Abstract
A collection of 147 isolates of Burkholderia pseudomallei, B. mallei, and B. thailandensis was characterized by multilocus sequence typing (MLST). The 128 isolates of B. pseudomallei, the causative agent of melioidosis, were obtained from diverse geographic locations, from humans and animals with disease, and from the environment and were resolved into 71 sequence types. The utility of the MLST scheme for epidemiological investigations was established by analyzing isolates from captive marine mammals and birds and from humans in Hong Kong with melioidosis. MLST gave a level of resolution similar to that given by pulsed-field gel electrophoresis and identified the same three clones causing disease in animals, each of which was also associated with disease in humans. The average divergence between the alleles of B. thailandensis and B. pseudomallei was 3.2%, and there was no sharing of alleles between these species. Trees constructed from differences in the allelic profiles of the isolates and from the concatenated sequences of the seven loci showed that the B. pseudomallei isolates formed a cluster of closely related lineages that were fully resolved from the cluster of B. thailandensis isolates, confirming their separate species status. However, isolates of B. mallei, the causative agent of glanders, recovered from three continents over a 30-year period had identical allelic profiles, and the B. mallei isolates clustered within the B. pseudomallei group of isolates. Alleles at six of the seven loci in B. mallei were also present within B. pseudomallei isolates, and B. mallei is a clone of B. pseudomallei that, on population genetics grounds, should not be given separate species status.
Burkholderia pseudomallei is the causative agent of melioidosis, an infectious disease that is largely restricted to Southeast Asia, northern Australia, and some other tropical and subtropical regions. B. pseudomallei is considered a saprophyte but occasionally causes serious invasive disease, including septicemia and pneumonia in susceptible individuals (6). The epidemiology of melioidosis is not entirely understood. Infections are mostly believed to occur by inoculation of the organism through puncture wounds in the skin or through cuts and abrasions from contaminated ground water (for example, in rice paddies), but infection by inhalation is also believed to occur, and infection by ingestion of contaminated drinking water has been described (5, 6). B. pseudomallei also causes melioidosis in a wide range of animal species, and at least in areas where the disease is endemic, the organism is commonly found in soil and groundwater (6, 21).
Environmental isolates previously considered to be B. pseudomallei have been shown to fall into two closely related groups on the basis of their different abilities to assimilate arabinose and differences in their DNA macrorestriction patterns and rRNA sequences (3, 4, 20, 24). Those that can assimilate arabinose (ara+) have been assigned a new species, Burkholderia thailandensis, and are considered avirulent, whereas the isolates that cannot assimilate arabinose (ara−) and that are associated with melioidosis are retained within the species B. pseudomallei (4, 20).
Burkholderia mallei is very closely related to B. pseudomallei and is the causative agent of glanders in horses and other equines and, occasionally, in humans and other animals (10, 12). Human infections with B. mallei are broadly similar in clinical presentation to those caused by B. pseudomallei. Glanders is now a very rare disease in Europe and North America and is largely an animal disease restricted to parts of Africa, Asia, the Middle East, and Central and South America. Both of these species cause serious disease which can be acquired by the aerosol route and which have been considered by some to be potential agents of biological warfare or bioterrorism and are in category B of the list of bioterrorism agents of the Centers for Disease Control and Prevention.
A number of molecular typing procedures have been used to investigate the epidemiology of melioidosis, including random amplified polymorphic DNA analysis, ribotyping, and pulsed-field gel electrophoresis (PFGE) (5, 9, 21, 24, 25). These methods are not well suited to interlaboratory comparisons, and molecular typing procedures that use nucleotide sequence data rather than DNA fragment patterns are increasingly being used. Of these, multilocus sequence typing (MLST) provides the most appropriate method, as it indexes variations within fragments of seven housekeeping genes that are expected mostly to be selectively neutral (16). The different sequences at each of the seven loci are assigned different allele numbers, and the series of seven integers that corresponds to the allele numbers at the seven loci define the allelic profile of a strain. Nucleotide sequences are unambiguous and are easily compared between laboratories, allowing strain characterization over the Internet by the interrogation of a website that holds the sequences of all of the known alleles at each locus and the allelic profiles of all previously characterized strains (22). MLST schemes and databases have been described for a number of important bacterial pathogens, including Neisseria meningitidis (16) Streptococcus pneumoniae (7), and Staphylococcus aureus (8); and MLST has become an established technique for the unambiguous and precise characterization of isolates and for epidemiological studies.
In this paper we describe the development of an MLST scheme for B. pseudomallei and closely related species. We demonstrate the utility of the MLST scheme for epidemiological studies of melioidosis and clarify the genetic relationships between B. pseudomallei, B. thailandensis, and B. mallei. MLST confirms that the first two species are distinct but that the last one is a clone of B. pseudomallei.
MATERIALS AND METHODS
Properties of bacterial strains.
A total of 147 Burkholderia isolates were characterized by MLST (including 1 B. pseudomallei isolate whose allelic profile was obtained from the genome sequence). The collection included 133 isolates assigned to the species B. pseudomallei; but from the MLST results, 2 of these isolates were subsequently reassigned to the species B. thailandensis-like and 3 of these isolates were reassigned to an unassigned Burkholderia species. Isolates with the SID prefix were received by the Central Public Health Laboratory, Colindale, London, United Kingdom, and mostly were from individuals who had acquired melioidosis abroad; in some cases, the country in which the disease was acquired was not known.
Thirty-seven isolates recovered between 1975 and 1999 were from an epidemiological investigation into cases of melioidosis among captive animals and humans in Hong Kong (unpublished data). Some of these isolates were expected to be the same since they were taken from soil or water in the cage of the same captive animal or from animals or humans at different times during the course of their disease. A subset of 17 of these isolates was chosen to compare the results obtained by MLST with those obtained by PFGE.
The 37 isolates from Hong Kong and a small number of duplicate isolates from the same patient were excluded, and 86 B. pseudomallei isolates were thus chosen to represent the diversity within this species. These were recovered from at least 21 countries (the sources of some isolates could not be ascertained) on five continents between 1949 and 2000 and were from cases of melioidosis in humans and animals and from the environment.
The five B. mallei strains were from the National Collection of Type Cultures and were recovered from cases of disease in humans and equines in the Far East, Asia, and Eastern Europe between 1932 and 1961. Nine environmental arabinose-assimilating isolates that had been assigned to the species B. thailandensis were included and were from Thailand, Laos, and Vietnam. Three isolates were from an investigation of an occurrence of invasive disease following soil contamination of wounds received during a tractor accident in Oklahoma and have been proposed to be B. pseudomallei (17, 26).
Growth of bacteria and preparation of chromosomal DNA.
All handling of live organisms was undertaken in a category III containment facility at the Central Public Health Laboratories. The bacteria were grown on Tryptone soy agar (Oxoid Ltd., Basingstoke, United Kingdom), and dense bacterial suspensions were prepared and boiled for 15 min. An aliquot of each boiled suspension was checked for the absence of living bacteria, and they were then transferred to Imperial College, London, where DNA was extracted from the suspensions by using a DNeasy tissue kit (Qiagen Inc., Valencia, Calif.).
PFGE.
PFGE was carried out with XbaI as described previously (25), and the relatedness among strains was determined from the number of DNA fragments in common and was displayed as a dendrogram by using Bionumerics software.
Multilocus sequence typing.
The following pairs of primers were used for the amplification of the housekeeping gene fragments: ace-up (5′-GAATCGCCTTCACCATGTC-3′) and ace-dn (5′-CCGCGCTTCTCAAAACGATA-3′), gltB-up (5′-ACGCTCGCGATCGCGATGAA-3′) and gltB-dn (5′-TTCAGCAGGAGCGTCTGCTG-3′), gmhD-up (5′-GCAGTTCCTGTATGCGTA-3′) and gmhD-dn (5′-GAAGCACTGGTACTTGCC-3′), lepA-up (5′-CATATTCGCAATTTCTCGATC-3′) and lepA-dn (5′-CACGAGCATCACGACGCCG-3′), lipA-up (5′-GGCACCGCGACGTTCATG-3′) and lipA-dn (5′-GACCATCAGGCCCGATTTCG-3′), narK-up (5′-CTACTCGTGCGCTGGGAT-3′) and narK-dn (5′-GACGATGACGGCACCCAC-3′), and ndh-up (5′-AGTCGCGACGTTCTACAC-3′) and ndh-dn (5′-CGAGTTGCAGACGAGATAT-3′). For each locus, DNA synthesis from the “up” primer occurs in the direction of transcription.
PCRs (reaction volumes, 50 μl) were carried out in a 96-well microtiter plate format with a PTC-200 DNA engine (MJ Research Inc., Waltham, Mass.), with initial denaturation at 95°C for 4 min, followed by 30 cycles of 95°C for 30 s, 62°C for 30 s, and 72°C for 60 s. The samples were then maintained at 72°C for a further 10 min, cooled to 4°C, and stored at −20°C. The amplified DNA fragments were precipitated with 20% polyethylene glycol 8000-2.5 M NaCl, washed twice in 70% ethanol, dried, and resuspended in sterile water. The DNA fragments on each strand were sequenced with the primers used in the initial PCR amplification by using an ABI PRISM BigDye Terminator Cycle Sequencing kit (Applied Biosystems, Foster City, Calif.) and an ABI 3700 DNA sequencer. The forward and reverse sequences were trimmed to the correct length and edited.
Data analysis.
For each housekeeping locus, the different sequences obtained from the Burkholderia isolates were assigned as distinct alleles by using the Macintosh program Sequence Output (available from www.mlst.net). Each isolate was defined by a string of seven integers (the allelic profile), which correspond to the allele numbers at the seven loci, in the order ace-gltB-gmhD-lepA-lipA-narK-ndh. Each unique allelic profile is considered a clone and is assigned a sequence type (ST), which also provides a convenient descriptor for the clone (16, 22). An MLST database containing the sequences of all alleles, the allelic profiles, and information about the Burkholderia isolates, together with analysis tools, is maintained at Imperial College (London, United Kingdom) and can be found on the Burkholderia pages of the MLST website (www.mlst.net).
The relatedness of isolates characterized by MLST was obtained by cluster analysis by using the matrix of pairwise differences in their allelic profiles and the unweighted pair group method with arithmetic averages (UPGMA; Statistica, StatSoft Inc., Tulsa, Okla.).
For each of the 81 STs resolved among the 147 Burkholderia isolates, the sequences at the seven loci were concatenated in the order of loci used to define the allelic profile. Since the gene fragments all started at position 1 of a codon and ended at position 3, the +1 reading frame was maintained throughout the concatenate. The ndh fragment spanned two overlapping reading frames, and for concatenation the nucleotides T and G within the junction sequence AAATGA were removed to maintain the +1 reading frame across the junction (see Results). A facility for concatenating the seven sequences (with removal of nucleotides T and G within ndh) is available at the Burkholderia pages of the MLST website (www.mlst.net).
A minimum-evolution tree was constructed from the concatenated sequences (3,399 bp) by using all nucleotide sites and the Kimura 2-parameter method for estimating pairwise genetic distances. An initial tree was obtained by the neighbor-joining method, and the minimum-evolution method was used to search for the tree which minimizes the sums of the branch length estimates by branch swapping by closest-neighbor interchange (23). The degree of statistical support for the nodes on the tree was evaluated by examining their percent recovery in 1,000 resampled trees by the bootstrap test (19). The minimum-evolution tree and measures of sequence diversity were obtained by using the MEGA program (version 2.1) (15).
RESULTS
Selection of housekeeping genes for the MLST scheme.
Candidate housekeeping genes were selected by using the sequences of B. pseudomallei K96243, a clinical isolate from Thailand, available at the Sanger Institute website (www.sanger.ac.uk/Projects/B_pseudomallei/). The contigs available at the time that this work was initiated were analyzed by using the BLASTX program, and housekeeping genes (those involved in essential metabolic processes) that were flanked by other housekeeping genes and that appeared to be devoid of nearby genes that might be under diversifying selection from the host immune system or that might have been subject to high rates of horizontal gene transfer were selected.
Primers that allowed the amplification by PCR of approximately 550-bp fragments from the candidate MLST loci were designed, and the same primers were used to sequence the fragments on each strand. For initial selection of MLST loci and primers, a set of 24 isolates that included 19 B. pseudomallei isolates and 5 B. thailandensis isolates was used, since primers that amplified both of these species were required. Several of the candidate housekeeping gene fragments gave good-quality sequences, but examination of the sequence traces showed that, for unknown reasons, there were two overlapping peaks, suggesting two different nucleotides at a few sites. Seven gene fragments that did not show this phenomenon were selected for use in the final MLST scheme (Table 1). One possible reason for the phenomenon described above would be the presence of two extremely similar copies of some genes on the B. pseudomallei genome. The sequences of the seven MLST loci selected were therefore compared to the recently completed genome sequence of B. pseudomallei. Six of the sequences gave the expected single perfect match, but the ace gene fragment detected a gene on chromosome II that had 69% nucleotide similarity (52% amino acid similarity), in addition to the perfect match on chromosome I. The primers used for PCR amplification and sequencing of the ace fragment did not amplify this divergent homolog of the ace gene.
TABLE 1.
Properties of the loci used in the B. pseudomallei MLST scheme
| Locus | Gene function | No. of allelesa | No. of variable sitesa | Genome location (kb) |
|---|---|---|---|---|
| ace | Acetyl coenzyme A reductase | 4 | 7 | 1,780 |
| gltB | Glutamate synthase | 7 | 12 | 3,761 |
| gmhD | ADP glycerol-mannoheptose epimerase | 15 | 19 | 3,023 |
| lepA | GTP-binding elongation factor | 6 | 10 | 2,938 |
| lipA | Lipoic acid synthetase | 7 | 12 | 448 |
| narK | Nitrite extrusion protein | 14 | 18 | 2,784 |
| ndh | NADH dehydrogenase | 7 | 12 | 1,400 |
Alleles and variable sites are those in the 128 isolates assigned to the species B. pseudomallei.
After completion of the genome sequence, the ndh fragment was found to include parts of two overlapping genes that encode 282 bp from the end of the E subunit and 162 bp from the start of the F subunit of NADH dehydrogenase I. The junction sequence AAATGA includes the final codon of the upstream gene (AAA; lysine) and its TGA termination codon, and the last nucleotide of the lysine codon is the first nucleotide of the ATG initiation codon of the downstream gene. The entire ndh fragment used in the MLST scheme therefore corresponds to a protein-coding region.
Following the completion of determination of the genome sequence of strain K96243, the locations of the seven housekeeping genes on the two circular chromosomes could be examined. All seven genes were located on chromosome I and were separated by at least 80 kb (Table 1).
Diversity and relatedness of alleles from the Burkholderia isolates.
The seven gene fragments were sequenced from the 147 Burkholderia isolates. Figure 1 shows the polymorphic sites within the different alleles at the seven loci. At each locus there were three distinct groups of alleles. One group of very similar alleles was found within those isolates assigned to the species B. pseudomallei, and a second group of similar alleles was found among isolates assigned to the species B. thailandensis. At each locus there was one allele that was divergent from both of these groups of alleles; these divergent alleles were present in only three identical isolates from Oklahoma, which tentatively (and it appears wrongly) had been assigned to the species B. pseudomallei (17, 26). The average divergence between the alleles of the B. pseudomallei and B. thailandensis isolates was 3.2%, and the average divergences between the alleles in the Oklahoma isolates and those of the B. pseudomallei and B. thailandensis isolates were 5.2 and 4.7%, respectively. Two additional isolates that had previously been assigned to the species B. pseudomallei (isolates 82172 and 1992/2572) possessed alleles that were very similar to, although distinct from, the alleles found in B. thailandensis; these isolates were also not considered to be members of the species B. pseudomallei.
FIG. 1.
Variable sites within the alleles at the seven MLST loci. The sequences of all of the alleles at each locus that are represented among the 147 Burkholderia isolates are shown. Only the variable sites are shown, and these are numbered in vertical format. For allele 1 at each locus, the nucleotide present at each variable site is shown. For other alleles, only those sites where the nucleotides differ from those in allele 1 are shown; sites that have the same nucleotide as that in allele 1 are shown by a dot. The alleles in normal font are from B. pseudomallei, those in boldface are from B. thailandensis, and the final allele at each locus (in italics) is from the Oklahoma isolates.
Among all Burkholderia isolates, the number of alleles per locus varied from 7 to 19. Among the 128 isolates, between 4 and 15 alleles were assigned to the species B. pseudomallei by MLST (average, 8.6), allowing about 3.4 million different allelic profiles to be distinguished within this species (Table 1). However, the level of sequence diversity within B. pseudomallei was low (average, 0.2%), and an average of 18% of all alleles at a locus differed at only a single nucleotide site.
Genetic diversity and relationships between Burkholderia isolates.
There were 81 different allelic profiles among the 147 Burkholderia isolates (Table 2). Figure 2 shows a UPGMA tree obtained by using the matrix of pairwise differences in the allelic profiles of the isolates. All B. pseudomallei isolates were grouped together and were resolved from isolates assigned to the species B. thailandensis and from a few isolates that were assigned as possibly belonging to the species B. pseudomallei. The alleles at all seven loci in the B. thailandensis isolates were different from those at the seven loci in all B. pseudomallei isolates. The three isolates from Oklahoma that had tentatively been assigned to the species B. pseudomallei were identical by MLST (ST81) but differed from all other isolates at all seven loci. Similarly, the allelic profiles of the two isolates of ST73 (isolates 82172 and 1992/2572; see above) differed from those of all other isolates at all loci.
TABLE 2.
Properties of the Burkholderia strains studied
| Strain | Source | Country or region | Species | Yr | ST | Allele at the following locus:
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ace | gltB | gmhD | lepA | lipA | narK | ndh | ||||||
| 7641 | Horse | France | B. pseudomallei | 1976 | 1 | 1 | 1 | 1 | 2 | 4 | 2 | 1 |
| Cam 70 | Human | Cambodia | B. pseudomallei | 1997 | 2 | 1 | 1 | 2 | 4 | 1 | 1 | 1 |
| SID 5278 | Human | Thailand | B. pseudomallei | 2000 | 3 | 1 | 1 | 2 | 2 | 5 | 3 | 1 |
| 2381f | Human | Thailand | B. pseudomallei | 1999 | 4 | 1 | 1 | 4 | 2 | 1 | 2 | 1 |
| AB2056 | Human | Kenya | B. pseudomallei | 1980 | 5 | 1 | 1 | 5 | 1 | 1 | 1 | 1 |
| 770429 | Environment | Madagascar | B. pseudomallei | 1977 | 6 | 1 | 1 | 7 | 2 | 4 | 5 | 1 |
| Ducrete | Human | Vietnam | B. pseudomallei | 1963 | 7 | 1 | 1 | 10 | 2 | 5 | 1 | 1 |
| E25 | Environment | Thailand | B. pseudomallei | 1990 | 8 | 1 | 1 | 11 | 6 | 1 | 3 | 1 |
| 5689/299 | Human | Papua New Guinea | B. pseudomallei | 1989 | 9 | 1 | 1 | 12 | 1 | 1 | 1 | 1 |
| K96243a | Human | Thailand | B. pseudomallei | 1999 | 10 | 1 | 1 | 13 | 1 | 1 | 1 | 1 |
| SID 4350a | Human | Thailand | B. pseudomallei | 1999 | 10 | 1 | 1 | 13 | 1 | 1 | 1 | 1 |
| 7894/300 | Ecuador | B. pseudomallei | 11 | 1 | 1 | 13 | 1 | 6 | 1 | 1 | ||
| D4899/303 | Venezuela | B. pseudomallei | 12 | 1 | 1 | 13 | 1 | 5 | 1 | 1 | ||
| 84-1097 | Sheep | Australia | B. pseudomallei | 1984 | 13 | 1 | 1 | 13 | 1 | 1 | 4 | 1 |
| 85-1097 | Cow | Australia | B. pseudomallei | 1985 | 13 | 1 | 1 | 13 | 1 | 1 | 4 | 1 |
| Cam Pus | Human | Cambodia | B. pseudomallei | 1997 | 14 | 1 | 2 | 2 | 2 | 5 | 2 | 1 |
| Hainan 4 | China | B. pseudomallei | 15 | 1 | 2 | 2 | 2 | 1 | 3 | 1 | ||
| KK 520 | Human | Thailand | B. pseudomallei | 1992 | 16 | 1 | 2 | 2 | 1 | 1 | 10 | 1 |
| Ln34170 | Human | Malaysia | B. pseudomallei | 17 | 1 | 2 | 3 | 1 | 1 | 1 | 1 | |
| AB2056 | Kenya | B. pseudomallei | 18 | 1 | 2 | 3 | 2 | 1 | 1 | 1 | ||
| NT20 | Environment | Burkina Faso | B. pseudomallei | 1973 | 19 | 1 | 2 | 6 | 1 | 1 | 3 | 1 |
| NT08 | Environment | Niger | B. pseudomallei | 1973 | 20 | 1 | 2 | 6 | 2 | 5 | 6 | 4 |
| S12 | Human | Singapore | B. pseudomallei | 1988 | 21 | 1 | 2 | 11 | 4 | 1 | 4 | 1 |
| OZ 431 | Human | Australia | B. pseudomallei | 22 | 1 | 2 | 12 | 2 | 1 | 8 | 1 | |
| X1003 | Goat | Australia | B. pseudomallei | 23 | 1 | 2 | 13 | 1 | 1 | 1 | 1 | |
| SID 4351 | Human | Thailand | B. pseudomallei | 1999 | 23 | 1 | 2 | 13 | 1 | 1 | 1 | 1 |
| No212 (dra) | B. pseudomallei | 24 | 1 | 2 | 13 | 2 | 1 | 8 | 1 | |||
| OZ 659 | Human | Australia | B. pseudomallei | 1998 | 25 | 1 | 3 | 3 | 1 | 1 | 8 | 1 |
| NT08 | Environment | Niger | B. pseudomallei | 26 | 1 | 3 | 10 | 2 | 5 | 1 | 1 | |
| Soil1977 | Soil | Madagascar | B. pseudomallei | 1997 | 27 | 1 | 3 | 19 | 4 | 5 | 1 | 1 |
| Thai 18-QM22 | Human | Thailand | B. pseudomallei | 1985 | 28 | 1 | 4 | 3 | 1 | 1 | 3 | 6 |
| SID 4352 | Human | Thailand | B. pseudomallei | 1999 | 29 | 1 | 4 | 3 | 1 | 1 | 4 | 1 |
| Hainan 2 | China | B. pseudomallei | 30 | 1 | 4 | 6 | 1 | 5 | 4 | 1 | ||
| WS-26G(Sal)/00 | Water from aviary | Hong Kong | B. pseudomallei | 2000 | 31 | 1 | 4 | 11 | 2 | 5 | 4 | 6 |
| A2b | Bottlenose dolphin | Hong Kong | B. pseudomallei | 1976 | 32 | 1 | 4 | 11 | 3 | 5 | 4 | 6 |
| NR1A3b | Human | Hong Kong | B. pseudomallei | 1999 | 32 | 1 | 4 | 11 | 3 | 5 | 4 | 6 |
| NR1A7 | Human | Hong Kong | B. pseudomallei | 1999 | 32 | 1 | 4 | 11 | 3 | 5 | 4 | 6 |
| NR1A8b | Human | Hong Kong | B. pseudomallei | 1996 | 32 | 1 | 4 | 11 | 3 | 5 | 4 | 6 |
| NR1A9 | Human | Hong Kong | B. pseudomallei | 1999 | 32 | 1 | 4 | 11 | 3 | 5 | 4 | 6 |
| Zebra Doveb | Zebra dove | Hong Kong | B. pseudomallei | 2000 | 32 | 1 | 4 | 11 | 3 | 5 | 4 | 6 |
| Gloria∗ | California sea lion | Hong Kong | B. pseudomallei | 2000 | 32 | 1 | 4 | 11 | 3 | 5 | 4 | 6 |
| A1 | Bottlenose dolphin | Hong Kong | B. pseudomallei | 1975 | 32 | 1 | 4 | 11 | 3 | 5 | 4 | 6 |
| A10b | Bottlenose dolphin | Hong Kong | B. pseudomallei | 1976 | 32 | 1 | 4 | 11 | 3 | 5 | 4 | 6 |
| A4b | Bottlenose dolphin | Hong Kong | B. pseudomallei | 1975 | 32 | 1 | 4 | 11 | 3 | 5 | 4 | 6 |
| VNs34A | Environment | Thailand | B. pseudomallei | 1997 | 33 | 1 | 4 | 12 | 1 | 1 | 2 | 1 |
| VN Hemo | Human | Vietnam | B. pseudomallei | 1995 | 33 | 1 | 4 | 12 | 1 | 1 | 2 | 1 |
| Ln29564 | Human | Malaysia | B. pseudomallei | 34 | 1 | 4 | 12 | 1 | 1 | 4 | 1 | |
| Ln31348 | Human | Malaysia | B. pseudomallei | 34 | 1 | 4 | 12 | 1 | 1 | 4 | 1 | |
| 7894 | Human | Ecuador | B. pseudomallei | 35 | 1 | 6 | 14 | 2 | 8 | 8 | 4 | |
| OZ 303 | Human | Australia | B. pseudomallei | 1994 | 36 | 1 | 7 | 14 | 7 | 1 | 12 | 11 |
| Whale 2b | Pilot whale | Hong Kong | B. pseudomallei | 1984 | 37 | 1 | 6 | 17 | 2 | 1 | 15 | 11 |
| A12 | Pilot whale | Hong Kong | B. pseudomallei | 1978 | 37 | 1 | 6 | 17 | 2 | 1 | 15 | 11 |
| HK20 | Human | Hong Kong | B. pseudomallei | 1986 | 37 | 1 | 6 | 17 | 2 | 1 | 15 | 11 |
| E1 | Soil | Hong Kong | B. pseudomallei | 1978 | 37 | 1 | 6 | 17 | 2 | 1 | 15 | 11 |
| HK26b | Human | Hong Kong | B. pseudomallei | 1986 | 37 | 1 | 6 | 17 | 2 | 1 | 15 | 11 |
| A3 | Pilot whale | Hong Kong | B. pseudomallei | 1978 | 37 | 1 | 6 | 17 | 2 | 1 | 15 | 11 |
| A5 | Bottlenose dolphin | Hong Kong | B. pseudomallei | 1975 | 37 | 1 | 6 | 17 | 2 | 1 | 15 | 11 |
| A6b | Bottlenose dolphin | Hong Kong | B. pseudomallei | 1976 | 37 | 1 | 6 | 17 | 2 | 1 | 15 | 11 |
| A7b | Bottlenose dolphin | Hong Kong | B. pseudomallei | 1976 | 37 | 1 | 6 | 17 | 2 | 1 | 15 | 11 |
| A8b | Harbour seal | Hong Kong | B. pseudomallei | 1975 | 37 | 1 | 6 | 17 | 2 | 1 | 15 | 11 |
| SID 3477 | Human | Thailand | B. pseudomallei | 1999 | 38 | 1 | 12 | 2 | 1 | 1 | 1 | 1 |
| SID 5752 | Human | Thailand | B. pseudomallei | 2000 | 39 | 1 | 12 | 3 | 2 | 1 | 6 | 1 |
| NCTC 03709 | Horse | India | B. mallei | 1932 | 40 | 1 | 12 | 3 | 4 | 1 | 18 | 1 |
| NCTC 10229 | Hungary | B. mallei | 1961 | 40 | 1 | 12 | 3 | 4 | 1 | 18 | 1 | |
| NCTC 10245 | Horse | China | B. mallei | 1942 | 40 | 1 | 12 | 3 | 4 | 1 | 18 | 1 |
| NCTC 10248 | Human | Turkey | B. mallei | 1950 | 40 | 1 | 12 | 3 | 4 | 1 | 18 | 1 |
| NCTC 10260 | Human | Turkey | B. mallei | 1949 | 40 | 1 | 12 | 3 | 4 | 1 | 18 | 1 |
| SID 5311 | Human | Southeast Asia | B. pseudomallei | 2000 | 41 | 1 | 12 | 6 | 1 | 5 | 2 | 1 |
| SID 3584 | Human | Vietnam | B. pseudomallei | 1998 | 41 | 1 | 12 | 6 | 1 | 5 | 2 | 1 |
| SID 3871c | Human | Bangladesh | B. pseudomallei | 1999 | 42 | 1 | 12 | 6 | 2 | 1 | 2 | 1 |
| SID 3811c | Human | Bangladesh | B. pseudomallei | 1999 | 43 | 1 | 12 | 10 | 2 | 1 | 2 | 1 |
| SID 4045c | Human | Bangladesh | B. pseudomallei | 1999 | 43 | 1 | 12 | 10 | 2 | 1 | 2 | 1 |
| NCTC 8016 | Sheep | Australia | B. pseudomallei | 1949 | 44 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| 2395a | Human | Thailand | B. pseudomallei | 1999 | 45 | 2 | 2 | 3 | 1 | 1 | 2 | 1 |
| NCTC 10276 | Human | Bangladesh | B. pseudomallei | 1960 | 46 | 3 | 1 | 2 | 1 | 1 | 3 | 3 |
| MK1867 | Monkey | Indonesia | B. pseudomallei | 1990 | 46 | 3 | 1 | 2 | 1 | 1 | 3 | 3 |
| SID 3783 | Human | Malaysia | B. pseudomallei | 1999 | 46 | 3 | 1 | 2 | 1 | 1 | 3 | 3 |
| SID 4151 | Human | Thailand | B. pseudomallei | 1999 | 46 | 3 | 1 | 2 | 1 | 1 | 3 | 3 |
| Thai20 | Human | Thailand | B. pseudomallei | 1986 | 46 | 3 | 1 | 2 | 1 | 1 | 3 | 3 |
| Thai19R | Human | Thailand | B. pseudomallei | 1986 | 46 | 3 | 1 | 2 | 1 | 1 | 3 | 3 |
| M7665/91 | Environment | Singapore | B. pseudomallei | 1991 | 47 | 3 | 1 | 2 | 1 | 1 | 3 | 1 |
| 2366a | Human | Thailand | B. pseudomallei | 1999 | 48 | 3 | 1 | 2 | 1 | 1 | 4 | 1 |
| SID 6025 | Human | Thailand | B. pseudomallei | 2000 | 49 | 3 | 1 | 2 | 1 | 6 | 4 | 3 |
| Hainan 106 | China | B. pseudomallei | 50 | 3 | 1 | 2 | 1 | 1 | 4 | 3 | ||
| 59 | Human | Singapore | B. pseudomallei | 1988 | 51 | 3 | 1 | 2 | 3 | 1 | 4 | 3 |
| NR1A2b,d | Human | Hong Kong | B. pseudomallei | 1998 | 51 | 3 | 1 | 2 | 3 | 1 | 4 | 3 |
| NR1A4d | Human | Hong Kong | B. pseudomallei | 1998 | 51 | 3 | 1 | 2 | 3 | 1 | 4 | 3 |
| NR1A6d | Human | Hong Kong | B. pseudomallei | 1998 | 51 | 3 | 1 | 2 | 3 | 1 | 4 | 3 |
| SID 4075 | Human | Thailand | B. pseudomallei | 1999 | 51 | 3 | 1 | 2 | 3 | 1 | 4 | 3 |
| E321 | Human | Thailand | B. pseudomallei | 1998 | 52 | 3 | 1 | 2 | 2 | 1 | 11 | 1 |
| Thai 9 | Human | Thailand | B. pseudomallei | 1986 | 53 | 3 | 1 | 2 | 1 | 1 | 16 | 1 |
| 204 | Human | Thailand | B. pseudomallei | 1988 | 54 | 3 | 1 | 3 | 3 | 1 | 2 | 1 |
| KK 454 | Human | Thailand | B. pseudomallei | 1992 | 54 | 3 | 1 | 3 | 3 | 1 | 2 | 1 |
| 956a | Human | Thailand | B. pseudomallei | 1992 | 54 | 3 | 1 | 3 | 3 | 1 | 2 | 1 |
| Hainan 55 | China | B. pseudomallei | 55 | 3 | 1 | 3 | 3 | 1 | 4 | 1 | ||
| SID 2889 | Human | Bangladesh | B. pseudomallei | 1999 | 56 | 3 | 1 | 4 | 1 | 1 | 4 | 1 |
| Thai 18-QM15 | Human | Thailand | B. pseudomallei | 1984 | 56 | 3 | 1 | 4 | 1 | 1 | 4 | 1 |
| MK 1900 | Monkey | Philippines | B. pseudomallei | 1990 | 57 | 3 | 1 | 5 | 1 | 1 | 1 | 1 |
| 7605 | Environment | France | B. pseudomallei | 1976 | 57 | 3 | 1 | 5 | 1 | 1 | 1 | 1 |
| 2396a | Human | Thailand | B. pseudomallei | 1999 | 58 | 3 | 1 | 5 | 1 | 1 | 4 | 1 |
| 34 | Environment | Kenya | B. pseudomallei | 1992 | 59 | 3 | 1 | 8 | 4 | 5 | 1 | 1 |
| 5892/339 | Human | Fiji | B. pseudomallei | 1992 | 60 | 3 | 1 | 12 | 1 | 1 | 3 | 1 |
| D228 | Environment | Australia | B. pseudomallei | 60 | 3 | 1 | 12 | 1 | 1 | 3 | 1 | |
| D260:53/30 | Environment | Australia | B. pseudomallei | 60 | 3 | 1 | 12 | 1 | 1 | 3 | 1 | |
| D304:S3/40 | Environment | Australia | B. pseudomallei | 60 | 3 | 1 | 12 | 1 | 1 | 3 | 1 | |
| OZ 373b | Human | Australia | B. pseudomallei | 1995 | 61 | 3 | 2 | 3 | 4 | 1 | 9 | 7 |
| MK441 | Monkey | Philippines | B. pseudomallei | 1990 | 62 | 3 | 3 | 4 | 1 | 3 | 1 | 1 |
| MK1831 | Monkey | Indonesia | B. pseudomallei | 1990 | 63 | 3 | 3 | 4 | 2 | 1 | 1 | 1 |
| 307a | Human | Thailand | B. pseudomallei | 1987 | 64 | 3 | 4 | 2 | 1 | 1 | 1 | 1 |
| Thai 7-3 | Human | Thailand | B. pseudomallei | 1984 | 65 | 3 | 4 | 3 | 3 | 1 | 3 | 1 |
| RAMAL22 | Human | Thailand | B. pseudomallei | 1990 | 66 | 3 | 4 | 3 | 4 | 1 | 4 | 1 |
| S6 | Human | Singapore | B. pseudomallei | 1988 | 67 | 3 | 4 | 3 | 4 | 1 | 4 | 6 |
| WS-22A(Sal)/00 | Water from aviary | Hong Kong | B. pseudomallei | 2000 | 68 | 3 | 4 | 11 | 1 | 5 | 4 | 6 |
| SS-35A(35G)/00 | Soil from aviary | Hong Kong | B. pseudomallei | 2000 | 69 | 3 | 4 | 11 | 2 | 5 | 4 | 6 |
| 383.5 | Human | Thailand | B. pseudomallei | 1992 | 70 | 3 | 4 | 11 | 3 | 5 | 4 | 6 |
| 383.9 | Human | Thailand | B. pseudomallei | 1992 | 70 | 3 | 4 | 11 | 3 | 5 | 4 | 6 |
| LE2 | Environment | Laos | B. pseudomallei | 1999 | 70 | 3 | 4 | 11 | 3 | 5 | 4 | 6 |
| 1986a | Human | Thailand | B. pseudomallei | 1998 | 70 | 3 | 4 | 11 | 3 | 5 | 4 | 6 |
| Margaretb | Scarlet macaw | Hong Kong | B. pseudomallei | 2000 | 70 | 3 | 4 | 11 | 3 | 5 | 4 | 6 |
| SS-32G/00 | Soil from aviary | Hong Kong | B. pseudomallei | 2000 | 70 | 3 | 4 | 11 | 3 | 5 | 4 | 6 |
| SS-34G/00 | Soil from aviary | Hong Kong | B. pseudomallei | 2000 | 70 | 3 | 4 | 11 | 3 | 5 | 4 | 6 |
| WS-20(Sal)/00b | Water from aviary | Hong Kong | B. pseudomallei | 2000 | 70 | 3 | 4 | 11 | 3 | 5 | 4 | 6 |
| WS-21(sal)/00 | Water from aviary | Hong Kong | B. pseudomallei | 2000 | 70 | 3 | 4 | 11 | 3 | 5 | 4 | 6 |
| WS-21G(Sal)/00 | Water from aviary | Hong Kong | B. pseudomallei | 2000 | 70 | 3 | 4 | 11 | 3 | 5 | 4 | 6 |
| WS-22(Sal)/00 | Water from aviary | Hong Kong | B. pseudomallei | 2000 | 70 | 3 | 4 | 11 | 3 | 5 | 4 | 6 |
| 102947b | Human | Hong Kong | B. pseudomallei | 1982 | 70 | 3 | 4 | 11 | 3 | 5 | 4 | 6 |
| 50562b | Human | Hong Kong | B. pseudomallei | 1982 | 70 | 3 | 4 | 11 | 3 | 5 | 4 | 6 |
| PS/102738 | Human | Hong Kong | B. pseudomallei | 1982 | 70 | 3 | 4 | 11 | 3 | 5 | 4 | 6 |
| 800498 | Human | Hong Kong | B. pseudomallei | 1991 | 70 | 3 | 4 | 11 | 3 | 5 | 4 | 6 |
| SID 4717e | Human | Bangladesh | B. pseudomallei | 1999 | 71 | 4 | 1 | 3 | 2 | 1 | 4 | 1 |
| SID 4935e | Human | Bangladesh | B. pseudomallei | 2000 | 71 | 4 | 1 | 3 | 2 | 1 | 4 | 1 |
| SID 4718e | Human | Bangladesh | B. pseudomallei | 1999 | 71 | 4 | 1 | 3 | 2 | 1 | 4 | 1 |
| 521 | Human | Pakistan | B. pseudomallei | 1988 | 72 | 4 | 1 | 4 | 1 | 1 | 2 | 1 |
| 82172 | Chicken | France | B. thailandensis? | 1982 | 73 | 5 | 5 | 9 | 5 | 7 | 7 | 5 |
| 1992/2572 | Water | Kenya | B. thailandensis? | 1992 | 73 | 5 | 5 | 9 | 5 | 7 | 7 | 5 |
| E27 | Environment | Thailand | B. thailandensis | 1990 | 74 | 6 | 8 | 15 | 8 | 9 | 13 | 8 |
| VN 534b | Environment | Vietnam | B. thailandensis | 1997 | 75 | 6 | 9 | 16 | 8 | 10 | 14 | 9 |
| LE1 | Environment | Laos | B. thailandensis | 1999 | 76 | 6 | 10 | 15 | 8 | 10 | 14 | 9 |
| E125 | Environment | Thailand | B. thailandensis | 1991 | 77 | 6 | 10 | 15 | 8 | 9 | 14 | 8 |
| E327 | Environment | Thailand | B. thailandensis | 1998 | 78 | 6 | 10 | 15 | 9 | 9 | 14 | 8 |
| E294 | Environment | Thailand | B. thailandensis | 1994 | 79 | 6 | 10 | 16 | 8 | 11 | 14 | 10 |
| E111 | Environment | Thailand | B. thailandensis | 1991 | 80 | 6 | 10 | 16 | 8 | 10 | 14 | 8 |
| E216 | Environment | Thailand | B. thailandensis | 1992 | 80 | 6 | 10 | 16 | 8 | 10 | 14 | 8 |
| G32 | Environment | B. thailandensis | 80 | 6 | 10 | 16 | 8 | 10 | 14 | 8 | ||
| C6756f | Human and environment | Oklahoma | Unassigned | 1970s | 81 | 7 | 11 | 18 | 10 | 12 | 17 | 12 |
| C7532f | Human and environment | Oklahoma | Unassigned | 1970s | 81 | 7 | 11 | 18 | 10 | 12 | 17 | 12 |
| C7552f | Human and environment | Oklahoma | Unassigned | 1970s | 81 | 7 | 11 | 18 | 10 | 12 | 17 | 12 |
K96243 is the strain used to obtain the genome sequence, and its allelic profile was obtained from the genome sequence; SID 4350 is another isolate of this strain that was analyzed by MLST.
Isolates studied by PFGE (Figure 4).
These three isolates were from a case of mother-to-child transmission of melioidosis (1); surprisingly, one isolate differed at a single locus.
These three isolates were from the same patient.
These three isolates were from the same patient: two during an initial disease episode and the third from a reoccurrence of melioidosis.
These three isolates were from invasive disease in a farm worker involved in an accident or from the soil that was considered to be the source of infection (17).
FIG. 2.
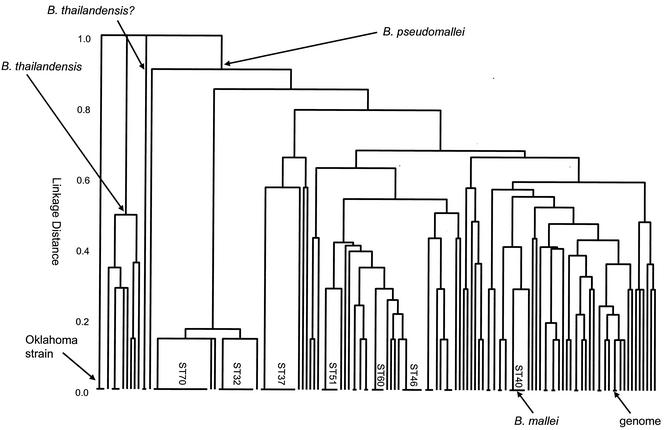
Relationships among Burkholderia isolates. A UPGMA tree was constructed from the matrix of pairwise differences in the allelic profiles of the 147 Burkholderia isolates. The nodes from which all B. pseudomallei and B. thailandensis isolates descend are marked. The five B. mallei isolates (ST40) have identical allelic profiles and cluster among the B. pseudomallei isolates. Two isolates that were assigned to the species B. pseudomallei but which in this study were found to be closely allied with B. thailandensis (shown as B. thailandensis?) and three isolates from Oklahoma that originally were tentatively assigned to the species B. pseudomallei had divergent allelic profiles and differed from all B. pseudomallei and B. thailandensis isolates at all seven loci. The STs that include at least four isolates and the strain used to obtain the genome sequence (K96243) are shown.
The five isolates of B. mallei had identical allelic profiles (ST40) and clustered with the B. pseudomallei isolates; for six of the seven MLST loci, the alleles in B. mallei were also found within B. pseudomallei isolates. The allele at the other locus (narK-18) was not found in any of the B. pseudomallei isolates, but it differed at only a single nucleotide site from one of the most common of the alleles in the latter species (narK-1). Inspection of the incomplete genome of B. mallei ATCC 23344, which was recovered in 1942 from a horse in China and which is being sequenced by The Institute for Genome Research (http://www.tigr.org/), showed that this strain also had an allelic profile identical to those of the other five B. mallei strains.
A total of 128 isolates were assigned to the species B. pseudomallei by MLST, and these were resolved into 71 STs. Among the B. pseudomallei isolates, there were 37 isolates from an epidemiological investigation of melioidosis in animals and humans in Hong Kong. These isolates and five further isolates that were from the same source as other isolates in the collection were removed, and the remaining 86 isolates provided a geographically and temporally diverse collection of B. pseudomallei isolates. There were 66 STs among these 86 isolates, and 12 of the STs included more than 1 isolate (range, 2 to 6 isolates).
The genome sequence of strain K96243, a clinical isolate from Thailand, has been obtained at the Sanger Institute, and the allelic profile was obtained from the genome sequence and was assigned to ST10 (Table 2). An isolate of the same strain that had been submitted to the Central Public Health Laboratory (SID 4350) was analyzed by MLST, and the sequences at the seven loci were identical to those obtained from the genome sequence.
Relationships among Burkholderia isolates and species using concatenated nucleotide sequences.
The UPGMA tree (Fig. 2) shows the clusters of identical and similar isolates, but it cannot resolve the deeper relationships between isolates (or between the species), since these typically differ at all or most MLST loci. The sequences at the seven loci were therefore concatenated (with removal of two nucleotides at the overlap between the two ndh reading frames; see above) to provide an in-frame sequence of 3,399 bp. A minimum-evolution tree was reconstructed from the concatenated sequences from all 81 STs (Fig. 3). The B. pseudomallei isolates were tightly clustered on the tree and were well resolved from the ara+ isolates assigned to the species B. thailandensis, which also formed a distinct group. The node separating the B. pseudomallei isolates from the B. thailandensis isolates was recovered in 100% of the bootstrap replicates. The B. mallei clone (ST40) was unambiguously placed within the cluster of B. pseudomallei isolates.
FIG. 3.
Tree constructed from the concatenated sequences of the seven MLST loci. The concatenated sequences from each of the 81 STs represented among the 147 Burkholderia isolates were used to construct a minimum-evolution tree. The percent recoveries of the major nodes in 1,000 bootstrap replicates are shown. The positions of the B. mallei clone (ST40) within the B. pseudomallei STs and of the isolate used for genome sequencing (ST10) are shown. The bar indicates differences at 0.5% of nucleotide sites. The position of B. cepacia genomovar III on the tree is shown by a dotted line, since a homolog of narK was not identified in the B. cepacia genome sequence (www.sanger.ac.uk/Projects/B_cepacia/), and its relationship to the other Burkholderia isolates was obtained on a tree (data not shown) constructed by using the concatenated sequences of only six of the seven MLST loci.
ST73, which included two identical isolates that appeared to be distantly related to all other isolates on the UPGMA tree (Fig. 2), was closely allied with the B. thailandensis isolates on the minimum-evolution tree; although they shared no alleles in common, the sequences of their alleles were very similar, resulting in their close association on the minimum-evolution tree but not on the UPGMA tree. These isolates were tentatively assigned to the species B. thailandensis. By using the concatenated sequences, the average sequence diversity among both the B. pseudomallei and the B. thailandensis STs (including ST73 in the latter species) was 0.2%, whereas the average divergence between the STs of the two species was 3.1%.
The minimum-evolution tree showed that the three identical isolates from Oklahoma (ST81), which had tentatively been assigned to the species B. pseudomallei (17, 26), were distantly related to all isolates of both B. pseudomallei (5.2% divergence) and B. thailandensis (4.7% divergence). The homologs of six of the seven MLST loci could be identified in the contigs available from the Burkholderia cepacia genomovar III genome project (www.sanger.ac.uk/Projects/B_cepacia/), and concatenation of these sequences showed that this genomovar of B. cepacia was more distantly related to B. pseudomallei than were the Oklahoma isolates (Fig. 3).
Analysis of isolates from cases of melioidosis in animals and humans in Hong Kong.
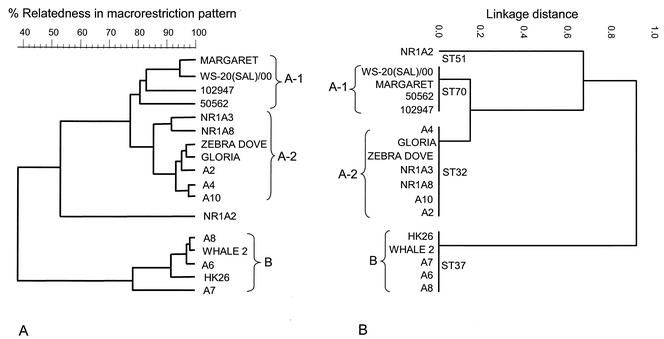
The validity of the MLST scheme and its utility for epidemiological studies of melioidosis were examined by characterizing by both MLST and PFGE a group of 17 B. pseudomallei isolates recovered in Hong Kong from cases of melioidosis in marine mammals at an Oceanarium, in birds at a nearby aviary, and in humans (Table 2). Figure 4 shows the clustering of this group of B. pseudomallei isolates obtained by MLST compared with that obtained by PFGE. Both methods divided the 17 isolates into the same two major clusters. Isolates of cluster B were very similar by PFGE (≥75% identity of DNA fragment patterns), and all these isolates had identical allelic profiles by MLST (ST37). Isolates of cluster A were distantly related to those of cluster B. By MLST, the isolates in cluster A were resolved into two clones (ST32 and ST70) that were very closely related, differing at only one of the seven MLST loci. The isolates of cluster A were also closely related by PFGE (≥75% identity of DNA fragment patterns), and the division into two closely related subclusters (A-1 and A-2) was also apparent, with the former corresponding precisely to ST70 and the latter corresponding precisely to ST32. One isolate from a case of disease in Hong Kong (NR1A2; ST51) was distantly related to the isolates of clusters A and B by both MLST and PFGE.
FIG. 4.
Correlation between molecular typing results obtained by MLST and PFGE. A set of 17 isolates of B. pseudomallei from captive animals with melioidosis and their environments and from cases of human disease in Hong Kong were analyzed by both PFGE (A) and MLST (B). The STs are shown on the dendrogram produced from the MLST data.
Each of the three major STs recovered from animals in Hong Kong (STs 32, 37, and 70) were also recovered from cases of human melioidosis in Hong Kong, and ST70 was also recovered from cases of human disease in Thailand (Table 2). ST51 was also obtained from a case of human disease in Singapore and Thailand.
DISCUSSION
The MLST scheme was developed primarily as a tool for epidemiological studies of melioidosis, and the primers were designed to amplify seven gene fragments from all B. pseudomallei isolates. The primers were also designed to amplify the seven fragments from B. thailandensis, since a genotypic method for reliably distinguishing between these two closely related species was considered valuable, but the fragments were not amplified from the other significant pathogen within the genus, B. cepacia (data not shown). Interestingly, all of the loci selected were found to be located on chromosome I. The reason for this is unknown, but it is not expected to be of any consequence for a molecular typing scheme.
The UPGMA tree based on differences in the allelic profiles of the isolates shows that all B. pseudomallei isolates are clustered and descend from a single node. The alleles at each locus in these isolates were very similar in sequence, and consequently, the minimum-evolution tree based on differences in the concatenated sequences showed a very tight clustering of these isolates. On the minimum-evolution tree, the B. pseudomallei isolates were fully resolved from the isolates of B. thailandensis, which is considered to be its closest relative, and B. pseudomallei appears to be a well-defined and genetically uniform species. On average, only 1 in every 500 nucleotides within the housekeeping genes differed between STs.
The lack of sequence diversity among B. pseudomallei isolates results in an MLST scheme that has fewer alleles per locus, and thus less discriminatory power, than the very highly discriminatory MLST schemes that have been reported previously. On the basis of the sequences in the initial database of 128 B. pseudomallei isolates, the MLST scheme can resolve over 3 million different STs, but the number of alleles per locus depends on the number of isolates that are characterized and takes no account of the frequency of each allele. A more appropriate measure of the discriminatory power of an MLST scheme is the probability of obtaining, by chance, an isolate with the most common allele at each locus (22). For the 86 geographically and temporally diverse isolates of B. pseudomallei, this probability is 0.005; no isolate with the most common allele at each locus was present in the MLST database. Thus, even in this relatively uniform species, two unrelated isolates with the same allelic profile are very unlikely to be found by chance. The MLST scheme is therefore considered to have adequate resolving power for epidemiological studies, and 66 different STs were resolved among the collection of 86 B. pseudomallei isolates from at least 21 countries that were chosen as a diverse sample of this species.
The utility of the scheme for epidemiological investigations was evaluated by examining a series of isolates recovered from animals and their environments and from humans with disease in Hong Kong. The results obtained by MLST were completely consistent with those obtained by PFGE, and both methods identified three major clones associated with disease in the animals. PFGE showed minor differences within the isolates assigned to the same ST by MLST, but it is doubtful that these very minor differences have epidemiological significance or utility for epidemiological studies. MLST provides data that are much more easily compared than PFGE data and is ideal for comparison of isolates characterized in different laboratories and for the detection of strains with international distributions. However, more detailed studies with isolates from a restricted geographic region are required to evaluate further the discriminatory ability of MLST compared to that of PFGE and their relative utilities for detailed epidemiological studies.
The three major STs represented among the isolates from cases of melioidosis in animals in Hong Kong were also recovered from cases of human melioidosis. Three further STs were recovered from different animal species or from both animals and humans. At least some B. pseudomallei clones that cause disease appear to be able to do so in both humans and animals. Most examples of the same ST recovered from different species with disease is likely to be due to the independent acquisition of the same virulent strain from the environment, although direct transmission from infected animals to humans has been proposed (11). Infection of both animals and humans is also well established in the case of B. mallei, which, as discussed below, is a clone of B. pseudomallei; and in this case, direct transmission from equines to humans is well documented (10, 12).
The geographic origins of some of the isolates from humans with disease were not available; but STs 46, 51, 56, and 70 included isolates recovered from humans with melioidosis in different countries. ST46 and ST51 were the most widespread STs. Isolates of ST46 were recovered from cases of melioidosis in Bangladesh, Malaysia, and Thailand and also from a monkey with melioidosis in Indonesia; isolates of ST51 were recovered from cases of melioidosis in Singapore, Hong Kong, and Thailand. More detailed studies of isolates from cases of melioidosis in different countries and studies that relate the STs of isolates in the local environment with those from cases of human disease are required to establish whether there are major clones with a wide geographic distribution that appear to have an enhanced ability to cause melioidosis or whether the diversity of isolates from cases of human disease in a particular region reflects their diversity in the local environment.
The nine isolates of B. thailandensis were closely related and formed a distinct cluster of genotypes. None of the alleles in B. thailandensis are present in B. pseudomallei isolates, and consequently, all of the isolates of these two species differ at all seven MLST loci. The degree of sequence divergence between these two species and their nonoverlapping alleles confirm their status as separate species. Two isolates (ST73) were not clustered with the B. pseudomallei or B. thailandensis isolates on the UPGMA dendrogram, as they differed from all other Burkholderia isolates at all seven loci; but the alleles at all seven loci were very similar to those in B. thailandensis, and they were closely allied to the isolates of this species on the minimum-evolution tree. Isolates of ST73 were tentatively assigned to a divergent lineage of B. thailandensis.
The construction of a minimum-evolution tree from the concatenated sequences of the seven MLST loci provides an excellent and unambiguous way of determining whether a query isolate is B. pseudomallei. A facility for concatenating the sequences at the seven loci from a query isolate and for displaying its position on the minimum-evolution tree shown in Fig. 3 is available at the MLST website.
The five B. mallei isolates that were examined by MLST and the isolate being used for genome sequencing all had identical allelic profiles, although they were recovered from cases of glanders in horses or humans in four different countries (Hungary, Turkey, India, and China) between 1932 and 1961. The alleles at six of the seven loci in B. mallei were also present in B. pseudomallei isolates, and the allele at the seventh locus in B. mallei differed at only a single nucleotide site from an allele in B. pseudomallei. Consequently, the B. mallei isolates clustered within the B. pseudomallei isolates on the minimum-evolution tree reconstructed from the concatenated sequences and on the UPGMA tree constructed from the pairwise differences in the allelic profiles of the isolates. B. mallei is unambiguously a clone of B. pseudomallei, and on population genetic grounds it should not be given a separate species status. B. mallei (or, more appropriately, the Mallei clone of B. pseudomallei) therefore joins the growing list of important pathogens, which includes Salmonella enterica serovar Typhi (14, 18), Bacillus anthracis (13), and Yersinia pestis (2), that represent clones (or clusters of very closely related clones) that have been raised to species status due to the distinctiveness of the diseases that they cause.
In conclusion, the MLST scheme presented here will be a useful new tool for the precise characterization of isolates of B. pseudomallei and for the unambiguous assignment of isolates to this species. Although B. pseudomallei isolates have a low level of sequence diversity, the MLST scheme appears to have sufficient discriminatory power for epidemiological investigations of melioidosis and should allow comparison of the isolates of this species that cause disease in different localities by a means much easier and more precise than the typing procedures used at present.
Acknowledgments
This work was supported by the Wellcome Trust and the University Grants Council, Hong Kong.
We thank P. L. Ho for providing the human isolates from Hong Kong and some of the animal strains.
REFERENCES
- 1.Abbink, F. C., J. M. Orendi, and A. J. de Beaufort. 2001. Mother-to-child transmission of Burkholderia pseudomallei. N. Engl. J. Med. 344:1171-1172. [DOI] [PubMed] [Google Scholar]
- 2.Achtman, M., K. Zurth, G. Morelli, G. Torrea, A. Guiyoule, and E. Carniel. 1999. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. USA 96:14043-14048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brett, P. J., D. DeShazer, and D. E. Woods. 1997. Characterization of Burkholderia pseudomallei and Burkholderia pseudomallei-like strains. Epidemiol. Infect. 118:137-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brett, P. J., D. DeShazer, and D. E. Woods. 1998. Burkholderia thailandensis sp. nov., a Burkholderia pseudomallei-like species. Int. J. Syst. Bacteriol. 48:317-320. [DOI] [PubMed] [Google Scholar]
- 5.Currie, B. J., M. Mayo, N. M. Anstey, P. Donohoe, A. Haase, and D. J. Kemp. 2001. A cluster of melioidosis cases from an endemic region is clonal and is linked to the water supply using molecular typing of Burkholderia pseudomallei isolates. Am. J. Trop. Med. Hyg. 65:177-179. [DOI] [PubMed] [Google Scholar]
- 6.Dance, D. A. B. 2002. Melioidosis. Curr. Opin. Infect. Dis. 15:127-132. [DOI] [PubMed] [Google Scholar]
- 7.Enright, M. C., and B. G. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144:3049-3060. [DOI] [PubMed] [Google Scholar]
- 8.Enright, M. C., N. P. J. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for the characterization of methicillin-resistant (MRSA) and methicillin-susceptible (MSSA) clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haase, A., H. Smith-Vaughan, A. Melder, Y. Wood, A. Janmaat, J. Gilfedder, D. J. Kemp, and B. J. Currie. 1995. Subdivision of Burkholderia pseudomallei ribotypes into multiple types by random amplified polymorphic DNA analysis provides new insights into epidemiology. J. Clin. Microbiol. 33:1687-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Howe, C. 1950. Glanders, p. 185-202. In H. A. Christian (ed.), The Oxford textbook of medicine. Oxford University Press, New York, N.Y.
- 11.Idris, A., R. F. N. Rachmat, and S. M. M. Ali. 1998. Melioidosis: a case of sheep to human transmission. J. Vet. Malaysia 10:77-79. [Google Scholar]
- 12.Jennings, W. E. 1963. Glanders, p. 264-292. In T. G. Hull (ed.), Diseases transmitted from animals to man. Charles C Thomas, Publisher, Springfield, Ill.
- 13.Keim, P., and K. L. Smith. 2002. Bacillus anthracis evolution and epidemiology. Curr. Top. Microbiol. Immunol. 271:21-32. [DOI] [PubMed] [Google Scholar]
- 14.Kidgell, C., U. Reichard, J. Wain, B. Linz, M. Torpdahl, G. Dougan, and M. Achtman. 2002. Salmonella typhi, the causative agent of typhoid fever, is approximately 50,0000 years old. Infect. Genet. Evol. 2:39-45. [DOI] [PubMed] [Google Scholar]
- 15.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: Molecular Evolutionary Genetics Analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 16.Maiden, M. C. J., J. A. Bygraves, E. J. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCormick, J. B., R. E. Weaver, P. S. Hayes, J. M. Boyce, and R. A. Feldman. 1977. Wound infection by an indigenous Pseudomonas pseudomallei-like organism isolated from the soil: case report and epidemiologic study. J. Infect. Dis. 135:103-107. [DOI] [PubMed] [Google Scholar]
- 18.Selander, R. K., P. Beltran, N. H. Smith, R. Helmuth, F. A. Rubin, D. J. Kopecko, K. Ferris, B. D. Tall, A. Cravioto, and J. M. Musser. 1990. Evolutionary genetic relationships of clones in Salmonella serovars that cause human typhoid and other enteric fevers. Infect. Immun. 58:2262-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sitnikova, T., A. Rzhetsky, and M. Nei. 1995. Interior-branch and bootstrap tests of phylogenetic trees. Mol. Biol. Evol. 12:319-333. [DOI] [PubMed] [Google Scholar]
- 20.Smith, M. D., B. J. Angus, V. Wuthiekanun, and N. J. White. 1997. Arabinose assimilation defines a nonvirulent biotype of Burkholderia pseudomallei. Infect. Immun. 65:4319-4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith, M. D., V. Wuthiekanun, A. L. Walsh, and N. J. White. 1995. Quantitative recovery of Burkholderia pseudomallei from soil in Thailand. Trans. R. Soc. Trop. Med. Hyg. 89:488-490. [DOI] [PubMed] [Google Scholar]
- 22.Spratt, B. G. 1999. Multilocus sequence typing: molecular typing of bacterial pathogens in an era of rapid DNA sequencing and the Internet. Curr. Opin. Microbiol. 2:312-316. [DOI] [PubMed] [Google Scholar]
- 23.Takahashi, K., and M. Nei. 2000. Efficiencies of fast algorithms of phylogenetic inference under the criteria of maximum parsimony, minimum evolution, and maximum likelihood when a large number of sequences are used. Mol. Biol. Evol. 17:1251-1258. [DOI] [PubMed] [Google Scholar]
- 24.Trakulsomboon, S., D. A. B. Dance, M. D. Smith, N. J. White, and T. L. Pitt. 1997. Ribotype differences between clinical and environmental isolates of Burkholderia pseudomallei. J. Med. Microbiol. 46:565-570. [DOI] [PubMed] [Google Scholar]
- 25.Vadivelu, J., S. D. Puthucheary, A. Mifsud, B. S. Drasar, D. A. B. Dance, and T. L. Pitt. 1997. Ribotyping and DNA macrorestriction analysis of isolates of Burkholderia pseudomallei from cases of melioidosis in Malaysia. Trans. R. Soc. Trop. Med. Hyg. 91:358-360. [DOI] [PubMed] [Google Scholar]
- 26.Yabuuchi, E., Y. Kosako, M. Arakawa, H. Hotta, and I. Yano. 1992. Identification of Oklahoma isolate as Pseudomonas pseudomallei. Microbiol. Immunol. 36:1239-1249. [DOI] [PubMed] [Google Scholar]