Abstract
The complex relation between the genotype of Helicobacter pylori and its association with clinical outcome is not well understood. Studies in the West have showed that strains expressing certain virulence factors (vacAs1, vacAm1, and cagA) are associated with duodenal ulcer disease. However, the H. pylori genotype is known to vary with geographic region. In the present study, we compared several virulence markers (cagA, vacA, and iceA) and neutral markers (IS605, IS606, and IS608) in H. pylori strains isolated from 65 adult patients with peptic ulcer (PU) and 50 patients with nonulcer dyspepsia (NUD). PCR tests indicated that cagA is present in 75% of the strains from patients with PU compared to 55% in patients with NUD, and 80% of the isolates from patients with PU carried potentially toxigenic vacAs1 alleles of the vacuolating cytotoxin gene (vacA) compared to 60% in isolates from patients with NUD. However, no significant difference in any other virulence marker was observed in isolates from both groups. Phylogenetic analysis of the vacA middle region and the 5′ end of the cagA gene indicates that Bangladeshi isolates are more closely related to H. pylori isolates from India and are different from isolates from East Asia.
Helicobacter pylori is a spiral, gram-negative microaerophilic bacterium that chronically infects the gastric mucosa of more than half of all people worldwide (30) and is a major cause of gastritis and peptic ulcer (PU) disease (37), and a risk factor for gastric adenocarcinoma and mucosa-associated lymphoid tissue lymphoma (29). Infection by H. pylori usually starts early in life and, unless eradicated by specific antibiotic treatment, persists for the rest of life. H. pylori produces a number virulence factors that are essential for colonization of the stomach and survival in the hostile gastric environment (27). Well known virulence factors are urease, which plays an important role in the neutralization of gastric acid secretion (19); flagella, which are essential for swimming through the mucous layer (21); vacuolating cytotoxin, which induces massive vacuolar degeneration of various epithelial cell lines (20); gene products encoded by cag pathogenicity island (PAI), which causes up-regulation of cytokines, including interleukin-8 (2); iceA, a homologue of a gene for restriction endonuclease, induced by contact with gastric epithelium (31); and OipA, a proinflammatory protein that contributes to interleukin-8 induction independent of cag PAI (38).
An intriguing aspect of H. pylori is an extraordinary diversity among strains that is seen in DNA fingerprinting and in tests of gene content and chromosomal gene order (1, 3). Together with this are indications of differences at certain loci between strains from different parts of the world or human ethnic groups. In particular, the DNA sequence motifs and disease association of two virulence genes, vacA and cagA, in strains from the United States and Europe were found to differ from those predominating in southern coastal China and Japan (14, 28, 34, 35), although less phylogenetic clustering was found in sequences of several housekeeping genes (1). However, most of our present understanding of H. pylori genome organization and bacterial traits are based on studies of strains from the West, and it is believed that further studies of H. pylori genotypes from different well-separated human populations may help to increase our understanding of bacterium-host interactions in colonization and disease.
Prevalence studies have indicated that H. pylori infection is extremely common in Bangladesh (22, 32) as in other developing countries. Although the available reports from this country have focused on the clinical patterns of gastroduodenal disease, detection of H. pylori infection, and effectiveness (or ineffectiveness) of anti-H. pylori therapies in the local population (6, 7, 13, 24), none, to our knowledge, provide information on the genotypes of the Bangladeshi H. pylori strains. In this context we have characterized H. pylori strains isolated from Dhaka, the capital of Bangladesh, in order to gain new insights into the population genetic structure of this important human pathogen and to learn if genotypes implicated in the disease in the West are similarly disease associated in Bangladesh.
MATERIALS AND METHODS
Patient samples.
Consecutive patients attending the gastroenterology Department of Dhaka Medical College hospital for routine endoscopy were enrolled in the study. Each subject was informed about the study and written informed consent was obtained under a protocol approved by the Ethical Review Committee of International Centre for Diarrheal Disease Research, Bangladesh. Endoscopy of the subjects was done by using well-washed and disinfected fiber optic endoscopes (model GIF XQ 30; Olympus, Japan). Diagnosis of PU was based on endoscopic examination of the stomach and duodenum during examination. If no evidence of ulcer was found the patient was considered to have nonulcer dyspepsia (NUD) or gastritis, which is a more benign infection. Two biopsy samples were taken from each patient for culture, one from the gastric antrum and one from the corpus. Immediately after collection the biopsy specimen was transported to the laboratory in Stuart transport medium at 4°C.
Bacterial culture.
Biopsy samples were vortexed vigorously for 5 min and plated on brain heart infusion agar (Oxoid, Ltd, Basingstoke, Hampshire, United Kingdom) supplemented with 7% sheep blood, 0.4% IsoVitaleX, and H. pylori Dent supplement (Oxoid). Plates were incubated at 37°C in an atmosphere of 5% O2, 10% CO2, and 85% N2 for 3 to 6 days. H. pylori colonies were identified based on their typical morphology, characteristic appearance on Gram staining, a positive urease test, and subsequent gene-specific PCR tests. Pure culture of pooled isolates was stored at −70°C in 0.5 ml of brucella broth with 15% glycerol until further study.
DNA extraction and PCR.
Chromosomal DNA was prepared by the CTAB (hexadecyl-trimethyl ammonium bromide) extraction method (4) from confluent brain heart infusion agar plate cultures. Specific PCR was carried out in a 20-μl volume using 10 ng of DNA, 1 U of Taq polymerase (Promega, Madison, Wis.), 10 pmol of each primer per reaction, 0.25 mM (each) deoxynucleoside triphosphate, and a 2 to 3 mM concentration of MgCl2 in standard PCR buffer for 30 cycles using the following conditions: 94°C for 40 s, 55°C for 40 s, and 72°C for a time chosen based on the size of the expected fragment (1 min/kb). PCR primers used for genotyping are listed in Table 1.
TABLE 1.
Primers used in this study
| Region | Primer | Nucleotide sequence |
|---|---|---|
| vacAs1 or vacAs2 | VA1-F | 5′-ATGGAAATACAACAAACACAC |
| VA1-R | 5′-CTGCTTGAATGCGCCAAAC | |
| vac m1b | Vam-F3 | 5′-GGCCCCAATGCAGTCATGGAT |
| Vam-R3 | 5′-GCTGTTAGTGCCTAAAGAAGCAT | |
| vacA m2 | VA4-F | 5′-GGAGCCCCAGGAAACATTG |
| VA4-R | 5′-CATAACTAGCGCCTTGCAC | |
| vacA 0.7-kb middle | VAm-F | 5′-GCTCATTACGGCTTCCACTAATGT |
| VAm-R | 5′-GCGGTTATTGTTGTTATAAAGGGCTA | |
| cagA (5′ end) | cagA5 | 5′-GGCAATGGTGGTCCTGGAGCTAGGC |
| cag A2 | 5′-GGAAATCTTTAATCTCAGTTCGG | |
| cag PAI empty site | Luni 1 | 5′-ACATTTTGGCTAAATAAACGCTG |
| R5280 | 5′-GGTTGCACGCATTTTCCCTTAATC | |
| iceA1 | iceA1F | 5′-TATTTCTGGAACTTGCGCAACCTGAT |
| M.Hpy1R | 5′-GGCCTACAACCGCATGGATAT | |
| iceA2 | cycSF | 5′-CGGCTGTAGGCACTAAAGCTA |
| IceA2R | 5′-TCAATCCTATGTGAAACAATGATCGTT | |
| iceA1 Δ94 bp | A1F673 | 5′-GGTGAGTCGTTGGGTAAGCGTTACAGAATT |
| A1R1174 | 5′-CACAACCATCATATTCAGCCTCCCCCTCATA | |
| IS605orfA | ORF18F | 5′-CGCCTTGATCGTTTCAGGATTAGC |
| ORF18R | 5′-CAACCAACCGAAGCAAGCATAATC | |
| IS605orfB | ORF19F | 5′-GGCTGTTCTAGGGTCGTGTATAAC |
| ORF19R | 5′-CAAGCTAGATGCAATCTAGCTACC | |
| IS606 | FB1 | 5′-GAATGTAATTCTACCTAATCCATTC |
| Rb8 | 5′-GAGAAACCTTGATTGTTCCATG | |
| IS608 | 608-F1 | 5′-CCATAACGCCTTAATAGTGTGTC |
| 608-R | 5′-CAAGCTTTGGAGTGATGAAGTTC | |
| Type I | cagF4856 | 5′-GCGATGAGAAGAATATCTTTAGCG |
| cagR5280 | 5′-GGTTGCACGCATTTTCCCTTAATC | |
| Type II | IS606-1692 | 5′-CTAACAATTTGCCATTATGCTGT |
| cagF4856 | 5′-GCGATGAGAAGAATATCTTTAGCG | |
| Type III | fcn unk | 5′-TGGATTAAATCTTAATGAATTATCG |
| cagF4856 | 5′-GCGATGAGAAGAATATCTTTAGCG |
DNA sequencing and analysis.
The PCR products of the 700-bp middle region of the vacA gene from six strains and 219-bp fragment from the 5′ end of the cagA gene of seven strains were purified with a QIAquick gel extraction kit (Qiagen, Valencia, Calif.). DNA sequencing was performed by the dideoxynucleotide chain termination method with an ABI PRISM Dye Termination Cycle Sequencing Ready Reaction kit (Perkin-Elmer-Applied Biosystems, Foster City, Calif.) in an ABI PRISM automated sequencer. DNA sequence editing and analysis were performed with programs in the Genetics Computer Group (Madison, Wis.) package, programs and data in the TIGR H. pylori genome database (http://www.tigr.org/tdb/mdb/hpdb.html), and the PHYLIP program of J. Felsenstein (http://evolution.genetics.washington.edu/phylip.html) (33). Phylogenetic trees were visualized using Treeview (version 1.61; http://www .taxonomy.zoology.gla.ac.uk/rod/rod.html).
Nucleotide sequence accession numbers.
Sequences obtained during this study were assigned the following GenBank accession numbers: accession no. AY169292 to AY169299 for the 219-bp cagA gene of DH102, DH140, DH200, DH29, DH37, DH60, DH93, and G31 and accession no. AY169286 to AY169291 for the 648-bp vacAm1 allele of DH153, DH131, DH60, DH92, DH99, and DH114, respectively.
RESULTS
Incidence of H. pylori.
A total of 115 consecutive patients between 20 and 55 years old were enrolled in the study. Of these 115 patients, 97 (85%) were male and 18 (15%) were female. Among the patients, 65 were diagnosed with PU and 50 were diagnosed with NUD. A total of 57 isolates were obtained from the enrolled subjects. H. pylori was isolated in 37 (56%) of the 65 patients presenting with PU and 20 (40%) of the 50 patients with NUD.
Genotype of H. pylori isolates. (i) cagA gene.
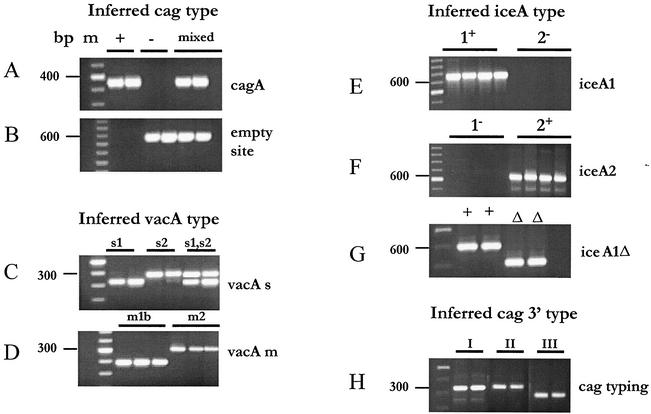
The genotype distributions of the strains form patients with PU and NUD is shown in Table 2. The presence or absence of the cagA gene and the empty-site-specific 550-bp amplicon was scored by PCR with specific primers using DNA extracted from cultured strains (Table 1). A 324-bp fragment indicative of the cagA gene (Fig. 1A) was obtained with primers specific for the cagA gene from the great majority of strains: 28 of 37 cultures from patients with PU and 11 of 20 patients with NUD. Eleven of 18 cultures from which no cagA gene-specific PCR product was obtained yielded an empty-site product of the expected size, 550 bp (Fig. 1B), indicating that they truly lacked the cagA gene (4). In addition, products corresponding to the empty site (Fig. 1A and B, lanes labeled mixed) were also obtained from seven individuals found to be infected with strains carrying the cagA gene, indicating that they had mixed infections, i.e., a mixture of strains with and without the cagA gene.
TABLE 2.
Genotypic characteristics of H. pylori isolates from Dhaka, Bangladesh, and relation to disease status
| Trait or markera | No. of patients with indicated disease status
|
|
|---|---|---|
| PU (n = 37 [64%]) | NUD (n = 20 [36%]) | |
| cag PAI+ onlyb | 28 | 11 |
| cag PAI+, cag PAI− mixedb | 3 | 4 |
| cag PAI− onlyb | 6 | 5 |
| vacAs1 only | 30 | 12 |
| vacAs2 | 6 | 6 |
| vacAs1, vacAs2 mixed | 1 | 2 |
| vacAm1c | 22 | 8 |
| vacAm2 only | 13 | 8 |
| vacAm1c, vacAm2 mixed | 2 | 4 |
| iceA1c | 19 | 9 |
| iceA2 | 17 | 10 |
| iceA1, iceA2 mixed | 1 | 1 |
| IS605 | 16 | 7 |
| IS606 | 11 | 7 |
| IS608 | 6 | 3 |
| cag type IIId | 26 | 11 |
| cag type IId | 1 | 0 |
| cag type Id | 4 | 3 |
Distribution of DNA markers was determined by PCR, as illustrated in the figures.
cag PAI+ only, infection with strains carrying cag PAI only; cag PAI+, cag PAI− mixed infection with strains carrying the cag PAI and strains lacking the cag PAI; cag PAI− only, infection with strains lacking the cag PAI only.
A 94-bp deletion was found in two isolates, both from patients with PU.
The 3′ end of the cag gene was typed and scored as type I, II, or III. Isolates from European origin usually belong to type I.
FIG. 1.
Ethidium bromide-stained agarose gel of representative PCR amplicon using primers mentioned in Table 1. (A) Tests for presence and absence of cagA gene using CagA5 and CagA2 primers. (B) Tests for presence of cag PAI empty site using Luni1 and R5280 primers. (C) Tests for presence of vacAs1 (259-bp amplicon), vacAs2 (286-bp amplicon), and both vacAs1 and vacAs2 alleles (both 259- and 298-bp amplicon) using primers VA1-F and VA1-R. (D) Tests for presence of vacAm1 allele using VAm-F and VAm-R primers and vacAm2 allele using VA4-F and VA4-R primers. (E) PCR amplicon of iceA1 obtained with primers iceAF1 and M.Hpy1R. (F) PCR amplicon of iceA2 obtained with cysF and iceA2R primers. (G) PCR amplicon demonstrating a 94-bp deletion in iceA1, obtained with primers A1F673 and A1R1174. (H) PCR typing of isolates by 3′ end of cag gene.
Phylogenetic relationship between cagA 5′ end of Bangladeshi isolates and those from other geographic regions.
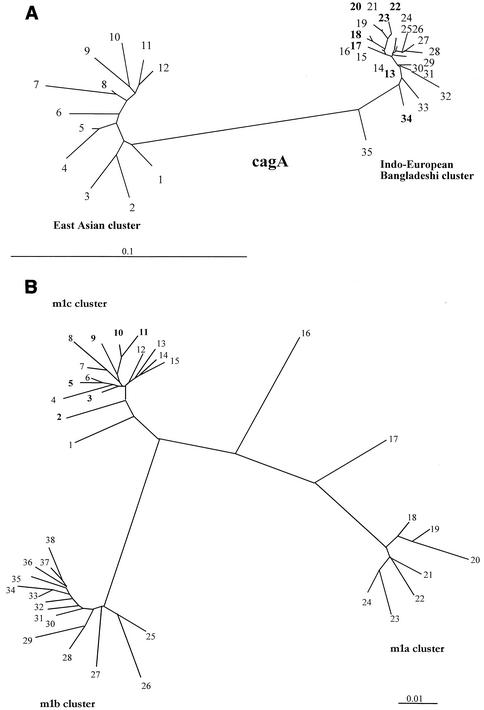
To assess the phylogenetic relationship between cag PAIs of Bangladeshi isolates and those from other regions, we sequenced a 219-bp fragment near the 5′ end of the cagA gene that had been used to distinguish east Asian and U.S. and European strains. The sequences were closely related to each other and also to those from Western strains, but not to those from Chinese and Japanese strains (Fig. 2A).
FIG. 2.
Phylogenetic trees of sequences within the cagA gene and vacAm1 alleles. Sequences from non-Bangladeshi strains that were used here were from public databases, as indicated. (A) Phylogenetic tree based on informative 219-bp segment of cagA of H. pylori strains determined in this study. The tree was generated by using PHYLIP (Phylogeny Inference Package), version 3.5c, of J. Felsenstein. The strains used were as follows (GenBank accession number in parentheses): 1, CHINAR48 (AJ252983); 2, JAPANF32 (AJ239726); 3, JAPANGC4 (AF198484); 4, CHINAR29 (AJ252980); 5, CHINAR59 (AJ252986); 6, CHINAR47 (AJ252985); 7, HK77 (AF198485); 8, THI88-28 (AJ239722); 9, CHINAR29 (AJ252980); 10, HK97-42 (AJ239726); 11, CHINAR40 (AJ252982); 12, HK81 (AF198486); 13, DH93 (AY169298); 14, S-AFR19 (AF198470); 15, I-9A (AF202225); 16, PERU35B (AF202221); 17, DH140 (AY169293); 18, DH37 (AY169296); 19, India 18 (AF202224); 20, DH60 (AY169297); 21, India19 (AF202225); 22, DH29 (AY169295); 23, DH200 (AY169294); 24, DUT79 (AJ252970); 25, DUT25 (AJ252968); 26, Guatemala88 (AF198472); 27, Gambia4797 (AF198472); 28, South Africa19 (AF198470); 29, Peru4A (AF198477); 30, Gambia4659 (AF198468); 31, Dutch107 (AJ252963); 32, 26695 (AE000569); 33, G31 (AY169299); 34, DH102 (AY196292); 35, Peru34B (AF198475). Isolates from Bangladesh are shown in boldface type. (B) Phylogenetic trees of sequences within the vacAm1 alleles. Sequences from non-Bangladeshi strains used here were from public databases as indicated. The tree is based on a 648-bp segment of vacA gene containing the vacAm1 allele of Bangladeshi H. pylori strains determined in this study. The tree was generated by using PHYLIP (Phylogeny Inference Package), version 3.5c, of J. Felsenstein. The sequences of East Asian, European, and Indian vacAm1 alleles were taken from GenBank (GenBank accession number in parentheses): 1, GERM19 (AJ006967); 2, DH131 (AY16928); 3, DH92 (AY169289); 4, India48 (AF220112); 5, DH60 (AY169288); 6, India89 (AF220114); 7, India19 (AFAF220111); 8, India226 (AF220115); 9, DH153 (AY169286); 10, DH 9 (DH169290); 11, DH114 (AY169291); 12, India18 (AF220110); 13, India227 (AF220116); 14, India66 (AF220113); 15, India230 (AF220117); 16, 26695 (AE000598); 17, NCTC11638 (U07145); 18, Poland492 (AF097570); 19, NCTC11637 (AF049653); 20, Poland278 (AF097571); 21 Japan99 (AE001511); 22, Kenya AFN4847 (AF191644); 23, Chile-CH1 (AF479031); 24 Mex4467 (AF159855); 25, JapanF63 (AF049635); 26, China13 (AF035610); 27, ChinaR59 (AF035611); 28, JapanF52 (AF049631); 29, JapanF55 (AF049632); 30, JapanF72 (AF049651); 31, JapanF61 (AF049645); 32, JapanF42 (AF049626); 33, JapanF36 (AF049462); 34, JapanF64 (AF049647); 35, JapanF35 (AF049625); 36, JapanF45 (AF049628); 37, JapanF47 (AF049629); 38, JapanF57 (AF049634). Isolates from Bangladesh are shown in boldface type.
(ii) vacA alleles.
The presence of the toxigenic vacAs1 or the nontoxigenic vacAs2 allele at the 5′ end of vacA was determined on the basis of the sizes of PCR products generated with specific primers. Of the 37 cultures tested from patients with PU, 30 yielded only a 259-bp fragment, indicating the s1 allele; 6 yielded only a 286-bp fragment, indicating the s2 allele; and the remaining 1 yielded both the 259- and 286-bp fragments, indicating an s1 and s2 mixed infection (Table 2; Fig. 1C).
In equivalent PCR tests with DNA from strains of 20 patients with NUD, 12 yielded only the vacAs1 PCR product, 6 yielded the vacAs2 product, and 2 yielded both vacAs1 and vacAs2 products, again indicating mixed infections (Table 2). The general association of cag+ strains and vacAs1 and of strains lacking cag and vacAs2 observed here matches with that seen in many strain collections from the United States and Europe.
The prevalence of the alleles of middle (m) region of vacA, a part that affects host specificity, was also determined by PCR. Products were obtained only with vacAm1b primers in strains of 22 patients with PU and 8 patients with NUD (Table 2; Fig. 1D) whereas PCR amplification with only vacAm2 primers occurred in the cases of 13 of 37 patients with PU and 8 patients with NUD. Both m1b and m2 products were observed in two patients with PU and four patients with NUD.
Sequence analysis of mid-region alleles of vacA.
DNAs of 0.7-kb PCR fragments amplified with vacA middle region-specific primers containing the vacAmlb region were sequenced for further analysis. The sequence was then compared with available vacAm1a, -m1b, and recently described -m1c sequences in GenBank. Multiple sequence analysis showed all sequences from Bangladeshi isolates are closely related to each other and also closely related (97 to 98% identity) to a recently described vacAm1c allele from Calcutta, India (25). However, only 90 to 91% identity was obtained with both vacAm1b alleles from East Asia and vacAm1a of ethnic European strains. Phylogenetic analysis of the same sequence showed that Bangladeshi isolates are closely clustered to each other and are distantly related to East Asian and ethnic European clusters (Fig. 2B).
(iii) iceA alleles.
PCR was used to test for iceA1, which is virulence associated in the West, and the completely unrelated iceA2 gene, which can occupy the same chromosomal locus in strains lacking iceA1. The iceA1 gene was in isolates from 19 of 37 patients with PU disease and in nine strains from patients with NUD. In contrast, only the iceA2 gene was present in strains from 17 patients with PU and in 10 strains from patients with NUD. A mixture of iceA1 and iceA2 PCR products, indicating mixed infection, was found in 2 cultures out of a total of 57 (Table 2; Fig. 1E and F). A 94-bp deletion in the iceA1 gene was found in two isolates, both from patients with PU (Fig. 1G).
cag right junction motifs.
The type III motif at the extreme right end of cag PAI, characterized by the presence of a function-unknown sequence replacing the hel gene fragment (17), was found in 40 of 50 (∼80%) cag PAI-positive Bangladeshi strains as determined by PCR with diagnostic primers (Fig. 1H). By PCR, 7 of the remaining 10 strains were found to carry the type I motif, which is common in strains from the United States, Peru, South Africa, and Spain and usually contains a 130-bp remnant of IS606 and about 423 bp of helicase gene fragment (17). One Bangladeshi strain was positive for the type II motif that is predominant in strains of coastal China and Japan and characterized by the presence of 320-bp fragment of IS606 and only a 9-bp remnant of helicase gene (Table 2).
Distribution of neutral markers.
The prevalence of several mobile DNAs, which should have no effect on colonization or disease, was also studied, since their distribution may be far less affected by selection and thus serve better as a marker of ancient phylogenetic lineages than alleles of virulence-associated genes. PCR tests identified sequences from IS606 in 18 strains and IS605 in 23 strains of the 57 studied (Table 2). Further, eight cultures contained both these elements, and 23 strains lacked both of them. IS605 was present in 43% (17 of 39), 42% (3 of 7), and 27% (3 of 11) of the cagA gene-positive cultures, mixed cultures, and cultures lacking cagA, respectively. Thus, each element appears to be inherited independently of the others. IS608 was identified in 6 of the 37 isolates from patients with PU and 3 of the 20 patients with NUD.
DISCUSSION
H. pylori strains from Bangladesh were studied to gain new insight into the population genetic structure of the pathogen and to study the implication of genotype in disease. Although H. pylori infection occurs worldwide, there are significant differences in its prevalence both within and between countries (11, 23). In developing countries, including Bangladesh, the majority of children usually get infected during their first 1 to 2 years of life, and most others acquire H. pylori even before adulthood (18). An age-related prevalence study in Bangladesh showed that 61% of infants (1 to 3 months old) were positive for H. pylori, and the figure rose to 84% in 6- to 9-year-olds (22). Otherwise, the reported seroprevalence of H. pylori in the hospitalized Bangladeshi population was 77.4% (26). A high association of H. pylori with PU (duodenal ulcer, 77%; gastric ulcer, 75%) and gastritis (74%) was observed in a previous study (12). However, in the present study, H. pylori could be cultured from 56% of PU and 40% of patients with NUD reporting at Dhaka Medical College Hospital, Bangladesh. This may be due to a difference in methods for detection of H. pylori (other studies used serology, rapid-urease test or histology) and uneven distribution of the organism throughout the gastric mucosa. Since the main focus of this study was to determine the genetic makeup in relation to virulence of Bangladeshi H. pylori strains and since culture of H. pylori from gastric biopsy specimens undoubtedly constitutes the most specific way to establish the presence of the bacteria and to study the genotype, it was not deemed necessary to employ all other methods for detection of H. pylori in the study.
H. pylori strains carrying the cag PAI and the potentially toxigenic s1 alleles of the vacuolating cytotoxin (vacA) gene were found to predominate in Bangladesh, but at the same time, some strains carrying the vacAs2 allele and lacking the cag PAI were recovered either as a sole pathogen infecting a patient or coresident with cag+ strains. The frequency at which strains carrying vacAs2 allele and lacking cag PAI were found in Bangladesh was higher than that in East Asia, where essentially all strains studied to date have carried the cag PAI and vacAs1 allele (14, 28). It is also noteworthy that strains lacking cag PAI were present in cases of both overt disease and more benign infections in Bangladesh, as opposed to the West, where such strains were associated only with benign infections (5). This, however, is similar to the situation in India, where cag+ H. pylori strains were harbored by both diseased patients and asymptomatic carriers (25). Mutational tests of strains from the West had shown that cag PAI genes are needed to strongly induce synthesis of proinflammatory cytokines in cell culture, and thus they are inferred to contribute to the strong and potentially damaging human host inflammatory responses that underlie overt disease (2). Hence, our results were unexpected. One explanation could be that infection by such strains in Bangladesh are often mixed with infections by strains with cag PAI, the latter being chiefly responsible for the observed severe pathology. A second explanation for the results invokes host or environmental factors or other bacterial genetic determinants that might be more important than cag+ vacAs1 status as determinants of disease in Bangladesh. A third possibility is suggested by indications that expression of cag PAI specified virulence functions may be highly controlled, with the severity of down-regulation varying among strains (8). Conceivably, this negative regulation might be more severe in most strains recovered from healthy individuals, effectively converting such nominally cag+ strains into phenocopies of strains lacking cag.
Similar to the Indian strains, iceA2 was found to be present in a large fraction of H. pylori strains isolated from patients with PU in Bangladesh, although iceA1 had been more strongly associated with disease in the U.S. and European populations studied to date (36). Expression of iceA1 is induced by gastric epithelial cell contact; its protein product is a homologue of a restriction endonuclease and is believed to cleave DNA in vitro (31). iceA2 is completely unrelated to iceA1 in sequence, although the iceA1 and iceA2 genes occupy the same locus in different strains and antibodies against iceA2 are found in sera of some infected patients, implying that this protein is also produced during infection (10). Beyond these observations, however, little is known about the actual roles (if any) of iceA1 and iceA2 in human colonization and disease that would explain the basis of high iceA2 disease associations in Bangladesh.
DNA sequence phylogenetic analysis of the vacA middle region indicates that the strains from Bangladesh harbor a unique m1c allele that is common among isolates from the ethnic Bengali population in Calcutta (25). Studies on the distribution of neutral markers showed a significantly higher incidence of IS606, a 2-kb transposable element, in Bangladeshi strains compared to strains from China and Japan. IS606, which is found in one-third of strains from the United States and Europe but rarely in strains from East Asia (15), was present in 31.5% of the H. pylori strains from Bangladesh. This percentage is also higher than that observed for Indian strains, 17 to 18% of which normally carry IS606 (25).
A similarity between Bangladeshi and Indian strains was also reflected in the predominance of the type III motif at the cag PAI right junction. This motif was found in 85% of strains from Calcutta, whereas ∼80% of Bangladeshi strains were positive for type III. Otherwise, the type III motif was uncommon elsewhere in the world.
The present study detected cases of mixed infection, i.e., the coexistence of genetically different H. pylori strains in a single infection in persons with overt disease as well as benign infections. This is important in the context that we humans differ in traits important to individual H. pylori strains, and that as a consequence, a given strain may not always be well adapted to the person who happens to ingest it. In this framework, infections by strains of different and complementary genotypes should allow formation of recombinants that are better adapted than either parental strain to the given host (9, 16). Based on the presence or absence of cag PAI, seven subjects were found to have mixed infection; the number of mixed infections reported here may underestimate the true frequency in the Bangladeshi population. Because of the patchy distribution of the organism throughout the gastric mucosa, there exists considerable variation in the number of bacteria in biopsy specimens from different gastric sites. At the same time, H. pylori strains with different genotypes may predominate at different gastric sites, and any given biopsy specimen may contain only one strain or a subset of the mix of infecting strains that do not reflect the actual microenvironment of the host's stomach. Mixed H. pylori infection is also common in India.
Thus, the overall genotypes of H. pylori from Bangladesh bear a very close resemblance to those of the strains from Calcutta, India. The high frequency of cag+ vacAs1 strains, the sequence homology in vacAm1, the predominance of the type III motif, the occurrence of mixed infection, and the high frequency of IS606 were observed in strains of both the countries. This, however, is quite likely considering the close proximity of the two countries and the similar physiological environments, culinary practices, and lifestyles of the hosts.
Acknowledgments
This research was funded by the ICDDR, B: Centre for Health and Population Research, which is supported by countries and agencies that share its concern for the health problems of developing countries. Current donors providing unrestricted support include the aid agencies of the governments of Australia, Bangladesh, Belgium, Canada, Saudi Arabia, Sweden, Switzerland, the United Kingdom, and the United States of America; international organizations include United Nations Children's Fund (UNICEF) and support from SIDA/SAREC. The research was also supported by grants from the U.S. Public Health Service (AI38166, AI49161, DK53727, and P30 DK52574) and the INDO-US Vaccine Action Program to G. B. Nair and Douglas E. Berg.
REFERENCES
- 1.Achtman, M., T. Azuma, D. E. Berg, Y. Ito, G. Morelli, Z. J. Pan, S. Suerbaum, S. A. Thompson, A. van der Ende, and L. J. van Doorn. 1999. Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol. Microbiol. 32:459-470. [DOI] [PubMed] [Google Scholar]
- 2.Akopyants, N. S., S. W. Clifton, D. Kersulyte, J. E. Crabtree, B. E. Youree, C. A. Reece, N. O. Bukanov, E. S. Drazek, B. A. Roe, and D. E. Berg. 1998. Analyses of the cag pathogenicity island of Helicobacter pylori. Mol. Microbiol. 28:37-54. [DOI] [PubMed] [Google Scholar]
- 3.Akopyanz, N., N. O. Bukanov, T. U. Westblom, S. Kresovich, and D. E. Berg. 1992. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 20:5137-5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1993. Current protocols in molecular biology. Greene Publishing and Wiley-Interscience, New York, N.Y.
- 5.Atherton, J. C. 1998. H. pylori virulence factors. Br. Med. Bull. 54:105-120. [DOI] [PubMed]
- 6.Casswall, T. H., H. O. Nilsson, M. Bergstrom, P. Aleljung, T. Wadstrom, A. K. Dahlstrom, M. J. Albert, and S. A. Sarker. 1999. Evaluation of serology, 13C-urea breath test, and polymerase chain reaction of stool samples to detect Helicobacter pylori in Bangladeshi children. J. Pediatr. Gastroenterol. Nutr. 28:31-36. [DOI] [PubMed] [Google Scholar]
- 7.Clemens, J., M. J. Albert, M. Rao, S. Huda, F. Qadri, F. P. Van Loon, B. Pradhan, A. Naficy, and A. Banik. 1996. Sociodemographic, hygienic and nutritional correlates of Helicobacter pylori infection of young Bangladeshi children. Pediatr. Infect. Dis. J. 15:1113-1118. [DOI] [PubMed] [Google Scholar]
- 8.Crabtree, J. E., D. Kersulyte, S. D. Li, I. J. Lindley, and D. E. Berg. 1999. Modulation of Helicobacter pylori induced interleukin-8 synthesis in gastric epithelial cells mediated by cag PAI encoded VirD4 homologue. J. Clin. Pathol 52:653-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dubois, A., D. E. Berg, E. T. Incecik, N. Fiala, L. M. Heman-Ackah, J. Del Valle, M. Yang, H. P. Wirth, G. I. Perez-Perez, and M. J. Blaser. 1999. Host specificity of Helicobacter pylori strains and host responses in experimentally challenged nonhuman primates. Gastroenterology 116:90-96. [DOI] [PubMed] [Google Scholar]
- 10.Figueiredo, C., W. G. Quint, R. Sanna, E. Sablon, J. P. Donahue, Q. Xu, G. G. Miller, R. M. Peek, Jr., M. J. Blaser, and L. J. van Doorn. 2000. Genetic organization and heterogeneity of the iceA locus of Helicobacter pylori. Gene 246:59-68. [DOI] [PubMed] [Google Scholar]
- 11.Goodman, K. J., and P. Correa. 1995. The transmission of Helicobacter pylori. A critical review of the evidence. Int. J. Epidemiol. 24:875-887. [DOI] [PubMed] [Google Scholar]
- 12.Haque, M., K. M. Rahman, A. A. Khan, M. Hasan, M. R. A. Miah, T. Rahman, A. K. K. Alam, and F. Qadri. 1995. Isolation and characterization of Helicobacter pylori strains from peptic ulcer patients in Dhaka, Bangladesh. Indian J. Gastroenterol. 14:128-130. [PubMed] [Google Scholar]
- 13.Hildebrand, P., P. Bardhan, L. Rossi, S. Parvin, A. Rahman, M. S. Arefin, M. Hasan, M. M. Ahmad, K. Glatz-Krieger, L. Terracciano, P. Bauerfeind, C. Beglinger, N. Gyr, and A. K. Khan. 2001. Recrudescence and reinfection with Helicobacter pylori after eradication therapy in Bangladeshi adults. Gastroenterology 121:792-798. [DOI] [PubMed] [Google Scholar]
- 14.Ito, Y., T. Azuma, S. Ito, H. Suto, H. Miyaji, Y. Yamazaki, Y. Kohli, and M. Kuriyama. 1998. Full-length sequence analysis of the vacA gene from cytotoxic and noncytotoxic Helicobacter pylori. J. Infect. Dis. 178:1391-1398. [DOI] [PubMed] [Google Scholar]
- 15.Kersulyte, D., N. S. Akopyants, S. W. Clifton, B. A. Roe, and D. E. Berg. 1998. Novel sequence organization and insertion specificity of IS605 and IS606: chimaeric transposable elements of Helicobacter pylori. Gene 223:175-186. [DOI] [PubMed] [Google Scholar]
- 16.Kersulyte, D., H. Chalkauskas, and D. E. Berg. 1999. Emergence of recombinant strains of Helicobacter pylori during human infection. Mol. Microbiol. 31:31-43. [DOI] [PubMed] [Google Scholar]
- 17.Kersulyte, D., A. K. Mukhopadhyay, B. Velapatiño, W. W. Su, Z. J. Pan, C. Garcia, V. Hernandez, Y. Valdez, R. S. Mistry, R. H. Gilman, Y. Yuan, H. Gao, T. Alarcón, M. López-Brea, G. B. Nair, A. Chowdhury, S. Datta, M. Shirai, T. Nakazawa, R. Ally, I. Segal, B. C. Y. Wong, S. K. Lam, F. Olfat, T. Borén, L. Engstrand, O. Torres, R. Schneider, J. E. Thomas, S. Czinn, and D. E. Berg. 2000. Differences in genotypes of Helicobacter pylori from different human populations. J. Bacteriol. 182:3210-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klein, P. D., R. H. Gilman, R. Leon-Barua, F. Diaz, E. O. Smith, and D. Y. Graham. 1994. The epidemiology of Helicobacter pylori in Peruvian children between 6 and 30 months of age. Am. J. Gastroenterol. 89:2196-2200. [PubMed] [Google Scholar]
- 19.Labigne, A., V. Cussac, and P. Courcoux. 1991. Shuttle cloning and nucleotide sequences of Helicobacter pylori genes responsible for urease activity. J. Bacteriol. 173:1920-1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leunk, R. D. 1991. Production of a cytotoxin by Helicobacter pylori. Rev. Infect. Dis. 13:S686-S689. [DOI] [PubMed]
- 21.Leying, H., S. Suerbaum, G. Geis, and R. Haas. 1992. Cloning and genetic characterization of a Helicobacter pylori flagellin gene. Mol. Microbiol. 6:2863-2874. [DOI] [PubMed] [Google Scholar]
- 22.Mahalanabis, D., M. M. Rahman, S. A. Sarker, P. K. Bardhan, P. Hildebrand, C. Beglinger, and K. Gyr. 1996. Helicobacter pylori infection in the young in Bangladesh: prevalence, socioeconomic and nutritional aspects. Int. J. Epidemiol. 25:894-898. [DOI] [PubMed] [Google Scholar]
- 23.Mendall, M. A. 1997. Transmission of Helicobacter pylori. Semin. Gastrointest. Dis. 8:113-123. [PubMed] [Google Scholar]
- 24.Morshed, M. G., F. Jinnah, M. S. Islam, M. A. Rumi, S. Ahmed, M. M. Ahmed, M. Sadeque, and M. F. Chowdhury. 1997. Evaluation of culture, histological examination, serology and the rapid urease test for diagnosis of Helicobacter pylori in patients with dyspepsia in Bangladesh. Jpn. J. Med. Sci. Biol. 50:55-62. [DOI] [PubMed] [Google Scholar]
- 25.Mukhopadhyay, A. K., D. Kersulyte, J.-Y. Jeong, S. Datta, Y. Ito, A. Chowdhury, S. Chowdhury, A. Santra, S. K. Bhattacharya, T. Azuma, G. B. Nair, and D. E. Berg. 2000. Distinctiveness of genotypes of Helicobacter pylori in Calcutta, India. J. Bacteriol. 182:3219-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nessa, J., H. Chart, R. J. Owen, and B. Drasar. 2001. Human serum antibody response to Helicobacter pylori whole cell antigen in an institutionalized Bangladeshi population. J. Appl. Microbiol. 90:68-72. [DOI] [PubMed] [Google Scholar]
- 27.NIH Consensus Development Panel on Helicobacter pylori in Peptic Ulcer Disease. 1994. Helicobacter pylori in peptic ulcer disease. JAMA 272:65-69. [PubMed] [Google Scholar]
- 28.Pan, Z. J., D. E. Berg, R. W. van der Hulst, W. W. Su, A. Raudonikiene, S. D. Xiao, J. Dankert, G. N. Tytgat, and A. van der Ende. 1998. Prevalence of vacuolating cytotoxin production and distribution of distinct vacA alleles in Helicobacter pylori from China. J. Infect. Dis. 178:220-226. [DOI] [PubMed] [Google Scholar]
- 29.Parsonnet, J. (ed.). 1999. Microbes and malignancy: infection as a cause of human cancers, p. 372-408. Oxford University Press, New York, N.Y.
- 30.Parsonnet, J. 1995. Incidence of Helicobater pylori infection. Aliment. Ther. Pharm. 9:45-51. [PubMed] [Google Scholar]
- 31.Peek, R. M., Jr., S. A. Thompson, J. P. Donahue, K. T. Tham, J. C. Atherton, M. J. Blaser, and G. G. Miller. 1998. Adherence to gastric epithelial cells induces expression of a Helicobacter pylori gene, iceA, that is associated with clinical outcome. Proc. Assoc. Am. Phys. 110:531-544. [PubMed] [Google Scholar]
- 32.Sarker, S. A., M. M. Rahman, D. Mahalanabis, P. K. Bardhan, P. Hildebrand, C. Beglinger, and K. Gyr. 1995. Prevalence of Helicobacter pylori infection in infants and family contacts in a poor Bangladesh community. Dig. Dis. Sci. 40:2669-2672. [DOI] [PubMed] [Google Scholar]
- 33.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, J. C. Venter, et al. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 34.van der Ende, A., Z.-J. Pan, A. Bart, R. W. M. van der Hulst, M. Feller, S.-D. Xiao, G. N. J. Tytgat, and J. Dankert. 1998. cagA-positive Helicobacter pylori populations in China and The Netherlands are distinct. Infect. Immun. 66:1822-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Doorn, L. J., C. Figueiredo, F. Megraud, S. Pena, P. Midolo, D. M. Queiroz, F. Carneiro, B. Vanderborght, M. D. Pegado, R. Sanna, W. De Boer, P. M. Schneeberger, P. Correa, E. K. Ng, J. Atherton, M. J. Blaser, and W. G. Quint. 1999. Geographic distribution of vacA allelic types of Helicobacter pylori. Gastroenterology 116:823-830. [DOI] [PubMed] [Google Scholar]
- 36.van Doorn, L. J., C. Figueiredo, R. Sanna, A. Plaisier, P. Schneeberger, W. de Boer, and W. Quint. 1998. Clinical relevance of the cagA, vacA, and iceA status of Helicobacter pylori. Gastroenterology 115:58-66. [DOI] [PubMed] [Google Scholar]
- 37.Veldhuyzen van Zanten, S. J. O., and A. Lee. 1999. The role of Helicobacter pylori infection in duodenal and gastric ulcer. p. 47-56. In T. U. Westblom, S. J. Czinn, and J. G. Nedrud (ed.), Gastroduodenal disease and Helicobacter pylori: pathophysiology, diagnosis and treatment. Springer-Verlag, Berlin, Germany. [DOI] [PubMed]
- 38.Yamaoka, Y. D., D. H. Kwon, and D. Y. Graham. 2000. A Mr 34,000 proinflammatory outer membrane protein (oipA) of Helicobacter pylori. Proc. Natl. Acad. Sci. USA 97:7533-7538. [DOI] [PMC free article] [PubMed] [Google Scholar]