentration of 0.2 μM except the common primer, which was used at a concentration of 1 μM in a reaction volume of 50 μl that contained 5 U of Taq polymerase, 1× PCR buffer containing 1.5 mM MgCl2 (Qiagen), a 200 μM concentration of deoxyribonucleotide triphosphates (Amersham Pharmacia Biotech, Inc., Piscataway, N.J.), 10% dimethyl sulfoxide (DMSO), and 1 μl (ca. 50 ng/μl) of genomic DNA. The thermal cycler parameters were as described previously (5). The PCR products were electrophoresed at 100 V for 4 h in a 2% agarose gel prestained with ethidium bromide and visualized on a UV transilluminator (Alpha Innotech Corporation, San Leandro, Calif.).
Analysis of MPIL fingerprints.
The resulting fingerprints were converted into binary data where “1” indicated the presence and “2” indicated the absence of a specific band. Cluster analysis was done by using Molecular Evolutionary Genetics Analysis (MEGA; version 2.1) by the unweighted pair group method with averages (UPGMA) (39). The distance matrix for input into the MEGA program was created from the binary data by using ETDIV and ETMEGA (http://foodsafe.msu.edu/whittam/programs/).
AFLP of M. avium subsp. paratuberculosis.
AFLP was performed as described in the AFLP Microbial Fingerprinting Kit protocol (Perkin-Elmer Applied Biosystems Division, Foster City, Calif.). In brief, the M. avium subsp. paratuberculosis DNA was restricted with endonucleases EcoRI and MseI. The restricted fragments were then ligated to EcoRI and MseI adapters. The restriction-ligation reaction mixture included 100 ng of genomic DNA, 5 U of EcoRI (New England Biolabs, Inc. [NEB], Beverly, Mass.), 5 U of MseI (NEB), 1 U of DNA ligase (NEB), and a 0.4 μM concentration of adapter pairs in T4 DNA ligase buffer (50 mM Tris-HCl [pH 7.8], 10 mM dithiothreitol, 1 mM ATP, 25 μg of bovine serum albumin). Preselective amplification was carried out in a final reaction volume of 20 μl that contained 4-μl portions of the products of the restriction-ligation reaction, 125 pmol of AFLP preselective primer pairs (EcoRI preselective primer [5′-GACTGCGTACCAATTC] and MseI preselective primer [5′-GATGAGTCCTGAGTAA]), and 15 μl of AFLP core mix (PE-ABD). PCRs were performed in a thermal cycler (GeneAmp PCR system 9700; Perkin-Elmer) with an initial denaturation at 72°C for 2 min, followed by 20 PCR cycles of denaturation at 94°C for 1 s, annealing at 56°C for 30 s, an extension step at 72°C for 2 min, and a final extension step at 60°C for 30 min. Selective amplification was performed with the EcoRI selective primer containing a fluorescent label on the 5′ end with additional base C (carboxyfluorescein dye, FAM-EcoRI-C [FAM-5′-GACTGCGTACCAATTCC]) and MseI selective primer with no additional base (MseI-O [5′-GATGAGTCCTGAGTAA]) in a final reaction volume of 10 μl containing 1.5 μl of the preselective amplification products, 125 pmol of MseI-O primer, 25 pmol of FAM-EcoRI-C primer, and 7.5 μl of AFLP core mix (PE-ABD) in a thermal cycler (GeneAmp PCR system 9700). Aliquots (1 μl) of each amplification product were mixed with 2 μl of sterile water and subjected to electrophoresis in 5% Long Ranger gels (LMC Bioproducts, Rockland, Maine) by using the ABI model 377 automated DNA fragment analyzer (Perkin-Elmer). An internal size standard (GS-500, ROX size standard) was included in each lane. The AFLP fingerprint profiles were then automatically analyzed with GeneScan (Perkin-Elmer). Sizes of amplified products were then tabulated and exported for cluster analysis by using Molecular Analyst software (Bio-Rad, Hercules, Calif.). To analyze AFLP fingerprints, DNA fragments of from 50 to 500 bp were included for cluster analysis.
Computer assisted analysis of AFLP fingerprints.
Molecular Analyst software (Bio-Rad) was used to compare the AFLP fingerprint profiles among M. avium subsp. paratuberculosis isolates. The program automatically computed the similarity for each pair of fingerprints on the basis of band positions by using the Dice coefficient (SD). Pairs of isolates with a similarity coefficient (SD) of 0.95 were considered similar and not distinguishable. Cluster analysis among isolates was performed by the UPGMA linkage method.
Concordance analysis.
Kappa statistics (27) were calculated for each concordant pair of the isolates (n = 100) that were analyzed by both MPIL and AFLP techniques to assess the degree of agreement between the two fingerprinting techniques.
PCR amplification and hybridization for IS900 integration loci L1 and L9.
Two IS900 integration sites designated locus L1 (left-side integration site of IS900 into an unknown open reading frame [ORF]) and locus L9 (left-side integration of IS900 into alkA) were amplified by using PCR. The primers used were L1 (5′-CCCGTGACAAGGCCGAAGA-3′), L9 (5′-CGGCCCTGGCGTTCCTATG-3′), and the biotinylated common forward primer 900R (5′-/5Bio/ACGCTGTCTACCACCCCGCA-3′) (5). Primers were used at a final concentration of 0.2 μM in a 50-μl PCR master mix also including 1.25 U of Taq polymerase, 1× PCR buffer containing 1.5 mM MgCl2 (Qiagen), and a 200 μM concentration of deoxyribonucleotide triphosphates (Amersham Pharmacia Biotech). A total of 5 μl (ca. 50 ng/μl) of genomic DNA was used in each reaction. The thermal cycler parameters used for amplification included an initial 95°C incubation step for 15 min for Taq polymerase activation, followed by 35 cycles of denaturation at 94°C for 15 s, annealing at 58°C for 20 s, and extension at 72°C for 20 s, with a final extension step at 72°C for 7 min. A PCR blank was included for each batch and used as negative control for molecular analyses. The amplification products were detected in two independent ways. First, they were visualized on a UV transilluminator (Alpha Innotech) after electrophoresis at 125 V for 45 min in 1.5% agarose gels prestained with 5% ethidium bromide. Second, the biotinylated amplicons were detected in a reverse hybridization reaction on a microtiter plate as described previously (40). The probes were designed to target the unique integration regions (spanning IS900 into the genes of insertion) located within the amplicons. The probes used were PrL1 (spanning the IS900-unknown ORF region; 5′-TACACACAGCCGCCATACACTTCGCTTCATGCCCTTACG-3′) and PrL9 (spanning the IS900-alkA region; 5′-AGCCATCGAAGGGGATTCTCAATGACGTTGTCAA-3′). The two integration loci were detected simultaneously in microtiter wells (Corning Incorporated, Corning, N.Y.) coated with a mixture of both probes as described previously (40). DNA extraction blanks, PCR amplification negatives, and hybridization negatives were used in all analyses as negative controls. The cutoff optical density was set at 0.2 on the basis of analysis of several known negatives and M. avium subsp. paratuberculosis positives.
PCR amplification of M. avium subsp. paratuberculosis unique sequence 251.
The primers used to amplify the entire 417-bp ORF of M. avium subsp. paratuberculosis unique sequence 251 have been published elsewhere (2). PCRs were performed in 25-μl volumes, which included 0.625 U of Taq polymerase, 1× PCR buffer, 2.5 mM MgCl2, a 200 μM concentration of deoxyribonucleotide triphosphates (Amersham Pharmacia Biotech), a 0.2 μM concentration of each primer, 5% DMSO, and 2 μl (ca. 50 ng/μl) of genomic DNA. The thermal cycler parameters used for amplification included an initial 95°C incubation step for 15 min for Taq polymerase activation, followed by 35 cycles of denaturation at 94°C for 20 s, annealing at 53°C for 30 s, and extension at 72°C for 30 s, with a final extension step at 72°C for 7 min. Amplicons were visualized after electrophoresis at 125 V for 45 min in a 1.5% agarose gel prestained with ethidium bromide.
PCR-REA for polymorphisms in hsp65 and IS1311 genes.
New primers were designed to target previously described (15, 46) polymorphic regions of hsp65 and IS1311. A 248-bp fragment of hsp65 was amplified with the primers hsp3 (5′-GCCGCTGCTGATCATCGCCGA--3′) and hsp4 (5′-CCTTGGTGACGACGACCTT--3′), whereas a 749-bp region of IS1311 was amplified with the primers IS1311F (5′-GCGTGAGGCTCTGTGGTGAA--3′) and IS1311R (5′-TCAGAGATCACCAGCTGCAC--3′). PCR was carried out in a reaction volume of 50 μl. The master mix included 1.25 U of Taq polymerase, a 0.2 μM concentration of each primer, 1× PCR buffer containing 1.5 mM MgCl2 (Qiagen), a 200 μM concentration of deoxyribonucleotide triphosphates (Amersham Pharmacia Biotech), 5% DMSO, and 2 μl (ca. 50 ng/μl) of genomic DNA. The thermal cycler parameters used were as described for integration loci PCR. A PCR blank was included in each batch and used as negative control for molecular analyses. After the amplification, 10 μl of each PCR product was digested with 4 U of restriction enzyme at 37°C for 1 h according to the supplier's instructions. PstI (New England Biolabs) was used to detect polymorphism in nucleotide 861 of the hsp65 ORF (GenBank accession no. X74518, ORF from 125 to 1750), and HinfI (New England Biolabs) was used to detect the IS1311 polymorphism in nucleotide 167 of the IS1311 ORF (GenBank accession no. AJ223974, ORF from 35 to 1219). Both digested and undigested amplicons were electrophoresed in adjacent wells of 1.5% agarose gels and visualized on a UV transilluminator (Alpha Innotech).
DNA sequencing.
PCR products were sequenced by using standard dye terminator chemistry and analyzed on an automated DNA sequencer (Perkin-Elmer ABI 377). Sequences were aligned against sequences in GenBank database and analyzed by using Editseq and MegAlign programs (DNASTAR, Inc., Madison, Wis.).
RESULTS
Fingerprinting by MPIL.
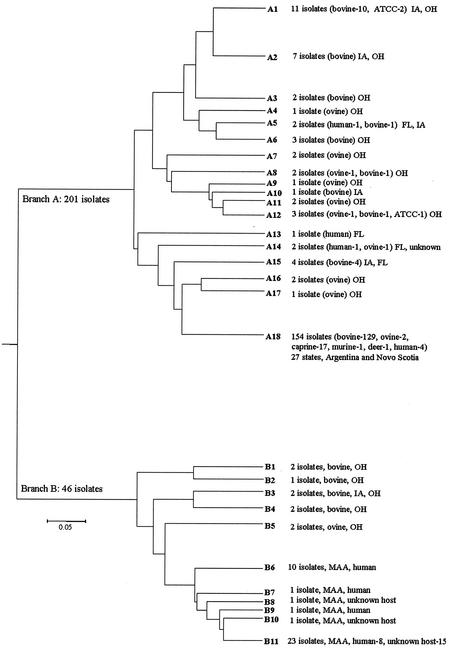
Of the 390 bacterial isolates fingerprinted by MPIL (Fig. 2), a panel of 247 isolates, including 210 M. avium subsp. paratuberculosis strains from a variety of species and geographic localities and 37 MAC strains, were subjected to cluster analysis (Table 1). Negative control strains were excluded from the analysis due to the absence of a fingerprint. These included extraction blank (n = 1), nonmycobacterial isolate (n = 1), atypical mycobacteria (n = 6), uncharacterized mycobacterial (BACTEC positive) controls (n = 14), and MAC strains with no or one band (n = 64). M. avium subsp. paratuberculosis isolates fingerprinted in duplicate (for technique validation and amplification quality control) either from the same (n = 21) or different (n = 30) culture tubes were also excluded from the cluster analysis. Six of the M. avium subsp. paratuberculosis isolates were excluded due to the absence of sufficient DNA for fingerprinting. Cluster analysis by UPGMA sorted the fingerprints into 90 electrophoretic types (ETs). The distances were generated as proportion differences. ET clusters at a distance of <0.1 U were grouped together and assigned a fingerprint. The major branches were arbitrarily designated A and B. Branch A had 18 different fingerprints; branch B had 11 fingerprints (Fig. 3). A majority of the M. avium subsp. paratuberculosis isolates (201 of 210 analyzed) clustered in branch A. A total of 78% of the total bovine isolates clustered in A18. Isolates from Ohio showed limited variation across the counties (Fig. 1). Fourteen of the ovine isolates were scattered throughout the dendrogram, with 50% clustering in the same node (A7 to A12) along with bovine isolates from Ohio and Iowa. Four of the human isolates also fell into the major cluster A18, whereas two appeared in close proximity to a bovine and an ovine strain (A5 and A14). The last human isolate appeared to have a distinct fingerprint (A13). All of the MAC strains (isolated from CD patients and other sources) clustered in branch B. This branch also included two ovine strains and seven bovine strains. On the basis of distance, both of the ovine strains were closer to the MAC strains than the bovine isolates.
FIG. 2.
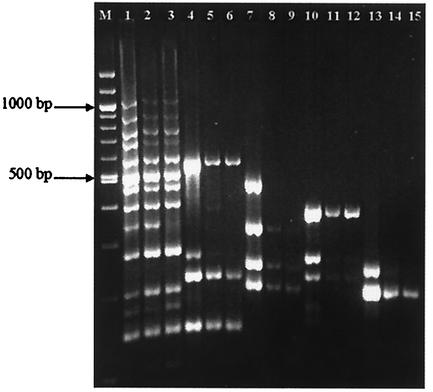
Multiplex PCR of IS900 loci of M. avium subsp. paratuberculosis isolates selected from bovine (lanes 1, 4, 7, 10, and 13), ovine (lanes 2, 5, 8, 11, and 14), and human (lanes 3, 6, 9, 12, and 15) hosts. Lanes 1, 2, and 3 depict the IS900 integrations amplified from the right end of the insertion. The left-end integration sites were analyzed as separate sets termed as 4L1 (lanes 4 to 6), 4L2 (lanes 7 to 9), 3L3 (lanes 10 to 12), and 3L4 (lanes 13 to 15).
FIG. 3.
MPIL cluster analysis by UPGMA. Cluster analysis of 247 isolates identified 90 ETs. Clusters at a proportion distance of <0.1 U were grouped together and assigned a fingerprint. Most of the bovine isolates clustered into a major group (A18) containing 154 isolates that included the caprine, murine, and deer isolates, indicating limited diversity. On the other hand, the sheep isolates were more polymorphic in that they were distributed in several nodes of the tree. Similarly, human isolates were distributed without clustering into any specific nodes. Unless specified, all isolates are M. avium subsp. paratuberculosis.
Fingerprinting by AFLP.
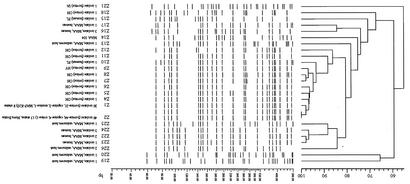
AFLP analysis was performed for 104 isolates from our culture collection. Isolates identified as distinct ETs and representing all of the MPIL fingerprint profiles were selected for AFLP analysis. These included 86 M. avium subsp. paratuberculosis isolates (bovine, n = 72; ovine, n = 4; caprine, n = 7; murine, n = 1; human, n = 2) and 16 MAC isolates (human host, n = 10; unknown host, n = 6). Type strains M. avium subsp. paratuberculosis strain K-10 and M. avium subsp. avium TIGR strain 104 were also included as positive controls. The dendrogram clustered 90% of the bovine M. avium subsp. paratuberculosis isolates into two major branches designated Z1 and Z2 (Fig. 4). These branches also included the goat and mouse isolates. Only one of the sheep isolates clustered into the major branch Z2. The other two Ohio ovine isolates clustered together in close proximity on one major node with a bovine isolate from New York and were assigned to the Z7, Z8, and Z9 genetic types (97% similarity), respectively. The MAC isolates from CD patients and unknown hosts clustered into distinct branches, suggesting a clear segregation between the strains. Both of the human M. avium subsp. paratuberculosis isolates analyzed appeared on independent branches. One bovine M. avium subsp. paratuberculosis isolate from Iowa appeared on a distinct branch and was assigned a unique fingerprint (Z21) (Fig. 4).
FIG. 4.
AFLP cluster analysis. A total of 104 isolates including type strains M. avium subsp. paratuberculosis strain K-10 and M. avium subsp. avium strain 104 were fingerprinted by using AFLP and analyzed by UPGMA. Analysis separated the isolates into 25 different fingerprints, which were arbitrarily designated Z1, Z2, and Z4 to Z26. Isolates with identical fingerprints (100% match) were assigned the same fingerprint. Unless otherwise specified, all isolates are M. avium subsp. paratuberculosis. Majority of the bovine isolates, including the caprine and murine isolates, clustered into Z1 and Z2. One of the four ovine isolates analyzed clustered with the bovine isolates in Z2, while the other two were in close proximity to a New York bovine isolate, and the fourth ovine isolate fell on a distinct branch. One of the human isolates appeared in closer proximity to the node containing most of the animal M. avium subsp. paratuberculosis isolates (Z1 to Z9), whereas the other one had a diverse fingerprint. This illustrates the limited variation in bovine isolates from a variety of hosts and the high degree of diversity among the ovine and human isolates. The MAC isolates from CD patients and unknown hosts fell into distinct branches.
Concordance analysis was performed for isolates (n = 100) that were fingerprinted by both MPIL and AFLP. The major MPIL cluster A18 containing 73% of the isolates could be further divided into different Z fingerprints by AFLP analysis as follows: Z1 (n = 23), Z2 (n = 42), Z4 (n = 1), Z5 (n = 1), Z6 (n = 1), Z9 (n = 1), Z10 (n = 1), Z11 (n = 1), Z12 (n = 1), and Z21 (n = 1). Isolates (n = 7) that had the B11 fingerprint could be divided into Z13 (n = 1), Z17 (n = 1), Z19 (n = 1), Z24 (n = 1), Z25 (n = 1), and Z26 (n = 2). Isolates (n = 8) that had the B6 fingerprint could be divided into Z16 (n = 3), Z22 (n = 2), Z23 (n = 1), and Z24 (n = 2). Similarly, the major AFLP cluster Z2 (n = 49) could be divided into A4 (n = 1), A6 (n = 1), A15 (n = 3), A18 (n = 42), and B2 (n = 1). Kappa coefficients of the major MPIL-AFLP pairs were A18-Z1 (κ = 0.2) and A18-Z2 (κ = 0.25).
PCR-REA for polymorphisms in IS1311 gene.
A C→T polymorphism in nucleotide 167 in ORF (GenBank accession no. AJ223974, ORF from 35 to 1219) of insertion element IS1311 is unique to bovine M. avium subsp. paratuberculosis isolates, whereas ovine M. avium subsp. paratuberculosis strains are identical to M. avium subsp. avium in this locus (46). This results in the gain of an HinfI restriction site in M. avium subsp. paratuberculosis of bovine origin. A panel of 190 isolates (Table 1) was analyzed for polymorphism in the IS1311 gene. Undigested amplicons from all MAC isolates and an atypical Mycobacterium (M. xenopi) showed the presence of a single 749-bp band after PCR. After digestion with HinfI all cattle, goat, mouse, and deer M. avium subsp. paratuberculosis isolates and five MAC isolates showed presence of the predicted four bands (67, 85, 218, and 379 bp), while all ovine M. avium subsp. paratuberculosis isolates and 50 of 55 MAC isolates showed the presence of three bands (85, 285, and 379 bp), indicating the absence of the restriction site (Fig. 5). The other five atypical mycobacterium and nonmycobacterium isolates did not amplify the IS1311 segment. Amplification specificity was confirmed by sequencing a bovine, an ovine, and a MAC isolate. All three isolates showed homology to the published sequences (GenBank accession numbers AJ308375, AJ223975, and U16276), as well as the expected polymorphism. One bovine isolate, which was L1-L9 and 251 positive, showed a different pattern when the restriction enzyme products were electrophoresed (Fig. 5). The undigested product was of the expected band length (749 bp). The nucleotide sequence of this product revealed that this isolate had numerous mutations resulting in a total of six restriction sites, only one of which was in the expected position. Restriction digestion resulted in seven fragments (31, 63, 85, 95, 130, 148, and 195 bp) instead of the expected three or four.
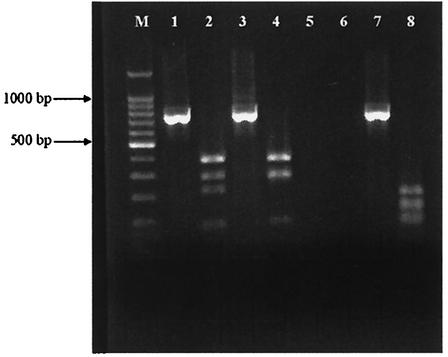
FIG. 5.
Typical IS1311 PCR-REA patterns with undigested and HinfI-digested amplicons in adjacent lanes. Shown are bovine M. avium subsp. paratuberculosis (lanes 1 and 2), ovine M. avium subsp. paratuberculosis (lanes 3 and 4), and PCR-negative control (lanes 5 and 6) isolates and a bovine isolate with several mutations resulting in multiple restriction sites (lanes 7 and 8).
PCR amplification of two IS900 integration sites.
A panel of 600 bacterial isolates including M. avium subsp. paratuberculosis (n = 303), MAC (n = 85), M. tuberculosis complex (n = 38), atypical mycobacteria (n = 6), nonmycobacterial negative controls (n = 35), and other uncharacterized non-M. avium subsp. paratuberculosis control isolates (n = 133) from a variety of geographic locations and hosts were analyzed for the presence of two integration sequences (Table 1). PCR amplified two regions corresponding to the integration sites of IS900 at loci L1 (206 bp) and L9 (147 bp). Gel electrophoresis showed the presence of two bands of expected sizes in 296 M. avium subsp. paratuberculosis and 5 of the 85 isolates from the MAC group. Microtiter plate hybridization results were positive for all of the known M. avium subsp. paratuberculosis isolates (n = 303). All other isolates including DNA extraction, PCR, and hybridization negatives were below the cutoff value of an optical density at 450 nm of 0.2.
Presence of M. avium subsp. paratuberculosis-specific target 251 in known M. avium subsp. paratuberculosis isolates.
A subset of 328 isolates from the culture collection was analyzed for the presence of M. avium subsp. paratuberculosis unique sequence 251 (Table 1). Agarose gel electrophoresis showed the presence of a 417-bp band for all 113 known M. avium subsp. paratuberculosis isolates and 5 MAC isolates that were also positive by L1-L9 PCR. The locus was absent in all other mycobacterial (n = 27), atypical mycobacterial (n = 6), and nonmycobacterial bacterial (n = 121) isolates analyzed. The PCR products from nine bovine (Ohio), three human (CD), and three ovine (Ohio) isolates were sequenced. The nucleotide sequences had 100% identity with the published ORF of locus 251 (GenBank accession no. AF445445, ORF from 123 to 539), indicating the stability of this target and its usefulness in JD diagnostics.
PCR-REA for polymorphisms in hsp65 gene.
A subset of 272 isolates (Table 1) that were analyzed for the presence of L1 and L9 integration sites were reflexed for identity confirmation by PCR-REA analysis of this locus. A G→T polymorphism in nucleotide 861 in the ORF (GenBank accession no. X74518) of the hsp65 gene is unique to M. avium subsp. paratuberculosis and leads to the loss of a PstI restriction site in the species. Prior to digestion, all mycobacterial isolates analyzed showed the presence of a single 248-bp band when visualized by agarose gel electrophoresis. After digestion with PstI, all M. avium subsp. paratuberculosis isolates retained the full 248-bp band, while all MAC and other bacterial isolates showed the presence of two bands (135 and 113 bp), indicating the presence of the restriction site. All six atypical mycobacterium and 10 M. tuberculosis complex isolates showed a MAC-like pattern. The nonmycobacterial isolates included in the analysis did not amplify the region of interest.
DISCUSSION
The efficiency of preventive and prophylactic measures in JD is restricted due to the lack of M. avium subsp. paratuberculosis-specific diagnostic tools and, in particular, the lack of knowledge of strain diversity that has been limited due to its slow growth rate and restricted allelic variation. It is desirable to differentiate among the isolates of M. avium subsp. paratuberculosis to better understand the epidemiology of M. avium subsp. paratuberculosis infections, its host specificity, distribution, and prevalence. In the present study we sought to establish the degree of molecular diversity among M. avium subsp. paratuberculosis strains from a variety of hosts and locations, to identify any strain sharing among various host species and to identify M. avium subsp. paratuberculosis-specific diagnostic molecular markers to aid in disease prevention and control.
Molecular diversity in M. avium subsp. paratuberculosis.
Mutations in structural genes (24) or antigenic variations (16) can serve as useful molecular markers to study epidemiology and natural history of an organism. However, in well-conserved genomes such as that of mycobacteria (41), these markers provide limited information in strain typing. In mycobacterial genomes, the degree of diversity in the number and site of integration of transposable genetic elements or single nucleotide polymorphisms in genes associated with drug resistance or host immunological pressure may be more useful than those that rely on genome-wide single nucleotide polymorphisms as indicators of strain variation. Several molecular epidemiological studies have demonstrated the usefulness of insertion sequences in determining strain distribution of mycobacteria such as IS1245 in M. avium (45) and IS6110 in M. tuberculosis (44) and IS711 in Brucella sp. (4). Other repetitive elements have also been used in strain typing (12). Seminal studies that identified IS900 in M. avium subsp. paratuberculosis strains suggested that it was exclusively present in M. avium subsp. paratuberculosis (19). Hence, IS900 has been the marker of choice for most fingerprinting studies reported (11, 34, 47). This exploits the fact that the IS900 elements show a high degree of target sequence specificity resulting in similar fingerprints in epidemiologically related isolates (19). IS900-RFLP analyses have broadly divided the M. avium subsp. paratuberculosis strains into either sheep or cattle and other ruminant subtypes, designated S and C, respectively (3, 8, 47). In contrast, isolation of an S strain from cattle has been reported (49). Most investigations have identified limited strain sharing between cattle and sheep hosts, whereas little variation in fingerprints is evident between cattle and other animals, including rabbits and goats, and M. avium subsp. paratuberculosis isolates from a human with CD. Evidence of strain sharing between bovine and human M. avium subsp. paratuberculosis isolates is of special interest since this implies the existence of a potential animal reservoir for CD. Several studies have implicated an inability to compare fingerprints from sheep isolates due to difficulties encountered in culturing them. Thus, an indication of limited strain sharing between bovine and ovine isolates does not rule out interspecies transmission since there may be a bias in the analysis due to under-representation of the slow-growing ovine strains. In summary, there has been inadequate data to support the idea of extensive strain sharing and interspecies transmission. Besides being elaborate and expensive, fingerprinting by RFLP requires large amounts of unsheared genomic DNA extracted from relatively large cultures. There are multiple reports (10, 14, 28, 30) of the presence of IS900-like elements in mycobacteria other than M. avium subsp. paratuberculosis. Sequence analysis of a few MAC isolates with the commonly used IS900 primers (3) showed the presence of IS900 elements (data not shown). Nucleotide sequences indicated significant homology between these regions and the IS900 element in M. avium subsp. paratuberculosis. This supports the notion that the use of IS900 probe as a tool in molecular typing could result in false positives corresponding to IS900-related elements in mycobacteria other than M. avium subsp. paratuberculosis (8, 38).
Both techniques reported in the present study require very small quantities of genomic DNA and are applicable to degraded DNA samples not suitable for RFLP analysis. AFLP analysis results with 104 isolates from distinct geographic regions and hosts showed that 72% of the M. avium subsp. paratuberculosis, including bovine, caprine, and murine strains, fell into either one of the two major clusters reiterating the low degree of heterogeneity at the genomic level. This suggests that only a few strains may be responsible for disease in bovine hosts and that interspecies transmission could have occurred since these clusters also included M. avium subsp. paratuberculosis strains isolated from other hosts. In agreement with previous studies, the two major branches included only one of the four ovine isolates, while the other two ovine strains clustered into distinct branches in close proximity to a bovine isolate. In contrast to earlier reports (33, 47) and our MPIL data, AFLP fingerprints of the human M. avium subsp. paratuberculosis isolates were unique and did not cluster with either the bovine or ovine strains, suggesting that they were relatively more heterogeneous at the genome level.
In MPIL analysis 78% of the U.S. bovine isolates, including those from Argentina and Nova Scotia, fell within one major cluster, indicating a significant degree of uniformity within isolates infecting bovine hosts. M. avium subsp. paratuberculosis strains from other ruminants, such as goat and deer, and one mouse strain also fell within this cluster. Similar to AFLP, this major cluster included only 2 of the 16 ovine isolates analyzed. The analysis identified clear genetic diversity between ovine isolates, whereas there were limited differences within strains from bovine hosts across several geographic localities. M. avium subsp. paratuberculosis isolates from the human CD patients were more diverse with phylogenetic proximities at various levels to strains isolated from both bovine and ovine host species, an indication of a greater level of diversity. Although this suggests a close association of the human and animal strains, it does not provide direct evidence for a causal role of M. avium subsp. paratuberculosis in CD. Comprehensive comparative genomics with larger numbers of human isolates and reproduction of JD in animals by the human strains will be required to demonstrate, albeit indirectly, a causal association between M. avium subsp. paratuberculosis and CD.
The possibility of nonspecific amplification of Loc5R and Loc5L primers in M. avium subsp. avium strains due to homology in desA1 and desA2 genes have been described (5). Similar nonspecific amplifications may have been the case in the low-copy strains of our collection. Conversion of this gel-based analysis into a microarray by using integration site probes would provide a more accurate picture of the fingerprinting by MPIL.
Concordance analysis showed that AFLP fingerprinting was able to discriminate between isolates clustered together by MPIL analysis. Similarly, MPIL fingerprinting discriminated between isolates in one cluster by AFLP analysis. Kappa scores for the major concordant pairs show that fingerprints generated by MPIL and AFLP analysis are independent. This indicates the need to use more than one fingerprinting technique to distinguish between the M. avium subsp. paratuberculosis strains. Restricted allelic variation within this mycobacterial subspecies may limit the application of any single fingerprinting method in epidemiological studies of JD. Alternately, the use of other combinations of restriction enzymes (such as ApaI and TaqI) or application of a multiplex-AFLP protocol (23) may provide a better indication of diversity.
Analysis of the isolates for presence of polymorphism in IS1311 by PCR-REA facilitated the classification of the M. avium subsp. paratuberculosis isolates into bovine or ovine strains. The gain of the HinfI restriction site due to a single nucleotide polymorphism, which is unique to the bovine strain, is indicative of limited strain sharing between the two hosts. One bovine isolate from an Ohio farm showed a difference in the density of the restricted bands compared to other isolates in our analysis. This is consistent with the speculation (46) that M. avium subsp. paratuberculosis isolates may vary in the number of IS1311 insertion elements carrying the polymorphism. The well-conserved nature of the mycobacterial genome indicates that the probability of several mutations within one gene is very low. Hence, the bovine M. avium subsp. paratuberculosis isolate with multiple HinfI restriction sites within IS1311 is of special interest. Although the absence of the C→T polymorphism indicates that it is an ovine strain, it clustered with other bovine isolates by MPIL analysis.
Integration loci as M. avium subsp. paratuberculosis unique targets.
IS900 is a 1,451-bp segment that lacks inverted repeats and has features characteristic of the IS900 family such as homologous transposases and particular insertion sites (19). Detection of a portion of IS900 sequence is routinely used to detect the presence of M. avium subsp. paratuberculosis in field and clinical samples (37, 50). However, the presence of IS900-like sequences has been demonstrated in non-M. avium subsp. paratuberculosis mycobacteria, including M. avium subsp. avium (30, 38), M. cookii (14), M. marinum, and M. scrofulaceum (10, 28). Because of the importance of differentiating these closely related mycobacteria in clinical specimens there is a need for molecular targets based on genes other than IS900 to confirm the presence of M. avium subsp. paratuberculosis.
The present study reports the development of a multiplex PCR to amplify two distinct integration sites of the insertion element IS900. This exploits the fact that the IS900 elements integrate into conserved loci in the M. avium subsp. paratuberculosis genome (19). Although geographically restricted exceptions cannot be ruled out, this large-scale analysis indicates that the integration sites of IS900 at loci L1 and L9 were unique to M. avium subsp. paratuberculosis and are consistently present in M. avium subsp. paratuberculosis from diverse locations and hosts. These two integration sequences were absent in non-M. avium subsp. paratuberculosis mycobacteria, nonspecific bacterial DNA, and in 80 of 85 MAC strains analyzed, 33 of which had low numbers (one to six) of IS900 integration sites as determined by multiplex PCR. The five MAC isolates that showed presence of both the integration sites were reclassified as M. avium subsp. paratuberculosis on the basis of M. avium subsp. paratuberculosis-like pattern for six distinct molecular targets: L1-L9, 251, hsp65 PCR-REA, IS1311 PCR-REA, MPIL, and AFLP (two isolates) cluster analysis. The isolates were confirmed to be mycobactin J dependent. This implies that these sites could serve as valuable tools in the molecular diagnostics of M. avium subsp. paratuberculosis and can significantly reduce the time to diagnosis of M. avium subsp. paratuberculosis from 16 to 23 weeks to 2 to 10 days in subclinical infections. Routine use of the integration sites to detect M. avium subsp. paratuberculosis will prevent the reporting of false-positive IS900 PCR results. A recent study in our laboratory of feces from field animals (A. Ozbek, F. C. Michel, M. Strother, A. S. Motiwala, B. Byrum, W. Shulaw, C. G. Thornton, and S. Sreevatsan, unpublished data) established the feasibility of application of this method in the development of a sensitive assay for primary clinical samples.
To confirm the stability of the integration loci as diagnostic targets and to evaluate the possibility of reflexing ambiguous isolates to hsp65 PCR-REA, we analyzed a subset of bacterial isolates for polymorphism in hsp65 gene. This subset included M. avium subsp. paratuberculosis strains obtained from a variety of host species. Although this technique proved to be very valuable for pure cultures, subsequent analysis of M. avium subsp. paratuberculosis from broth cultures showed that it is not as efficient for mixed cultures from feces or broth due to the highly conserved nature of this gene across many species. Hence, hsp65 analysis was restricted to pure cultures, whereas locus 251 was used as an additional marker for broth cultures.
Recently identified unique M. avium subsp. paratuberculosis sequences (2) show promise of serving as target sequences in molecular diagnosis of M. avium subsp. paratuberculosis. One of these sequences, locus 251, was consistently detected with the L1-L9 integration sites in 118 of 121 known positive isolates. Sequence analysis from bovine, ovine, and human isolates show 100% homology with the published sequence, indicating the possibility of applying this locus in a multiplex PCR-hybridization format, along with LI-L9 integration sites to aid in the unambiguous diagnosis of JD. Although the specificity in amplification of the locus from M. avium subsp. paratuberculosis isolates in our sample was absolute, we note that the forward primer (251R) used for the amplification of locus 251 revealed homology (15 of 20 nucleotides at the 3′ end) to plasmid sequences from the enteric bacterium Salmonella spp. on BLASTN searches with the public databases (GenBank accession numbers AF250878 and AL513383). Hence, positive results of amplification, as seen on a gel, must be interpreted with caution, especially when fecal samples and primer pairs are used as described in the present study. Postamplification detection on a solid phase (such as a microtiter plate) through hybridization to an internal sequence is likely to improve the specificity of the results.
In conclusion, the present study identified a high degree of genetic similarity within the bovine isolates, indicating that only a few very closely related clones of M. avium subsp. paratuberculosis may be responsible for widespread infection in cattle, other ruminants, and possibly wildlife. This study established strain sharing among isolates from a variety of hosts and geographic locations. In addition, we identified multiple M. avium subsp. paratuberculosis unique diagnostic targets and protocols that may provide a facile approach to unambiguously detect M. avium subsp. paratuberculosis. Future comparative genomics and pathogenesis studies with the bovine and ovine M. avium subsp. paratuberculosis strains and with human M. avium subsp. paratuberculosis and MAC isolates will be required to further elucidate the natural history of this organism.
Acknowledgments
This study was supported by state and federal funds provided to the Ohio Agricultural Research and Development Center (OARDC), including an OARDC Competitive Research Enhancement Seed Grant awarded to S. Sreevatsan. Research in the laboratory of V. Kapur is funded by competitive awards from the National Institutes of Health, the U.S. Department of Agriculture, and the Rapid Minnesota Agricultural Experiment Station.
REFERENCES
- 1.Ashford, D., I. B. Angulo, et al. 2001. October 2001. Over a half-million persons may have Crohn's disease in the United States. Infectious Diseases Society of America, San Francisco, Calif.
- 2.Bannantine, J. P., E. Baechler, Q. Zhang, L. Li, and V. Kapur. 2002. Genome scale comparison of Mycobacterium avium subsp. paratuberculosis with Mycobacterium avium subsp. avium reveals potential diagnostic sequences. J. Clin. Microbiol. 40:1303-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauerfeind, R., S. Benazzi, R. Weiss, T. Schliesser, H. Willems, and G. Baljer. 1996. Molecular characterization of Mycobacterium paratuberculosis isolates from sheep, goats, and cattle by hybridization with a DNA probe to insertion element IS900. J. Clin. Microbiol. 34:1617-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bricker, B. J., and S. M. Halling. 1994. Differentiation of Brucella abortus bv. 1, 2, and 4, Brucella melitensis, Brucella ovis, and Brucella suis bv. 1 by PCR. J. Clin. Microbiol. 32:2660-2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bull, T. J., J. Hermon-Taylor, I. Pavlik, F. El Zaatari, and M. Tizard. 2000. Characterization of IS900 loci in Mycobacterium avium subsp. paratuberculosis and development of multiplex PCR typing. Microbiology 146(Pt. 9):2185-2197. [DOI] [PubMed] [Google Scholar]
- 6.Bull, T. J., J. Hermon-Taylor, I. Pavlik, F. El-Zaatari, and M. Tizard. 2000. Characterization of IS900 loci in Mycobacterium avium subsp. paratuberculosis and development of multiplex PCR typing. Microbiology 146:3285. [DOI] [PubMed]
- 7.CAST. 2001. CAST Issue Paper: Johne's Disease in Cattle. Council on Agriculture, Science, and Technology Task Force, Ames, Iowa.
- 8.Collins, D. M., D. M. Gabric, and G. W. de Lisle. 1990. Identification of two groups of Mycobacterium paratuberculosis strains by restriction endonuclease analysis and DNA hybridization. J. Clin. Microbiol. 28:1591-1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corti, S., and R. Stephan. 2002. Detection of Mycobacterium avium subspecies paratuberculosis specific IS900 insertion sequences in bulk-tank milk samples obtained from different regions throughout Switzerland. BMC Microbiol. 2:15-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cousins, D. V., R. Whittington, I. Marsh, A. Masters, R. J. Evans, and P. Kluver. 1999. Mycobacteria distinct from Mycobacterium avium subsp. paratuberculosis isolated from the feces of ruminants possess IS900-like sequences detectable IS900 polymerase chain reaction: implications for diagnosis. Mol. Cell. Probes 13:431-442. [DOI] [PubMed] [Google Scholar]
- 11.Cousins, D. V., S. N. Williams, A. Hope, and G. J. Eamens. 2000. DNA fingerprinting of Australian isolates of Mycobacterium avium subsp paratuberculosis using IS900 RFLP. Aust. Vet. J. 78:184-190. [DOI] [PubMed] [Google Scholar]
- 12.Dombek, P. E., L. K. Johnson, S. T. Zimmerley, and M. J. Sadowsky. 2000. Use of repetitive DNA sequences and the PCR To differentiate Escherichia coli isolates from human and animal sources. Appl. Environ. Microbiol. 66:2572-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.El Zaatari, F. A., M. S. Osato, and D. Y. Graham. 2001. Etiology of Crohn's disease: the role of Mycobacterium avium paratuberculosis. Trends Mol. Med. 7:247-252. [DOI] [PubMed] [Google Scholar]
- 14.Englund, S., G. Bolske, and K. E. Johansson. 2002. An IS900-like sequence found in a Mycobacterium sp. other than Mycobacterium avium subsp. paratuberculosis. FEMS Microbiol. Lett. 209:267-271. [DOI] [PubMed] [Google Scholar]
- 15.Eriks, I. S., K. T. Munck, T. E. Besser, G. H. Cantor, and V. Kapur. 1996. Rapid differentiation of Mycobacterium avium and M. paratuberculosis by PCR and restriction enzyme analysis. J. Clin. Microbiol. 34:734-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Espul, C., H. Cuello, N. Martinez, O. Centorbi, M. O'Ryan, L. Jackson, F. Campos, and D. O. Matson. 2000. Genomic and antigenic variation among rotavirus strains circulating in a large city of Argentina. J. Med. Virol. 61:504-509. [PubMed] [Google Scholar]
- 17.Fang, Y., W. H. Wu, J. L. Pepper, J. L. Larsen, S. A. Marras, E. A. Nelson, W. B. Epperson, and J. Christopher-Hennings. 2002. Comparison of real-time, quantitative PCR with molecular beacons to nested PCR and culture methods for detection of Mycobacterium avium subsp. paratuberculosis in bovine fecal samples. J. Clin. Microbiol. 40:287-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferreira, R., L. S. Fonseca, and W. Lilenbaum. 2002. Agar gel immunodiffusion test (AGID) evaluation for detection of bovine paratuberculosis in Rio de Janeiro, Brazil. Lett. Appl. Microbiol. 35:173-175. [DOI] [PubMed] [Google Scholar]
- 19.Green, E. P., M. L. Tizard, M. T. Moss, J. Thompson, D. J. Winterbourne, J. J. McFadden, and J. Hermon-Taylor. 1989. Sequence and characteristics of IS900, an insertion element identified in a human Crohn's disease isolate of Mycobacterium paratuberculosis. Nucleic Acids Res. 17:9063-9073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greig, A., K. Stevenson, D. Henderson, V. Perez, V. Hughes, I. Pavlik, M. E. Hines, I. McKendrick, and J. M. Sharp. 1999. Epidemiological study of paratuberculosis in wild rabbits in Scotland. J. Clin. Microbiol. 37:1746-1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hermon-Taylor, J. 2001. Protagonist. Mycobacterium avium subspecies paratuberculosis is a cause of Crohn's disease. Gut 49:755-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hulten, K., H. M. El Zimaity, T. J. Karttunen, A. Almashhrawi, M. R. Schwartz, D. Y. Graham, and F. A. El Zaatari. 2001. Detection of Mycobacterium avium subspecies paratuberculosis in Crohn's diseased tissues by in situ hybridization. Am. J. Gastroenterol. 96:1529-1535. [DOI] [PubMed] [Google Scholar]
- 23.Huys, G., L. Rigouts, K. Chemlal, F. Portaels, and J. Swings. 2000. Evaluation of amplified fragment length polymorphism analysis for inter- and intraspecific differentiation of Mycobacterium bovis, M. tuberculosis, and M. ulcerans. J. Clin. Microbiol. 38:3675-3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones, M. E., M. B. Avison, E. Damdinsuren, A. P. MacGowan, and P. M. Bennett. 1994. Heterogeneity at the beta-lactamase structural gene ampC amongst Citrobacter spp. assessed by polymerase chain reaction analysis: potential for typing at a molecular level. J. Med. Microbiol. 41:209-214. [DOI] [PubMed] [Google Scholar]
- 25.Kalis, C. H., H. W. Barkema, J. W. Hesselink, C. van Maanen, and M. T. Collins. 2002. Evaluation of two absorbed enzyme-linked immunosorbent assays and a complement fixation test as replacements for fecal culture in the detection of cows shedding Mycobacterium avium subspecies paratuberculosis. J. Vet. Diagn. Investig. 14:219-224. [DOI] [PubMed] [Google Scholar]
- 26.Kapur, V., L. L. Li, M. R. Hamrick, B. B. Plikaytis, T. M. Shinnick, A. Telenti, W. R. Jacobs, Jr., A. Banerjee, S. Cole, K. Y. Yuen, et al. 1995. Rapid Mycobacterium species assignment and unambiguous identification of mutations associated with antimicrobial resistance in Mycobacterium tuberculosis by automated DNA sequencing. Arch. Pathol. Lab. Med. 119:131-138. [PubMed] [Google Scholar]
- 27.Kelsey, J. L., W. D. Thompson, and A. S. Evans. 1986. Measurement error, p. 285-307. In J. L. Kelsey, W. D. Thompson, and A. S. Evans (ed.), Methods in observational epidemiology. Oxford University Press, New York, N.Y.
- 28.Kim, S. G., S. J. Shin, R. H. Jacobson, L. J. Miller, P. R. Harpending, S. M. Stehman, C. A. Rossiter, and D. A. Lein. 2002. Development and application of quantitative polymerase chain reaction assay based on the ABI 7700 system (TaqMan) for detection and quantification of Mycobacterium avium subsp. paratuberculosis. J. Vet. Diagn. Investig. 14:126-131. [DOI] [PubMed] [Google Scholar]
- 29.Miller, J. M., A. L. Jenny, and J. B. Payeur. 2002. Polymerase chain reaction detection of Mycobacterium tuberculosis complex and Mycobacterium avium organisms in formalin-fixed tissues from culture-negative ruminants. Vet. Microbiol. 87:15-23. [DOI] [PubMed] [Google Scholar]
- 30.Naser, S. A., J. Felix, H. Liping, C. Romero, N. Naser, A. Walsh, and W. Safranek. 1999. Occurrence of the IS900 gene in Mycobacterium avium complex derived from HIV patients. Mol. Cell. Probes 13:367-372. [DOI] [PubMed] [Google Scholar]
- 31.Naser, S. A., D. Schwartz, and I. Shafran. 2000. Isolation of Mycobacterium avium subsp. paratuberculosis from breast milk of Crohn's disease patients. Am. J. Gastroenterol. 95:1094-1095. [DOI] [PubMed] [Google Scholar]
- 32.Nielsen, S. S., C. Gronbaek, J. F. Agger, and H. Houe. 2002. Maximum-likelihood estimation of sensitivity and specificity of ELISAs and faecal culture for diagnosis of paratuberculosis. Prev. Vet. Med. 53:191-204. [DOI] [PubMed] [Google Scholar]
- 33.Pavlik, I., L. Bejckova, M. Pavlas, Z. Rozsypalova, and S. Koskova. 1995. Characterization by restriction endonuclease analysis and DNA hybridization using IS900 of bovine, ovine, caprine, and human dependent strains of Mycobacterium paratuberculosis isolated in various localities. Vet. Microbiol. 45:311-318. [DOI] [PubMed] [Google Scholar]
- 34.Pavlik, I., A. Horvathova, L. Dvorska, J. Bartl, P. Svastova, M. R. Du, and I. Rychlik. 1999. Standardisation of restriction fragment length polymorphism analysis for Mycobacterium avium subspecies paratuberculosis. J. Microbiol. Methods 38:155-167. [DOI] [PubMed] [Google Scholar]
- 35.Pillai, S. R., and B. M. Jayarao. 2002. Application of IS900 PCR for detection of Mycobacterium avium subsp. paratuberculosis directly from raw milk. J. Dairy Sci. 85:1052-1057. [DOI] [PubMed] [Google Scholar]
- 36.Quirke, P. 2001. Antagonist Mycobacterium avium subspecies paratuberculosis is a cause of Crohn's disease. Gut 49:757-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Richter, E., J. Wessling, N. Lugering, W. Domschke, and S. Rusch-Gerdes. 2002. Mycobacterium avium subsp. paratuberculosis infection in a patient with HIV in Germany. Emerg. Infect. Dis. 8:729-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roiz, M. P., E. Palenque, C. Guerrero, and M. J. Garcia. 1995. Use of restriction fragment length polymorphism as a genetic marker for typing Mycobacterium avium strains. J. Clin. Microbiol. 33:1389-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rzhetsky, A., S. Kumar, and M. Nei. 1995. Four-cluster analysis: a simple method to test phylogenetic hypotheses. Mol. Biol. Evol. 12:163-167. [DOI] [PubMed] [Google Scholar]
- 40.Sreevatsan, S., J. B. Bookout, F. Ringpis, V. S. Perumaalla, T. A. Ficht, L. G. Adams, S. D. Hagius, P. H. Elzer, B. J. Bricker, G. K. Kumar, M. Rajasekhar, S. Isloor, and R. R. Barathur. 2000. A multiplex approach to molecular detection of Brucella abortus and/or Mycobacterium bovis infection in cattle. J. Clin. Microbiol. 38:2602-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sreevatsan, S., X. Pan, K. E. Stockbauer, N. D. Connell, B. N. Kreiswirth, T. S. Whittam, and J. M. Musser. 1997. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc. Natl. Acad. Sci. USA 94:9869-9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stabel, J. R. 1998. Johne's disease: a hidden threat. J. Dairy Sci. 81:283-288. [DOI] [PubMed] [Google Scholar]
- 43.Stabel, J. R., S. J. Wells, and B. A. Wagner. 2002. Relationships between fecal culture, ELISA, and bulk tank milk test results for Johne's disease in US dairy herds. J. Dairy Sci. 85:525-531. [DOI] [PubMed] [Google Scholar]
- 44.van Embden, J. D., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, T. M. Shinnick, et al. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Soolingen, D., J. Bauer, V. Ritacco, S. C. Leao, I. Pavlik, V. Vincent, N. Rastogi, A. Gori, T. Bodmer, C. Garzelli, and M. J. Garcia. 1998. IS1245 restriction fragment length polymorphism typing of Mycobacterium avium isolates: proposal for standardization. J. Clin. Microbiol. 36:3051-3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whittington, R., I. Marsh, E. Choy, and D. Cousins. 1998. Polymorphisms in IS1311, an insertion sequence common to Mycobacterium avium and M. avium subsp. paratuberculosis, can be used to distinguish between and within these species. Mol. Cell. Probes 12:349-358. [DOI] [PubMed] [Google Scholar]
- 47.Whittington, R. J., A. F. Hope, D. J. Marshall, C. A. Taragel, and I. Marsh. 2000. Molecular epidemiology of Mycobacterium avium subsp. paratuberculosis: IS900 restriction fragment length polymorphism and IS1311 polymorphism analyses of isolates from animals and a human in Australia. J. Clin. Microbiol. 38:3240-3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whittington, R. J., and E. S. Sergeant. 2001. Progress towards understanding the spread, detection, and control of Mycobacterium avium subsp. paratuberculosis in animal populations. Aust. Vet. J. 79:267-278. [DOI] [PubMed] [Google Scholar]
- 49.Whittington, R. J., C. A. Taragel, S. Ottaway, I. Marsh, J. Seaman, and V. Fridriksdottir. 2001. Molecular epidemiological confirmation and circumstances of occurrence of sheep strains of Mycobacterium avium subsp. paratuberculosis in cases of paratuberculosis in cattle in Australia and sheep and cattle in Iceland. Vet. Microbiol. 79:311-322. [DOI] [PubMed] [Google Scholar]
- 50.Zwick, L. S., T. F. Walsh, R. Barbiers, M. T. Collins, M. J. Kinsel, and R. D. Murnane. 2002. Paratuberculosis in a mandrill (Papio sphinx). J. Vet. Diagn. Investig. 14:326-328. [DOI] [PubMed] [Google Scholar]