Abstract
We developed a novel multiplex PCR assay for enteroaggregative Escherichia coli (EAEC) detection, by using three plasmid-borne genes (the aggregative adherence [AA] probe, aap, and aggR). One or more of the loci were detected in 24 (86%) of 28 patient isolates analyzed. The multiplex PCR assay is a fast, convenient, and sensitive molecular test to detect EAEC.
The importance of enteroaggregative Escherichia coli (EAEC) strains in public health around the world is becoming increasingly clear. The pathogenicity of these strains has been proven in adult volunteers (12, 18) and in several studies from around the world (3, 4, 5, 9, 14). EAEC strains have been associated classically with persistent diarrhea (≥14 days), which represents a disproportionate share of diarrheal mortality. EAEC strains have been shown to elicit damage to the intestinal mucosa (C. Eslava, J. Villaseca, R. Morales, A. Navarro, and A. Cravioto, Abstr. 93rd Gen. Meet. Am. Soc. Microbiol. 1993, abstr. B-105, p. 44, 1993), and recent data link EAEC with growth retardation in infants (19, 24). Thus, detection of EAEC strains can make a significant contribution to public health in many areas.
EAEC diagnosis has long been problematic. The pathogen was initially defined by the presence of a characteristic stacked brick pattern, designated aggregative adherence (AA) in the HEp-2 cell adherence assay (14). Although this pattern provided the first EAEC epidemiologic associations with diarrhea, there has long been a desire to base detection on the presence of requisite virulence factors.
The pathogenesis of EAEC infection is only partly understood. EAEC adheres to the intestinal mucosa by means of aggregative adherence fimbriae (15) and then elicits damage to the mucosal epithelium, which is manifested as exfoliation of epithelial cells (19). A cytotoxin and several enterotoxins have been described (19), each of which may contribute to secretory diarrhea. EAEC diarrhea exhibits some features of an inflammatory enteritis, and the flagellin protein has been implicated in the release of interleukin-8 (IL-8) (24).
Many of the EAEC virulence factors have been localized to the 60- to 65-MDa AA plasmid, including the aggregative adherence fimbrial adhesin (16), the Pet (8) and EAST (19) enterotoxins, the transcriptional activator AggR (17), and a novel antiaggregation protein (dispersin) encoded by the aap gene (formerly known as aspU) (23). Furthermore, several other loci on the plasmid appear to be specific for EAEC, although they have not yet been implicated in virulence. The importance of the AA plasmid is underscored by the fact that an empirically derived gene probe, designated the AA or CVD432 probe, has been reported to be sensitive and specific for EAEC detection (2). In the initial publication, the AA probe hybridized with 89% of EAEC strains, although this number can vary dramatically from study to study (4, 20, 21).
It has also become apparent that strains carrying the AA plasmid represent an important subgroup of EAEC strains and that many strains which do not hybridize with the AA probe nevertheless share plasmid-borne and chromosomal virulence determinants. Indeed, it has recently been shown that adult travelers carrying EAEC strains which harbored one of more of the AA plasmid loci were significantly more likely to shed fecal IL-8 and IL-1 than if their EAEC strains lacked all plasmid-borne factors (10). Thus, developing improved methods for detecting AA plasmid-positive EAEC strains is a priority. Here, we have developed a multiplex PCR assay to detect three AA plasmid-borne genes and show that these loci are commonly but not inevitably linked.
For the sake of comparison, all EAEC reference and patients strains were first characterized by the HEp-2 assay, which was performed as described previously (6). Strains were then further characterized by the standard colony blot method (1) for the identification of the plasmid-borne genes aap and aggR and for the AA probe (7). We next developed a multiplex PCR with previously described primers for the AA probe (22). For aggR detection, we used a previously described aggR forward primer (7) and a newly chosen reverse primer (GenBank accession no. 232523). Aap primers were chosen (GenBank accession no. 232523) and optimized specifically for the present study (Table 1). For standardization purposes, we used three previously described EAEC reference strains: JM221, 042, 17-2, and 12 additional diarrheagenic E. coli reference strains (11) (three enterotoxigenic, three enteroinvasive, three Shiga-like toxin producing, and three enteropathogenic), plus two E. coli negative controls strains: 3030 and K-12. Bacterial lysates were prepared by resuspending a single colony in 1 ml of deionized water (MilliQ) in a polypropylene tube, followed by boiling for 1 min. Each PCR tube contained 23 μl of reaction mix comprised of (final concentrations): Tris-HCl (pH 8.3), 10 mM; KCl, 50 mM; MgCl2, 2 mM; gelatin, 100 μg/ml; glycerol, 5% (vol/vol); dATP, dCTP, dGTP, and dTTP (Roche), 200 μM each; AmpliTaq polymerase (Gibco-BRL), 1 U/25 μl. Also included were a mixture of the six primers (Table 1) and a 2-μl portion of the bacterial lysates. The solutions were then subjected to the following cycling conditions: 50°C (2 min, 1 cycle); 95°C (5 min, 1 cycle); 95, 55, and 72°C (45 s at each temperature, 40 cycles); and a final extension step (10 min, 72°C) in a thermal cycler (Bio-Rad). Then, 4 μl of the PCR mixture was visualized by ethidium bromide staining after electrophoresis in a 2.5% agarose gel in Tris-acetate-EDTA buffer (Gibco-BRL); the amplicon sizes are shown in Table 1 and Fig. 1. Only the presence of the correct sized gene product(s) was interpreted as a positive test.
TABLE 1.
Primers, concentrations in the PCR mix, and amplicon sizes
| Locus | Primers
|
Primer amt (pMol) in mix | Amplicon size (bp) | |
|---|---|---|---|---|
| Oa | Sequence | |||
| aap | F | 5′-CTT GGG TAT CAG CCT GAA TG-3′ | 10 | 310 |
| R | 5′-AAC CCA TTC GGT TAG AGC AC-3′ | |||
| aggR | F | 5′-CTA ATT GTA CAA TCG ATG TA-3′ | 15 | 457 |
| R | 5′-AGA GTC CAT CTC TTT GAT AAG-3′ | |||
| AA probe | F | 5′-CTG GCG AAA GAC TGT ATC AT-3′ | 20 | 629 |
| R | 5′-CAA TGT ATA GAA ATC CGC TGT T-3′ | |||
O, orientation: F, forward; R, reverse.
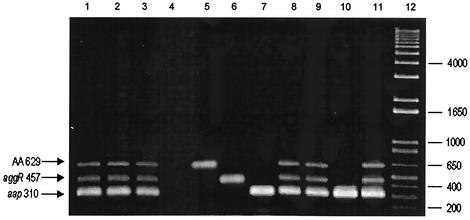
FIG. 1.
PCR products. Lanes 1 to 4, PCR products (in base pairs) obtained with DNA of reference strains mix 17-2, 042, JM221, and 3030, respectively, along with the primer mix; lanes 5 to 7, PCR products obtained with the 17-2 strain and primers for the AA probe, aggR, and aap genes, respectively; lanes 8 to 11, PCR products obtained with DNA of patient isolates; lane 12, 1-kb molecular weight marker in base pairs.
All three reference strains—JM221, 042, and 17-2—were found to be positive for the AA probe and aap and aggR genes (Table 2). In contrast, none of the diarrheagenic E. coli or negative controls were positive for any of the analyzed genes. The minimal concentrations of CFU detected by this assay were 322 CFU (AA probe), 471 CFU (aggR gene), and 70 CFU (aap gene)/ml of saline buffer.
TABLE 2.
Results of control, reference, and patient EAEC strains as analyzed by HEp-2 assay, colony blot, and multiplex PCR techniques
| Strain | HEp-2 assay result | Colony blot result
|
PCR multiplex result
|
||||
|---|---|---|---|---|---|---|---|
| AA probe | aap | aggR | AA probe | aap | aggR | ||
| EAEC strains | |||||||
| 101-1 | AA | (−) | + | (−) | (−) | + | (−) |
| Mex 60A | AA | + | + | + | + | + | + |
| 43977 | AA | (−) | (−) | (−) | (−) | (−) | (−) |
| Peru H32-1 | AA | + | + | + | + | + | + |
| Peru H38-1 | AA | + | + | + | + | + | + |
| Peru H46-2 | AA | + | + | + | + | + | + |
| Peru H72-2 | AA | (−) | (−) | (−) | (−) | (−) | (−) |
| Peru H92-1 | AA | + | + | + | + | + | + |
| Peru H133 | AA | + | (−) | (−) | + | + | + |
| Peru H145-1 | AA | + | + | + | + | + | + |
| Peru H191-1 | AA | (−) | (−) | (−) | (−) | (−) | (−) |
| Peru H194-2 | AA | + | + | + | + | + | + |
| Peru H223-1 | AA | + | + | + | + | + | + |
| Peru H232-1 | AA | + | + | + | + | + | + |
| Thai 6-1-1 | AA | + | + | + | + | + | + |
| Thai 44-1-1 | AA | + | + | + | + | + | + |
| Thai 103-1-1 | AA | (−) | + | (−) | + | + | + |
| Thai 144-1-1 | AA | + | + | + | + | + | + |
| Thai 199-1-4 | AA | + | + | + | + | + | + |
| Thai 216-1-1 | AA | + | + | + | + | + | + |
| Thai 232-1-1 | AA | + | + | + | + | + | + |
| Thai 239-1-1 | AA | + | + | (−) | + | + | + |
| Thai 253-1-1 | AA | + | + | + | + | + | + |
| Thai 278-1-1 | AA | + | + | + | + | + | + |
| Thai 309-1-1 | AA | + | + | + | + | + | + |
| Thai 435-1-1 | AA | + | + | + | + | + | + |
| Thai 495-1-1 | AA | + | + | + | + | + | + |
| Thai 501-1-1 | AA | (−) | (−) | (−) | (−) | (−) | (−) |
| Reference strains | |||||||
| 042 | AA | + | + | + | + | + | + |
| 17-2 | AA | + | + | + | + | + | + |
| JM221 | AA | + | + | + | + | + | + |
| Negative-control E. coli strains | |||||||
| m61655 (K-12) | Nonadherent | (−) | (−) | (−) | (−) | (−) | (−) |
| HS | Nonadherent | (−) | (−) | (−) | (−) | (−) | (−) |
The multiplex assay was further evaluated against 28 EAEC strains isolated from patients and two other E. coli-negative control strains (from the collections of the Center of Vaccine Development, University of Maryland School of Medicine) (Table 2). All strains had been tested for HEp-2 adherence and dot blot for the EAEC genes tested here. It was observed that 24 (86%) of 28 strains were positive for at least one gene, 23 (82%) of 28 strains were positive for the three genes, none of the strains were positive for just two genes, and 4 (14%) of 28 strains were negative for the three genes (Table 2). In the 28 strains analyzed, the PCR method identified 70 genes (23 AA probe, 24 aap, and 23 aggR), and the colony blot identified only 65 genes (22 AA probe, 23 aap, and 20 aggR) revealing that the PCR method is more sensitive. This PCR sensitivity was confirmed by the fact that 23 (82%) of 28 strains were positive for the three loci by PCR compared to 20 (71%) of 28 strains found to be positive by colony blot (Table 2).
There is a need for a convenient assay to detect EAEC strains based on molecular characteristics. The HEp-2 assay remains the “gold standard” for identification of EAEC when these strains exhibit the AA pattern. However, this technique requires special expertise and facilities and is time-consuming; thus, it is only performed in a few laboratories around the world, limiting both EAEC diagnosis and the epidemiological studies. Although a simplified version of the HEp-2 assay have been reported (13), the main problem remains that the strains exhibiting the AA pattern almost certainly encompass both pathogenic and nonpathogenic clones (20). Thus, we have developed a multiplex PCR that specifically and sensitively detects loci on the AA plasmid. The test appears to be more sensitive than the colony blot technique (for the same three genes) and is also less time-consuming than the colony blot and HEp-2 assay.
Our multiplex PCR has demonstrated that most of the EAEC strains isolated from patients with diarrhea 23 (82%) of 28 were positive for the three loci, i.e., aggR, aap, and the AA probe, revealing that these plasmid-borne genes may be more frequently linked than was previously demonstrated by colony blot. These three loci have been reported in a majority of strains (7), and it has been suggested that they may be phylogenetically or pathogenetically linked (7, 23). Of course, it is possible that all three genes are markers for truly virulent EAEC clones, and this hypothesis remains to be tested. Importantly, however, the use of the multiplex assay increases both the sensitivity and the specificity of EAEC detection, which may permit earlier diagnosis of this infection and improve epidemiologic understanding of this emerging pathogen.
Acknowledgments
Jorge F. Cerna was supported by the a CONACyT scholarship 71484. Work in the Nataro lab is supported by U.S. Public Health Service grant AI33096.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. A. Smith, J. G. Seidman, and K. Struhl (ed.). 1989. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 2.Baudry, B., S. J. Savarino, P. Vial, J. B. Kaper, and M. M. Levine. 1990. A sensitive and specific DNA probe to identify enteroaggregative Escherichia coli, a recently discovered diarrheal pathogen. J. Infect. Dis. 161:1249-1251. [DOI] [PubMed] [Google Scholar]
- 3.Bhan, M. K., V. Khoshoo, H. Sommerfelt, P. Raj, S. Sazawal, and R. Srivastava. 1989. Enteroaggregative Escherichia coli and Salmonella associated with nondysenteric persistent diarrhea. Pediatr. Infect. Dis. J. 8:499-502. [DOI] [PubMed] [Google Scholar]
- 4.Bouzari, A., A. Jafari, A. Azizi, M. Oloomi, and J. P. Nataro. 2001. Short report: characterization of enteroaggregative Escherichia coli isolates from Iranian children. Am. J. Trop. Med. Hyg. 65:13-14. [DOI] [PubMed] [Google Scholar]
- 5.Cravioto, A., A. Tello, A. Navarro, J. Ruiz, H. Villafan, F. Uribe, and C. Eslava. 1991. Association of Escherichia coli HEp-2 adherence patterns with type and duration of diarrhoea. Lancet 337:262-264. [DOI] [PubMed] [Google Scholar]
- 6.Cravioto, A., R. J. Gross, S. Scotland, and B. Rowe. 1979. An adhesive factor found in strains of Escherichia coli belonging to the traditional enteropathogenic serotypes. Curr. Microbiol. 3:95-99. [Google Scholar]
- 7.Czeczulin, J. R., T. S. Whittam, I. R. Henderson, F. Navarro-Garcia, and J. P. Nataro. 1999. Phylogenetic analysis of enteroaggregative and diffusely adherent Escherichia coli. Infect. Immun. 67:2692-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eslava, C., F. Navarro-Garcia, J. Czeczulin, I. A. Henderson, A. Cravioto, and J. P. Nataro. 1998. Pet, an autotransporter enterotoxin from enteroaggregative Escherichia coli. Infect. Immun. 66:3155-3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang, G. D., A. A. M. Lima, C. V. Martins, J. P. Nataro, and R. L. Guerrant. 1995. Etiology and epidemiology of persistent diarrhea in northeastern Brazil: a hospital-based, prospective, case-control study. J. Pediatr. Gastroenterol. Nutr. 21:137-144. [DOI] [PubMed] [Google Scholar]
- 10.Jiang, Z.-D., D. Greenberg, J. P. Nataro, R. Steffen, and H. L. DuPont. 2002. Rate of occurrence and pathogenic effect of enteroaggregative Escherichia coli virulence factors in international travelers. J. Clin. Microbiol. 40:4185-4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.López-Saucedo, C., J. F. Cerna, N. Villegas-Sepulveda, R. Thompson, F. R. Velazquez, J. Torres, P. I. Tarr, and T. Estrada-García. 2003. A single multiplex polymerase chain reaction to detect diverse loci associated with diarrheagenic Escherichia coli. Emerg. Infect. Dis. 9:127-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mathewson, J. J., P. C. Johnson, and H. L. DuPont. 1986. Pathogenicity of enteroadherent Escherichia coli in adult volunteers. J. Infect. Dis. 154:524-527. [DOI] [PubMed] [Google Scholar]
- 13.Miqdady, M. S., Zhi-Dong Jiang, J. P. Nataro, and H. L. DuPont. 2002. Detection of enteroaggregative Escherichia coli with formalin-preserved HEp-2 cells. J. Clin. Microbiol. 40:3066-3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nataro, J. P., J. B. Kaper, R. Robins Browne, V. Prado, P. Vial, and M. M. Levine. 1987. Patterns of adherence of diarrheagenic Escherichia coli to HEp-2 cells. Pediatr. Infect. Dis. J. 6:829-831. [DOI] [PubMed] [Google Scholar]
- 15.Nataro, J. P., Y. Deng, D. R. Maneval, A. L. German. W. C. Martin, and M. M. Levine. 1992. Aggregative adherence fimbriae I of enteroaggregative Escherichia coli mediate adherence to HEp-2 cells and hemagglutination of human erythrocytes. Infect. Immun. 60:2297-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nataro, J. P., D. Yikang, J. A. Giron, S. J. Savarino, M. H. Kothary, and R. Hall. 1993. Aggregative adherence fimbria I expression in enteroaggregative Escherichia coli requires two unliked plasmid regions. Infect. Immun. 61:1126-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nataro, J. P., D. Yikang, D. Yingkang, and K. Walter. 1994. AggR a transcriptional activator of aggregative adherence fimbria I expression in enteroaggregative Escherichia coli. J. Bacteriol. 176:4691-4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nataro, J. P., D. Yikang, S. Cookson, A. Cravioto, S. J. Savarino, L. D. Guers, M. M. Levine, and C. O. Tacket. 1995. Heterogeneity of enteroaggregative Escherichia coli virulence demonstrated in volunteers. J. Infect. Dis. 171:465-468. [DOI] [PubMed] [Google Scholar]
- 19.Nataro, J. P., and J. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okeke, I. N., A. Lamikanra, H. Steinrûck, and J. B. Kaper. 2000. Characterization of Escherichia coli strains from cases of childhood diarrhea in provincial southwestern Nigeria. J. Clin. Microbiol. 38:7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scaletsky, I. C. A., S. H. Fabbricotti, K. R. Aranda, M. B. Morais, and U. Fagundes-Neto. 2002. Comparison of DNA hybridization and PCR assays for detection of putative pathogenic enteroadherent Escherichia coli. J. Clin. Microbiol. 40:1254-1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidt, H., C. Knop, S. Franke, S. Aleksic, J. Heesemann, and H. Karch. 1995. Development of PCR for screening of enteroaggregative Escherichia coli. J. Clin. Microbiol. 33:700-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sheikh, J., J. R. Czeczulin, S. Harrington, S. Hicks, I. R. Henderson, C. Le Bouguénec, P. Guonon, A. Phillips, and J. P. Nataro. 2002. A novel dispersin in enteroaggregative Escherichia coli. J. Clin. Investig. 110:1329-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steiner, T. S., A. M. Lima, J. P. Nataro, and R. L. Guerrant. 1998. Enteroaggregative Escherichia coli produce intestinal inflammation and growth impairment and cause interleukin-8 release from intestinal epithelial cells. J. Infect. Dis. 177:88-96. [DOI] [PubMed] [Google Scholar]