Abstract
Mycoplasma conjunctivae is the etiological agent of infectious keratoconjunctivitis, a highly contagious ocular infection that affects both domestic and wild Caprinae species in the European Alps. In order to study the transmission and spread of M. conjunctivae across domestic and wild Caprinae populations, we developed a molecular method for subtyping and identifying strains of M. conjunctivae. This method is based on DNA sequence determination of a variable domain within the gene lppS, a gene that encodes an antigenic lipoprotein of M. conjunctivae. This domain of lppS shows variations among different strains but remains constant upon generations of individual strains on growth medium and thus allows identification of individual strains and estimation of their phylogenetic intercorrelations. The variable domain of lppS is amplified by PCR using primers that match conserved sequences of lppS flanking it. Sequence analysis of the amplified fragment enables fine subtyping of M. conjunctivae strains. The method is applicable both to isolated strains and to clinical samples directly without requiring the cultivation of the strain. Using this method, we show that M. conjunctivae was transmitted between domestic and wild animals that were grazing in proximate pastures. Certain animals also presented infections with two different strains simultaneously.
Infectious keratoconjunctivitis (IKC) is a common, contagious ocular disease known as pinkeye of domestic small ruminants, particularly sheep and free-ranging Caprinae mainly in the Alps (13). This disease is characterized by inflammation of the conjunctiva and cornea. In the most-advanced stage, the cornea is opaque or even perforated (18), and blind wild animals may fall from cliffs or die from starvation. Mycoplasma conjunctivae is considered as the major etiological agent of IKC in Caprinae species such as alpine ibex (Capra ibex ibex) (13), alpine chamois (Rupicapra rupicapra rupicapra) (10, 20), and mouflon (Ovis orientalis musimon) (23), as well as in domestic sheep and goat (15, 16, 25, 26). In Switzerland, the prevalence of M. conjunctivae antibodies in adult sheep at the individual level was 53%, and the domestic sheep population was shown to act as a reservoir of the M. conjunctivae infection (15). In contrast, the M. conjunctivae infection is not self-maintained in alpine chamois in eastern Switzerland and their infection may originate from domestic sheep living in proximity to chamois during the summer (12). Susceptibility of alpine ibex to sheep strains of M. conjunctivae was demonstrated by experimental infections, hence proving the possibility of transmission of mycoplasmal IKC between different species (13). This transmission of M. conjunctivae between domestic small ruminants and wild Caprinae may be caused by physical contacts and by flies acting as vectors between the species (11). In alpine regions, increases of IKC outbreaks are generally observed during the summer and autumn, which is coincident with the presence of domestic sheep grazing on alpine summer pasture (12).
Transmission of infectious agents across host species is common in nature. In particular, spillover from reservoir animal populations (often domesticated species) to wildlife underpins the appearances of a range of emerging infectious diseases in wildlife (9). Evidence of bacterial transmission between domestic and wild animals based on molecular techniques was reported by Chang et al. (7), when the transmission of Bartonella among cattle and wildlife in North America was described. In 1996, transmission of Mycobacterium bovis between wild boar and cattle in Spain was reported (1). Problems associated with spillover of infectious agents include a more complex surveillance of the flow of the pathogenic agent and a more difficult disease control. Spillover is facilitated by the presence of various hosts in the same area. However, if an agent is present in two different species in the same region, it cannot be assumed offhand that transfer between these species always occurs (24) unless evidence based on markers capable of distinguishing strain subtypes is provided.
In this study, we present a subtyping method for M. conjunctivae strains using a variable domain of the 3′ end of the lppS gene which encodes the lipoprotein S adhesin (LppS) of M. conjunctivae (5). We have used this method to analyze differences among strains of M. conjunctivae in sheep, goat, chamois, and ibex and to study inter species transmission of M. conjunctivae.
MATERIALS AND METHODS
Origin of M. conjunctivae strains and samples.
The origin of the different M. conjunctivae strains and isolates are given in Table 1. They were collected from different host species and various geographic places during 1994 and 2001. The type strain of M. conjunctivae, HRC/581T, was isolated in 1972 (4) and was obtained from the Mycoplasma Reference Center, Aarhus, Denmark, in 1973. A different sample of HRC/581T that was propagated for a large number of generations on growth medium was obtained from Agence Française de Sécurité Sanitaire des Aliments (AFSSA), Lyon, France, in 2001. M. conjunctivae strains were grown on standard mycoplasma PPLO Growth medium (Difco Laboratories, Detroit, Mich.) enriched with 20% horse serum, 2 to 5% yeast extract, and 1% glucose (3). Direct samples were taken from the conjunctiva of affected animals with cotton swabs and stored at −18°C. The hosts from which the different strains and samples were collected included domestic sheep and goats as well as free-ranging chamois and ibexes from different cantons of Switzerland. Furthermore, chamois from two different Italian provinces, sheep and chamois from the Salzach valley in Austria, and sheep and goats from Croatia were analyzed. The last of these groups included sheep that were imported from Australia, native sheep that were reported to have been infected by the introduction of Australian rams in the flocks, and a goat that originated from France. All strains and isolates were confirmed to belong to the species M. conjunctivae using a nested PCR method with the primer pairs MOLIGEN1-L/16SUNI-R and McoR1/McoF1 (14).
TABLE 1.
Strains of Mycoplasma used in this study
| Mycoplasma species and strain/isolate | Host | Date of isolation | Originl | PCR IppSk |
|---|---|---|---|---|
| M. conjunctivae | ||||
| HRC/581a | Sheep | 1972 | Type strain | + |
| 1 | Sheep | 08 Mar. 2001 | Austria (Salzach, Stubachtal) | + |
| 5 | Sheep | 08 Mar. 2001 | Austria (Salzach, Stubachtal) | + |
| 9 | Sheep | 08 Mar. 2001 | Austria (Salzach, Felbertal) | + |
| 2777 | Chamois | 08 Nov. 2000 | Austria (Salzach, Mittersill) | + |
| 2778 | Chamois | 07 Dec. 2000 | Austria (Salzach, Mittersill) | + |
| 2784 | Chamois | 19 Nov. 2000 | Austria (Salzach, Mittersill) | + |
| 2785 | Chamois | 20 Nov. 2000 | Austria (Salzach, Mittersill) | + |
| My-66/95b | Sheep | 1995 | Croatiac | + |
| My-86/95b | Sheep | 1995 | Croatiad | + |
| My-87/95b | Sheep | 1995 | Croatiad | + |
| My-88/95b | Sheep | 1995 | Croatiad | + |
| My-93/95b | Sheep | 1995 | Croatiad | + |
| My-94a/95b | Sheep | 1995 | Croatiad | + |
| My-7/96b | Goat | 1996 | Croatiae | + |
| 14706 | Chamois | Oct. 2000 | Italy (Bergamo province) | + |
| 15240 | Chamois | Oct. 2000 | Italy (Bergamo province) | + |
| 15244 | Chamois | Oct. 2000 | Italy (Bergamo province) | + |
| 15667/2 | Chamois | Oct. 2000 | Italy (Bergamo province) | + |
| 1727 (1) | Chamois | 21 Oct. 2000 | Italy (Sondrio province) | + |
| 1877 (3) | Chamois | 21 Oct. 2000 | Italy (Sondrio province) | + |
| G130b | Sheep | 04 Sep. 1995 | Switzerland (Curaglia, GR) | + |
| G131b | Sheep | 04 Sep. 1995 | Switzerland (Safien, GR) | + |
| N50 | Sheep | 04 Apr. 2000 | Switzerland (Kehrsatz, BE) | + |
| 38 | Sheep | 12 Apr. 2000 | Switzerland (Kehrsatz, BE) | + |
| 49-af | Sheepg | 22 May 2001 | Switzerland (Kehrsatz, BE) | + |
| 49-bf | Sheepg | 22 May 2001 | Switzerland (Kehrsatz, BE) | + |
| 50 | Sheepg | 22 May 2001 | Switzerland (Kehrsatz, BE) | + |
| 52f | Sheepg | 22 May 2001 | Switzerland (Kehrsatz, BE) | + |
| 53f | Sheepg | 22 May 2001 | Switzerland (Kehrsatz, BE) | + |
| 54 | Sheep | 24 Apr. 2001 | Switzerland (Kehrsatz, BE) | + |
| 95 | Sheep | 24 Apr. 2001 | Switzerland (Kehrsatz, BE) | + |
| 96 | Sheep | 24 Apr. 2001 | Switzerland (Kehrsatz, BE) | + |
| 2820 | Sheep (lamb) | 27 Feb. 2001 | Switzerland (Kehrsatz, BE) | + |
| 2821h | Sheep (lamb) | 27 Feb. 2001 | Switzerland (Kehrsatz, BE) | + |
| s2826 | Sheep (lamb) | 27 Feb. 2001 | Switzerland (Kehrsatz, BE) | + |
| 2833Lii | Sheep (lamb) | 14 Aug. 2001 | Switzerland (Mesocco, GR) | + |
| 2833Rei | Sheep (lamb) | 14 Aug. 2001 | Switzerland (Mesocco, GR) | + |
| 2834 | Sheep (lamb) | 14 Aug. 2001 | Switzerland (Mesocco, GR) | + |
| 2831 | Chamois | 14 Aug. 2001 | Switzerland (Mesocco, GR) | + |
| 2832 | Chamois | 14 Aug. 2001 | Switzerland (Mesocco, GR) | + |
| W00-5129-163 | Chamois | 13 Sep. 2000 | Switzerland (Poschiavo, GR) | + |
| G6b | Ibex | 24 Aug. 1994 | Switzerland (Matt, GL) | + |
| G9b | Ibex | 31 Aug. 1994 | Switzerland (Flims, GR) | + |
| M. hyopneumoniae RF4738 | Swine | − | ||
| M. agalactiae | − | |||
| PG2 | Goat | − | ||
| 3990 | − | |||
| M. bovoculi RF20391 | Bovine | − | ||
| M. mycoides subsp. capri PG3 | Goat | − | ||
| M. mycoides subsp. mycoides SC Afadé | Bovine | − | ||
| M. putrefaciens KS1T | − | |||
| M. arginini G230T | − | |||
| M. capricolum subsp. capricolum California kid | Goat | − |
Strain previously described by Barile et al. (4).
Strain previously described by Giacometti et al. (14).
Isolate from a sheep that was imported from Australia.
Imported from native sheep of a flock into which a ram from Australia was introduced.
Goat in quarantine imported from France.
Isolate originates from the same eye of a sheep that was initially free of M. conjunctivae and that was put in contact under controlled conditions with animals that were known to carry either isolate 54 or 95 (identical to isolate 96).
Controlled contact infection.
Sample was shown to contain two different M. conjunctivae strains and was not considered as an isolate for the epidemiological study.
2833Li, left eye of sheep 2833; 2833Re, right eye of sheep 2833.
PCR using oligonucleotide primers Ser_start1 and Ser_end (Table 2).
Abbreviations: GR, Grisons; BE, Berne; GL, Glarus.
Detection of M. conjunctivae infections.
Detection of M. conjunctivae from conjunctival swabs was done by nested PCR (14). Briefly, cotton swabs were placed into microcentrifuge tubes containing 0.5 ml of lysis buffer (100 mM Tris-HCl [pH 8.5], 0.05% Tween 20, proteinase K [0.24 mg/ml]) and mixed for 1 min. The buffer was incubated for 60 min at 60°C, and this was followed by an incubation for 15 min at 97°C to obtain the lysate as template for PCRs. In vitro amplifications from the lysates were performed by nested PCR with the primer pair MOLIGEN1-L-16SUNI-R (Table 2) in the first step and the primer pair McoR1-McoF1 (Table 2) in the second step (14).
TABLE 2.
Sequences of PCR primers used for detection of M. conjunctivae and amplification of the C-terminal part of lppS
| Name | Sequence (5′-3′) | Annealing temp (°C) | Positiona | Reference |
|---|---|---|---|---|
| MOLIGEN1-Lb | ACTCCTACGGGAGGCAGCA | 51 | 14 | |
| 16SUNI-Rb | GTGTGACGGGCGGTGTGTAC | 51 | 14 | |
| McoR1b | CAGCGTGCAGGATGAAATCCCTC | 54 | 14 | |
| McoF1b | GTATCTTTAGAGTCCTCGTCTTTCAC | 54 | 14 | |
| Ser_start1 | GCTCAAGAGCAAACTGACC | 49.1c | 3927-3945 | |
| Ser_start2 | CACTATACTTAACAGATAGTCC | 46.3c | 3781-3802 | |
| Ser_end | GCAGCAACTGCTGAAAGTC | 49.4c | 4728-4746 |
Position with reference to nucleotide sequence of lppS and lppT of M. conjunctivae HRC/581T (accession number AJ318939).
Used as diagnostic PCR for detection of M. conjunctivae infections.
Obtained with the “PCR primer annealing temperature calculator” developed by J. Boxall (http://www.iacr.bbsrc.ac.uk/res/depts/biochem/old-or-to-move/tcalculator.html) by using the parameters 30% as target GC content and 1,000 bp as target size.
Amplification of the 3′ part of lppS and sequence analysis.
The 3′ part of the lppS gene was amplified by PCR from lysates of M. conjunctivae cultures or directly from lysates of conjunctival swab samples by PCR with oligonucleotide primers Ser_start1 and Ser_end (Table 2) which were derived from the DNA sequence of M. conjunctivae HRC/581T lppS gene (EMBL/GenBank accession number AJ318939). PCR was carried out in a 50-μl reaction mix [50 mM Tris-HCl, pH 9.2, 1.75 mM MgCl2, 16 mM (NH4)2SO4, a 350 μM concentration of each deoxynucleoside triphosphate] using 3.5 μl of lysate as template. To each reaction, 1.75 U of a mixture of Taq DNA and Pwo DNA polymerases (Expand Long Template PCR System kit; Roche Diagnostics, Rotkreuz, Switzerland) and a 400 nM concentration of the respective forward and reverse primer couple (Table 2) were added. The samples were subjected to a denaturation step at 94°C for 2 min followed by 40 cycles of amplification consisting of 30 s at 94°C, 30 s at 51°C, and elongation at 68°C during 75 s. All PCRs were carried out in a GeneAmp 9600 DNA thermal cycler (Applied BioSystems, Norwalk, Conn.). The PCR amplification products were analyzed by electrophoresis through 0.7% agarose gels and visualized after staining with ethidium bromide on a UV Transilluminator (2).
For DNA sequence analysis PCR fragments were purified using the High Pure PCR product purification kit (Roche Diagnostics). The concentration of the purified DNA was determined spectrophotometrically with a GeneQuantII (Pharmacia Biotech, Cambridge, England) and 50 ng of purified PCR products were used for the sequencing reaction. Sequencing reactions were performed using the dRhodamine terminator cycle sequencing kit (Applied Biosystems), with the primers Ser_start1 and Ser_end used for the PCR amplification. Reaction products were analyzed with an ABI Prism 3100 genetic analyzer (Applied Biosystems).
Sequence analysis and editing were done with the software Sequencher (Gene Codes Corporation, Ann Arbor, Mich.). Alignment was done with the Wisconsin package (Genetics Computer Group, Inc., Madison, Wis.). A phylogenetic relationship was established with PILEUP from the Genetics Computer Group program package (gap creation penalty, 5; gap extension penalty, 1) and by further analysis with the Mega 1.02 program (by complete deletion of gaps and missing information). Corrections were calculated with the Jukes-Cantor algorithm (17), and a tree was derived by the neighbor-joining method (22).
DNA probe for lppS and Southern blot analysis.
A specific probe for lppS was prepared by PCR using 1 ng DNA of purified plasmid pJFF2E carrying the cloned lppS gene (5) as template with the oligonucleotide primers Ser_start2 and Ser_end and by supplementing the reaction mix by 40 μM digoxigenin-11-dUTP (DIG) (Roche Diagnostics). Genomic DNA of the different Mycoplasma species was digested, subjected to electrophoresis on a 0.7% (wt/vol) agarose gel, and transferred onto a positively charged nylon membrane (Roche Diagnostics) following standard protocol (2). The membrane was preincubated with 20 ml of hybridization buffer (5× SSC [1× SSC is 150 mM NaCl, 15 mM sodium citrate, pH 7.7], 0.1% N-lauroylsarcosine, 0.02% sodium dodecyl sulfate [SDS], 1% [wt/vol] blocking reagent [Roche Diagnostics]) per 100-cm2 membrane at 68°C for 2 h and then hybridized overnight at 60°C with 2.5 ml of hybridization buffer containing 1 μg DIG-labeled lppS probe per 100-cm2 membrane. The membrane was washed twice for 5 min at room temperature with 2× SSC containing 0.1% SDS and twice for 15 min at 25°C with 0.2× SSC containing 0.1% SDS. The digoxigenin-labeled probe was detected using phosphate-labeled antidigoxigenin antibodies (Roche Diagnostics) according to the manufacturer's instructions.
RESULTS
Variability of the 3′ end of the lppS gene in various M. conjunctivae strains.
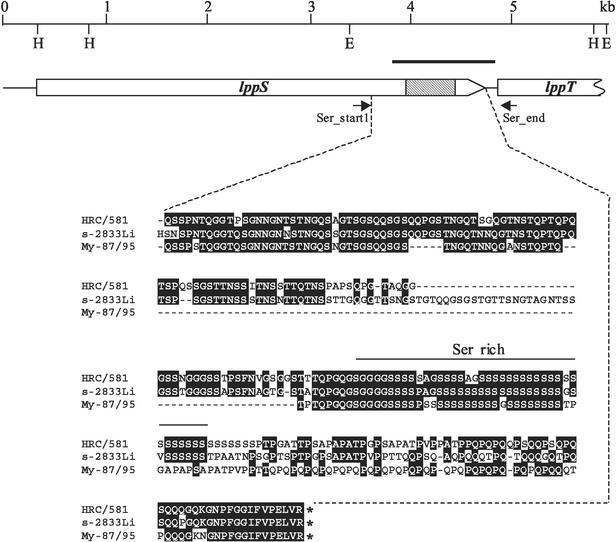
Sequence analysis of the lppS gene of different M. conjunctivae field stains revealed that the 3′-coding region which includes the serine rich domain of lipoprotein LppS (Fig. 1) shows significant variability among the field isolates compared to the HRC/581T, while the rest of the gene seemed to be stable. Sequence comparison of the lppS genes of the M. conjunctivae type strain HRC/581T isolated from a sheep in 1972 and from field strain 2784 (accession no. AJ514404) isolated from a chamois in 2000 showed 96% identity in the first 3,508 bp of the coding sequence for LppS and only 68% identity in the last 719 bp at the 3′ end. The 5′ part of the subsequent gene lppT showed no differences. We exploited these results to develop oligonucleotide primers matching conserved segments in the lppS gene and the 5′ end of lppT flanking the variable domain of lppS which allowed PCR amplification of this variable segment (Fig. 1). PCR amplifications were made using the primer pair Ser_start1-Ser_end (Table 2) from the different strains or directly from eye swabs of the affected animals (Table 1). Agarose gel analysis of the PCR products showed fragments of variable size as expected, ranging from 519 to 935 bp. Subsequent DNA sequence analysis of the PCR amplicons was performed using the same primers Ser_start1 and Ser_end. Sequence data were used 10 nucleotides downstream the sequence of the respective primers from strain HRC/581T. The nucleotide sequence data from the 44 individual strains or isolates listed in Table 1 showed differences up to 35%. This allowed a good resolution and identification of individual strains. A few of the sequences were identical, indicating that the corresponding isolates were identical strains. The sequence chromatograms of fragments from four samples (49, 52, 53 and 2821; Table 1) showed multiple double peaks referring to different nucleotides and could not be analyzed. This indicated the presence of two or more different M. conjunctivae strains in the same sample. In sample 49 two different isolates, 49-a and 49-b, were segregated and sequenced individually (Table 1). The other three samples (52, 53, and 2821) were not used any further in this study.
FIG. 1.
Genetic map of the adhesin gene lppS and part of the lppT gene. The upper line shows the scale in kilobase pairs (kb) and the position of the restriction enzyme cutting sites for HindIII (H) and EcoRI (E). The bold line represents the position of the lppS gene probe. The boxes represent the coding sequences for the proteins LppS and part of LppT. The arrowhead of the box gives the direction of transcription and translation. The grey part of the box represents the serine-rich domain of LppS. The region between the broken lines shows the amino acid sequence, derived from the DNA sequence, of the variable domain of LppS of type strain HRC/581 and of two field strains of M. conjunctivae. This part of the gene was used for the subtyping of M. conjunctivae strains. Identical amino acids are shown on black background.
In order to study the stability of the variable part of the lppS gene when M. conjunctivae is grown in culture medium, we have sequenced the same domain from two different samples of the type strain HRC/581T. One sample was obtained in the year 1973 from the Mycoplasma Reference Center and had a particularly low number of generations under in vitro growth. The second sample was obtained in the year 2001 from F. Poumarat, AFSSA, and was grown for approximately 40 generations in our laboratory. There were no differences in the nucleotide sequences of the lppS gene among these two samples, thus showing that lppS in a given strain is stable. We therefore used the differences in the variable part of lppS in order to differentiate individual strains of M. conjunctivae and to calculate the phylogenetic distance among the strains as shown in Fig. 2.
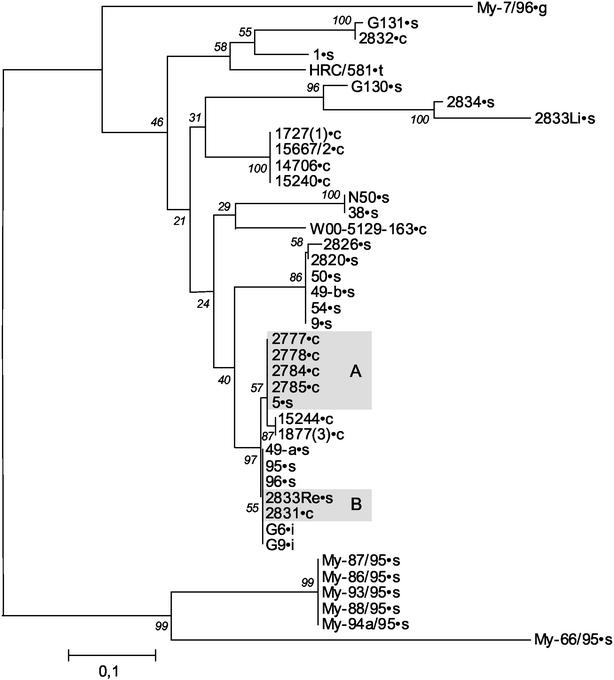
FIG. 2.
Representation of the phylogenetic relationship of the variable domain of lppS of all M. conjunctivae isolates analyzed. A distance matrix was calculated by the Jukes-Cantor algorithm (17), and a tree was built by the neighbor-joining method (22). Bootstrap values of 500 simulations are given at the branching points of the tree. The scale bar indicates the genetic distance of the variable segment of lppS as a ratio of different nucleotides. For clarity, we have labeled the different isolates with their origin by a point followed by a letter: c (chamois), g (goat), i (ibex), s (sheep), or t (type strain). The shaded boxes indicate isolates of the same strain found in sheep and neighboring chamois. “A” indicates the samples from animals from the Salzach valley, Austria, and “B” highlights the samples from animals from the San Bernardino region, Grisons, Switzerland.
In order to ascertain that the lppS gene is present in a single copy in M. conjunctivae and is absent in other related Mycoplasmas species, Southern blot analysis of HindIII digested genomic DNA of M. conjunctivae strains HRC/581T and 2784, M. mycoides subsp. mycoides SC strain Afadé, M. putrefaciens strain KS1T, M. agalactiae strain 3990, M. arginini strain G230T and strain M. mycoides subsp. capri PG3 was performed with the gene probe for lppS. This analysis showed a single 4.8-kb band hybridizing to the lppS probe with M. conjunctivae (as expected from the physical map shown in Fig. 1), and no signal with the other Mycoplasma species, thus revealing that lppS is specific to the species M. conjunctivae. This was confirmed further by PCR analysis using the primer pair Ser_start1-Ser_end (Table1). Southern blot analysis of genomic DNA of M. conjunctivae digested with BamHI, SalI, SmaI, and PvuI which cut outside the lppS gene resulted in single band, thus confirming that lppS is present in a single copy.
Molecular epidemiology of M. conjunctivae infections.
The variable part of the lppS gene of 40 different isolates and of the type strain of M. conjunctivae was sequenced and their phylogenetic relationship was established (Fig. 2). Several isolates showed identical nucleotide sequences in the variable part of lppS indicating that they represented the same strains. Isolates My-86/95, My-87/95, My-88/95, My-93/95, and My-94a/95 all represent the same strain (Fig. 2). They were isolated from native Croatian sheep in a flock that started to show symptoms of IKC after a ram had been introduced from Australia which was considered to have infected the flock (19) (T. Naglic, personal communication). In addition, strain My-66/95 which was isolated in Croatia from a sheep directly imported from Australia shows to be most closely related to the former four isolates. These strains form a distinct cluster that is different from all other isolates which originate from European Alpine countries (Fig. 2). Among the different isolates from sheep, goat, chamois and ibex from European Alpine regions, the typing method is able to distinguish 16 different strains which form the second cluster. Of particular interest in this cluster are isolates 2777, 2778, 2784, and 2785 (Fig. 2, shaded box A) which were all isolated from diseased chamois in the Salzach valley in Austria. After diagnosis of M. conjunctivae in these chamois, eye swabs from a few sheep that were grazing in these pastures and that showed signs of potential M. conjunctivae infections were analyzed. Among three positive sheep, one isolate, number 5, showed the same lppS sequence as the latter four isolates from chamois (Fig. 2, shaded box A). Hence, this sheep carried the same strain as that detected in the chamois in this valley. A second situation where the same M. conjunctivae strain was found in chamois and in sheep grazing in the vicinity is represented by isolate 2833 Re from a sheep, and isolate 2831 from a chamois that was found with IKC in the San Bernardino region, Switzerland (Fig. 2, shaded box B).
Two further cases show that sheep can become infected by two different strains simultaneously: the isolates 2833Li and 2833Re were found in the same sheep, one in the left eye and the other in the right eye. Furthermore, isolates 49-a and 49-b could be separated as two different strains originating from the same eye of a sheep that was initially free of M. conjunctivae and that was put in contact under controlled conditions with a flock that was known to be infected with M. conjunctivae isolate 54 (which is the same strain as isolate 49-b) and isolate 95 (which is the same strain as isolate 49-a). The latter strain was identified in three host species: sheep, chamois, and ibex. It must be noted that upon mixed infections with two or more different M. conjunctivae strains, the PCR method is normally expected to amplify the most abundant subtype. Therefore, the method is not designed to detect and/or analyze multiple infections with different strains simultaneously. Finally, we had the surprising finding that a female chamois and her 2 month old kid, both affected with IKC, carried 2 different strains (2831 and 2832, respectively). These two strains, however, were also found in sheep (2833 and G131, respectively) in the same canton (Grisons) (Fig. 2).
DISCUSSION
Molecular epidemiological studies of mycoplasmas are in general hampered by the difficulty to cultivate mycoplasmas and to subtype Mycoplasma species by means of phenotypic markers such as biochemical reactions or surface markers for serotyping. Furthermore, antigenic hyper-variability often makes strain differentiation by serotyping impossible. Recently, insertion sequence typing by Southern blot hybridization with a labeled probe of IS1296 was used to subtype strains of Mycoplasma mycoides subsp. mycoides SC. This method allowed the conclusion that the reemerging outbreaks of M. mycoides subsp. mycoides SC infections of cattle in Europe were due to an endogenous European strain and not by reimporting the disease from the African continent (8). Insertion sequence typing, however, required extracted DNA from approximately 10 to 20 ml of cultures of each strain which is rather expensive in labor and cost. For M. conjunctivae no insertion elements are known yet. Species identification of M. conjunctivae is currently performed by PCR based on specific segments of the rrs (16S rRNA) gene (14). Here we presented a new subtyping method that was developed on the basis of DNA sequence of the variable part of the adhesin lipoprotein S. This variable part which is conveniently flanked by stable gene sequences was shown to be stable within an isolated strain over many generations and hence can serve as accurate target for identification of individual strains. The variable domain of lppS seems to evolve slowly and does not share common features with hypervariable antigens known in M. bovis and M. agalactiae (6, 21). The subtyping method of M. conjunctivae by sequence analysis of the variable domain of lppS presented in this work has the advantage that it can be done directly from single colonies of primary cultures as well as from liquid cultures or from purified genomic DNA. The particular advantage, however, is that the method can be applied even directly to clinical samples such as eye swabs without prior cultivation. The latter, however, required that animals are infected by single strains, since double infections by different strains cannot be resolved by this method, unless they are separated, e.g., by cloning. Our study showed that in most cases infections with single strains were encountered in sheep and chamois that we analyzed. Only four double infections were found in our study whereof one was analyzed in particular. The DNA sequence heterogeneity in the variable part of lppS was also used as a phylogenetic marker for the different M. conjunctivae strains. Hence, the six M. conjunctivae isolates from Croatia that are presumed to originate from imported Australian sheep (19) clearly show a cluster that is distinct from the other strains that are generally found in the European Alpine region (Fig. 2). Since IKC was not detected in this geographical area earlier (Naglic, personal communication), this particular epidemiological situation explains the spread of a single strain in this flock. In addition, the distinct provenance of this strain and also of strain My-66/95 which was isolated in Croatia from a sheep directly imported from Australia explains the particular phylogenetic position of these strains (Fig. 2). Isolate My-7/96 which also was collected in Croatia, was isolated from a goat that was imported from France and that was kept in quarantine after the transport. This isolate represents a strain that belongs to the European Alpine cluster. It is the only goat isolate in this study and takes a particular phylogenetic position in the Alpine European cluster. The phylogenetic position of the type strain HRC/581T matches closely Alpine sheep and chamois strains, even though the origin of this strain is reportedly Maryland (4).
Most interestingly, our study revealed that the same M. conjunctivae strains could be isolated from chamois with IKC and from sheep that were grazing on the same pastures (Fig. 2, shaded boxes), showing that M. conjunctivae can be transmitted between domestic small ruminants and free-ranging wild Caprinae. This confirms previous speculations from seroepidemiological surveillance of M. conjunctivae infections in sheep (15). These studies showed the domestic sheep population to be an important reservoir of M. conjunctivae from which alpine chamois, which do not maintain the infection themselves, were considered to be infected by mainly neighboring sheep populations.
In summary, we have developed a molecular method for subtyping individual strains of M. conjunctivae based on the variable segment of the adhesin gene lppS. This method allowed us to perform a molecular epidemiological study of M. conjunctivae in Alpine regions and to demonstrate the possibility of transmission of M. conjunctivae between domestic sheep and wild Caprinae.
Acknowledgments
We are grateful to A. Pacher-Theinburg, Salzburg, Austria, and J. Steiner, Zell am See, Austria, who provided valuable samples from sheep and chamois; to Tomo Naglic, Zagreb, Croatia, for valuable strains from Croatia and epidemiological information; and to Alessandra Gaffuri, Bergamo, Italy, and Irene Bertoletti, Sondrio, Italy, for providing chamois samples from Italy. We also acknowledge François Poumarat, Lyon, France, for the gift of type strain HRC/581 and Yvonne Schlatter for valuable technical support. P. Ratti, G. Brosi, K. Jörger, H.-J. Blankenhorn, N. De Tann, and M. Nyffeler provided administrative and technical support for the study in Switzerland.
This research was funded by a research grant from the Institute for Veterinary Bacteriology, Berne, Switzerland; and by the Fund for Research on Infectious Keratoconjunctivitis, Chur, Switzerland.
REFERENCES
- 1.Aranaz, A., E. Liebana, A. Mateos, L. Dominguez, and D. Cousins. 1998. Restriction fragment length polymorphism and spacer oligonucleotide typing: a comparative analysis of fingerprinting strategies for Mycobacterium bovis. Vet. Microbiol. 61:311-324. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1999. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 3.Bannerman, E. S., and J. Nicolet. 1971. Isolation and identification of porcine Mycoplasma in Switzerland. Schweiz. Arch. Tierheilkd. 113:697-710. [PubMed] [Google Scholar]
- 4.Barile, M. F., R. A. Del Giudice, and J. G. Tully. 1972. Isolation and characterization of Mycoplasma conjunctivae sp. n. from sheep and goats with keratoconjunctivitis. Infect. Immun. 5:70-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belloy, L., E. M. Vilei, M. Giacometti, and J. Frey. 2003. Characterisation of LppS, an adhesin of Mycoplasma conjunctivae. Microbiology 149:185-193. [DOI] [PubMed]
- 6.Bergonier, D., F. De Simone, P. Russo, M. Solsona, M. Lambert, and F. Poumarat. 1996. Variable expression and geographic distribution of Mycoplasma agalactiae surface epitopes demonstrated with monoclonal antibodies. FEMS Microbiol. Lett. 143:159-165. [DOI] [PubMed] [Google Scholar]
- 7.Chang, C. C., R. W. Kasten, B. B. Chomel, D. C. Simpson, C. M. Hew, D. L. Kordick, R. Heller, Y. Piemont, and E. B. Breitschwerdt. 2000. Coyotes (Canis latrans) as the reservoir for a human pathogenic Bartonella sp.: molecular epidemiology of Bartonella vinsonii subsp. berkhoffii infection in coyotes from central coastal California. J. Clin. Microbiol. 38:4193-4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng, X., J. Nicolet, F. Poumarat, J. Regalla, F. Thiaucourt, and J. Frey. 1995. Insertion element IS1296 in Mycoplasma mycoides subsp. mycoides small colony identifies a European clonal line distinct from African and Australian strains. Microbiology 141:3221-3228. [DOI] [PubMed] [Google Scholar]
- 9.Daszak, P., A. A. Cunningham, and A. D. Hyatt. 2000. Emerging infectious diseases of wildlife—threats to biodiversity and human health. Science 287:443-449. [DOI] [PubMed] [Google Scholar]
- 10.Degiorgis, M. P., E.-M. Abdo, J. Nicolet, J. Frey, D. Mayer, and M. Giacometti. 2000. Immune responses to Mycoplasma conjunctivae in Alpine ibex, Alpine chamois, and domestic sheep in Switzerland. J. Wildl. Dis. 36:265-271. [DOI] [PubMed] [Google Scholar]
- 11.Degiorgis, M. P., E. Obrecht, A. Ryser, and M. Giacometti. 1999. The possible role of eye-frequenting flies in the transmission of Mycoplasma conjuntivae. Bull. Soc. Entomol. Suisse 72:189-194. [Google Scholar]
- 12.Giacometti, M., M. Janovsky, H. Jenny, J. Nicolet, L. Belloy, E. Goldschmidt-Clermont, and J. Frey. 2002. Mycoplasma conjunctivae infection is not maintained in alpine chamois in eastern Switzerland. J. Wildl. Dis. 38:297-304. [DOI] [PubMed] [Google Scholar]
- 13.Giacometti, M., J. Nicolet, J. Frey, M. Krawinkler, W. Meier, M. Welle, K. E. Johansson, and M. P. Degiorgis. 1998. Susceptibility of Alpine ibex to conjunctivitis caused by inoculation of a sheep-strain of Mycoplasma conjunctivae. Vet. Microbiol. 61:279-288. [DOI] [PubMed] [Google Scholar]
- 14.Giacometti, M., J. Nicolet, K. E. Johansson, T. Naglic, M. P. Degiorgis, and J. Frey. 1999. Detection and identification of Mycoplasma conjunctivae in infectious keratoconjunctivitis by PCR based on the 16S rRNA gene. J. Vet. Med. B 46:173-180. [DOI] [PubMed] [Google Scholar]
- 15.Janovsky, M., J. Frey, J. Nicolet, L. Belloy, E. Goldschmidt-Clermont, and M. Giacometti. 2001. Mycoplasma conjunctivae infection is self-maintained in the Swiss domestic sheep population. Vet. Microbiol. 83:11-22. [DOI] [PubMed] [Google Scholar]
- 16.Jones, G. E., A. Foggie, A. Sutherland, and D. B. Harker. 1976. Mycoplasmas and ovine keratoconjunctivitis. Vet. Rec. 99:137-141. [DOI] [PubMed] [Google Scholar]
- 17.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism, vol. 3. Academic Press AP, New York, N.Y.
- 18.Mayer, D., M. P. Degiorgis, W. Meier, J. Nicolet, and M. Giacometti. 1997. Lesions associated with infectious keratoconjunctivitis in Alpine ibex. J. Wildl. Dis. 33:413-419. [DOI] [PubMed] [Google Scholar]
- 19.Naglic, T., D. Hajsig, J. Frey, B. Seol, K. Busch, and M. Lojkic. 2000. Epidemiological and microbiological study of an outbreak of infectious keratoconjunctivitis in sheep. Vet. Rec. 147:72-75. [DOI] [PubMed] [Google Scholar]
- 20.Nicolet, J., and E. A. Freundt. 1975. Isolation of Mycoplasma conjunctivae from chamois and sheep affected with keratoconjunctivitis. Zentbl. Veterinarmed. B 22:302-307. [DOI] [PubMed] [Google Scholar]
- 21.Rosengarten, R., A. Behrens, A. Stetefeld, M. Heller, M. Ahrens, K. Sachse, D. Yogev, and H. Kirchhoff. 1994. Antigen heterogeneity among isolates of Mycoplasma bovis is generated by high-frequency variation of diverse membrane surface proteins. Infect. Immun. 62:5066-5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 23.Terrier, M.-E. 1998. La kératoconjonctivite des ongulés sauvages de montagne—reproduction expérimentale chez le mouflon (Ovis gmelini musimon). Ph.D. dissertation. University Claude Bernard of Lyon, Lyon, France.
- 24.Thrusfield, M. 1995. Veterinary epidemiology, 2nd ed. Blackwell Science, Oxford, United Kingdom.
- 25.Trotter, S. L., R. M. Franklin, E. J. Baas, and M. F. Barile. 1977. Epidemic caprine keratoconjunctivitis: experimentally induced disease with a pure culture of Mycoplasma conjunctivae. Infect. Immun. 18:816-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Halderen, A., W. J. J. Vanrensburg, A. Geyer, and J. H. Vorster. 1994. The Identification of Mycoplasma conjunctivae as an aetiological agent of infectious keratoconjunctivitis of sheep in South Africa. Onderstepoort. J. Vet. Res. 61:231-237. [PubMed] [Google Scholar]