Abstract
A collection of ten strains of Vibrio cholerae O139, comprising six isolates from Eichhornia crassipes, two from water of the River Ganga, and one each from a well and a hand pump, were characterized. All the strains carried the CTX genetic element (ctxA, zot, and ace) except for the st gene and carried structural and regulatory genes for toxin-coregulated pilus (tcpA, tcpI, and toxR), adherence factor (ompU), and accessory colonization factor (acfB); all produced cholera toxin (CT). These strains were resistant to trimethoprim, sulfamethoxazole, streptomycin, and to the vibriostatic agent pteridine. Results obtained by ribotyping and enterobacterial repetitive intergenic consensus sequence-PCR fingerprint analysis indicate that multiple clones of toxigenic-pathogenic V. cholerae O139 were present in the aquatic environment.
Until 1992, Vibrio cholerae belonging to serogroup O1 was considered to be the only causative agent of epidemic cholera (2). However, V. cholerae non-O1 serogroups were associated mostly with sporadic cases of diarrhea and extraintestinal infections (4, 24). It was found recently that a highly epidemic form of cholera-like disease on the Indian subcontinent was strongly associated with non-O1 strains of V. cholerae designated V. cholerae O139 Bengal (1, 21, 25), which subsequently spread to the different parts of the world (1, 6, 11). Studies of V. cholerae from environmental surface water indicated that O139 Bengal, like O1 vibrios and non-O1, non-O139 vibrios, may survive better in the aquatic environment in association with aquatic plants (13, 14, 26) and that environmental water may be a reservoir for infectious V. cholerae O139 (14, 29).
In the present study, we used molecular techniques to analyze V. cholerae O139 Bengal isolated between January 1992 and September 1994 from water and the aquatic plant Eichhornia crassipes (water hyacinth) from the river Ganga, Varanasi, India, to study genomic diversity and to examine the presence of virulence and regulatory genes, pathogenicity islands, and antibiotic resistance. Results obtained by ribotyping, enterobacterial repetitive intergenic consensus sequence (ERIC)-PCR, PCR analysis, and antibiotic susceptibility testing indicated that the cholera outbreaks caused by O139 vibrios between 1992 and 1994 were caused by toxigenic-pathogenic and antibiotic-resistant V. cholerae O139 strains identical to that found in the aquatic environment.
Bacterial strains.
A total of 10 strains of V. cholerae O139 were included in this study. Six isolates (EC1, EC2, EC3, EC6, EC7, and EC8) were from the aquatic plant Eichhornia crassipes and were collected between 28 January and 6 April 1992; two (GW3 and GW4) were from surface water of the River Ganga and were collected between 3 August and 29 August 1993 from different ghats on the bank of the River Ganga in Varanasi; and one each was from a well (WO4, collected on 10 September 1994 from Bhadaini, Varanasi) and a hand pump (HP11, collected on 5 August 1992 from Assi, Varanasi). The samples from the aquatic plants and water were processed by standard methods described elsewhere (26) and were identified earlier by following standard bacteriological techniques (33). These isolates, primarily identified as non-O1 V. cholerae, agglutinated with monoclonal O139 antiserum obtained from National Institute of Cholera and Enteric Diseases, Kolkata, and were confirmed to be V. cholerae O139. The V. cholerae O139 reference strain ATCC 51394, provided by G. B. Nair, was used as control. All strains were maintained in Tryptic soy broth (Difco) supplemented with 30% glycerol at −70°C. Before use, the identity of the cultures was confirmed by selected biochemical tests and serology (33).
Probes and hybridization.
A colony blot was prepared by using nylon filters (Hybond N+; Amersham International, Buckinghamshire, United Kingdom) and was processed by standard methods (18). Briefly, colonies were lysed with denaturing solution (0.5 M NaOH, 1.5 M NaCl) and neutralized in neutralizing solution (0.5 M Tris-HCl [pH 8.0], 1.5 M NaCl), and the liberated DNA was fixed to the nylon membrane by exposure to UV light for 2 min, in accordance with the supplier's instructions, and hybridized with the ctxB probe, a 1.4-kb KpnI fragment of pCVD621 (16).
Ribotyping was performed by the procedure described by Faruque et al. (10) in which the restriction enzyme BglI (Amersham Pharmacia Biotech, Piscataway, N.J.) was used to digest the chromosomal DNA, and hybridization was performed with [α-32P]dCTP (3,000 Ci/mmol) (BARC, Bombay, India)-labeled 16S and 23S rRNA probe (5). Autoradiographs were developed in PhosphorImager (Bio-Rad, Hercules, Calif.).
ERIC-PCR.
With the two oligonucleotide primers 5′-ATG TAA GCT CCT GGG GAT TCA C-3′ and 5′-AAG TAA GTG ACT GGG GTG AGC G-3′, ERIC-PCR was carried out as described by Rivera et al. (22). The amplicons were electrophoresed in 1.8% (wt/vol) agarose at 80 V for 6 h and stained in ethidium bromide. The fingerprint pattern was digitized in a Fluoro-S MultiImager (Bio-Rad) and analyzed by using Bionumerics fingerprint analyst (Applied Maths, Kortrijk, Belgium) software with a simple-match similarity matrix, and data were clustered by the unweighted pair group method with arithmetic means.
Hexaplex PCR assay.
Presence of the virulence and regulatory genes ctxA, zot, ace, tcpA, ompU, and toxR was determined by a hexaplex PCR assay (28). Oligonucleotide sequences for the primers for the virulence and regulatory genes have been described previously (23, 27). Amplified products were separated on 1.8% agarose gel, stained in ethidium bromide, and visualized by using a Fluoro-S MultiImager (Bio-Rad).
PCR assay.
Presence of the tcpI gene in V. cholerae O139 strains was determined by a PCR assay described previously (23). The acfB gene was detected by a PCR assay described by Faruque et al. (8), and the st gene was detected by another PCR assay described by Vicente et al. (31). All oligonucleotides were purchased either from GIBCO-BRL (Gaithersburg, Md.) or Sigma Genosys (Cambridgeshire, United Kingdom). The size of the amplicon for tcpI, acfB, and st genes was ascertained by electrophoresis in 1.5% agarose gels.
Assay for CT production.
The ability of the strains to produce cholera toxin (CT) in vitro was determined by GM1 ganglioside-dependent enzyme-linked immunosorbent assay (GM1-ELISA) as described by Svennerholm and Holmgren (30). Briefly, 100 μl of the samples was added to each well of microtiter plates precoated with GM1, and the plates were incubated at 37°C for 2 h. After the plates were washed with phosphate-buffered saline (PBS) containing 0.05% Tween 20 (Sigma), the GM1-bound CT was reacted with rabbit anti-CT antibody (Sigma) diluted 1:10,000 in PBS containing 0.05% Tween 20 and 0.5% bovine serum albumin. Antibody binding to CT was detected by reaction with horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (whole molecule) antibody (Sigma) diluted 1:5,000 in PBS containing 0.05% Tween 20 and 0.5% bovine serum albumin with the 3, 3′, 5, 5′-tetra methyl benzidine (TMB) substrate system (ready-to-use; Sigma) followed by measurement of the A405 in a microplate reader (Model 550; Bio-Rad).
Antibiotic resistance.
All the V. cholerae O139 strains were tested for antimicrobial resistance by the method of Bauer et al. (3) by using antibiotics discs (Hi-Media Laboratories, Bombay, India), such as discs with ampicillin (10 μg), chloramphenicol (30 μg), cefotaxime (30 μg), ciprofloxacin (5 μg), cephalexine (30 μg), co-trimoxazole (25 μg), furazolidone (100 μg), gentamicin (10 μg), neomycin (30 μg), norfloxacin (10 μg), polymyxin B (50 U), streptomycin (30 μg), nalidixic acid (30 μg), tetracycline (30 μg), and the vibriostatic agent pteridine (10 μg and 150 μg).
Detection of the SXT element by PCR and restriction fragment length polymorphism.
The presence of the SXT genetic element was determined by a PCR assay based on a published sequence of sxt gene (12) in which sets of primers were used (Table 1). PCR amplification of the target DNA was carried out in a thermal cycler (Bio-Rad) by using 200-μl PCR tubes with a reaction mixture volume of 25 μl. Each reaction mixture contained 2.5 μl of 10× PCR amplification buffer (500 mM KCl, 100 mM Tris-HCl [pH 9.0], 1.0% Triton X-100) (Promega, Madison, Wis.), 2.5 μl of MgCl2 (25 mM), 2.5 μl each of 2.5 mM dATP, dCTP, dGTP, and dTTP (Amersham Pharmacia Biotech), 1 μl each of forward and reverse primers (20 ng/μl), 0.125 μl of Taq DNA polymerase at 5U/μl (Promega), and Milli-Q water to a final volume of 22.5 μl and 2.5 μl of cell lysate (template DNA). PCR was programmed for an initial denaturation at 94°C for 2 min followed by 30 cycles consisting of denaturation at 94°C for 1 min, annealing at 60°C for 1 min, and extension at 72°C for 1 min and a final extension at 72°C for 10 min. The size of the PCR product was ascertained by electrophoresis in 1.0% agarose gel. The presence of the sxt gene in the PCR-positive strains was confirmed by dot blot hybridization by using PCR-generated amplicon obtained by using SXT1 and SXT4 primers from a positive control V. cholerae O139 strain MO45 (ATCC 51394). The identity of the PCR products was further verified by digesting the PCR products with the restriction enzymes HindIII or MseI (NEB, Beverly, Mass.) and by determining the sizes of the digestion fragments by agarose (3.0%) gel electrophoresis and comparing the sizes of the fragments with expected sizes based on the published sequence of the sxt gene (12).
TABLE 1.
Oligonucleotide primer sequences used for the detection of the SXT element by PCR in V. cholerae O139 strainsa
| Primer pair | Sequence position | Oligonucleotide sequence | Amplicon size (bp) | Fragment sizes (bp) with restriction enzyme:
|
|
|---|---|---|---|---|---|
| MseI | HindIII | ||||
| SXT1 | 1-22 | ATGGCGTTATCAGTTAGCTGGC | 1,240 | 392, 291, 204, 183 | 946, 184, 110 |
| SXT2 | 1218-1240 | ATTGTCCGAAACGCCAGTTGAAC | 136, 34 | ||
| SXT3 | 90-109 | TCGGGTATCGCCCAAGGGCA | 946 | 291, 204, 187, 136 | 741, 110, 95 |
| SXT4 | 1016-1035 | GCGAAGATCATGCATAGACC | 128 | ||
| SXT1 | 1-22 | ATGGCGTTATCAGTTAGCTGGC | 1,035 | 291, 204, 187, 183 | 741, 184, 110 |
| SXT4 | 1016-1035 | GCGAAGATCATGCATAGACC | 136, 34 | ||
| SXT3 | 90-109 | TCGGGTATCGCCCAAGGGCA | 1,151 | 392, 291, 204, 136 | 946, 110, 95 |
| SXT2 | 1218-1240 | ATTGTCCGAAACGCCAGTTGAAC | 128 | ||
SXT element GenBank accession no. AF099172.
Genomic diversity of Vibrio cholerae O139.
It was reported that O139 Bengal, like O1 vibrios and non-O1, non-O139 vibrios, survive better in the aquatic environment in association with plankton (7, 13, 14, 26) and that the aquatic environment serves as reservoir of toxigenic V. cholerae strains. The present study was designated to ascertain whether strains of V. cholerae O139 isolated from the environmental sources between January 1992 and September 1994 represented identical clones that caused an epidemic in 1992 through 1994.
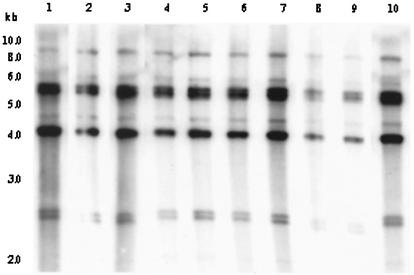
Analysis of rRNA genes with BglI produced three different restriction patterns (patterns B-I, B-II, and B-IV) among environmental V. cholerae O139 strains (Fig. 1). The rRNA gene restriction patterns were reproducible in repeated assays and consisted of 8 to 10 bands between 9.0 and 2.2 kb in size. Of the 10 isolates of V. cholerae O139, two belonged to ribotype I, seven belonged to ribotype II, and one belonged to ribotype IV. The restriction pattern representing ribotype IV contained a unique band of 7.0 kb which is not present in any other restriction patterns (Fig. 1). The O139 vibrios isolated from epidemic outbreaks between 1992 and 1994 were of two reported ribotypes designated B-I and B-II (9, 20). Results of this study thus suggest that strains of V. cholerae O139 that caused an epidemic between 1992 and 1994 had a clonal origin and may have originated in the aquatic environment.
FIG. 1.
Genomic DNA of environmental V. cholerae O139 strains were digested with BglI and probed with a 7.5-kb BamHI fragment of the Escherichia coli rRNA clone pkk3535. Restriction pattern I, produced by two strains (GW3 and WO4), is shown in lanes 1 and 2; restriction pattern II, produced by seven strains (EC1, EC2, EC3, EC6, EC7, EC8, and HP11), is shown in lanes 3 through 9; restriction pattern IV, produced by one strain (GW4) is shown in lane 10. The numbers indicating the molecular sizes of bands correspond to a 1-kb DNA ladder (NEB) that was used as molecular size marker.
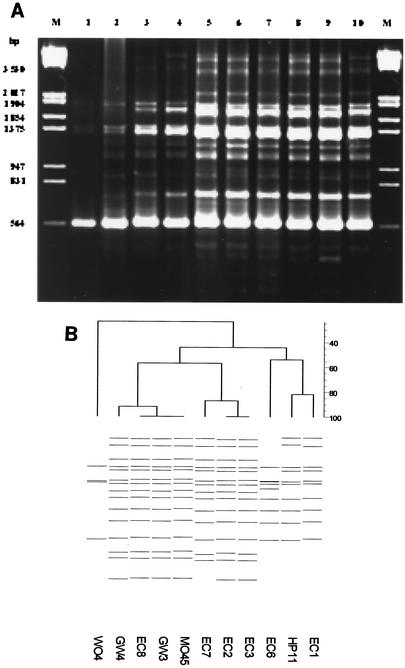
ERIC-PCR of genomic DNA from V. cholerae O139 yielded a total of four fingerprint profiles (profiles I through IV) consisting of 4 to 15 bands ranging in size between 0.31 and 3.5 kb (Fig. 2A). One of the 10 strains isolated from well water (WO4) yielded a fingerprint pattern consisting of four bands of 1.74, 1.37, 1.26, and 0.56 kb in size; this profile was designated E-I. Another strain (EC6) amplified four additional bands of 1.18, 1.02, 0.85, and 0.71 kb in size, and its profile was designated E-II. Two strains, one each from Eichhornia crassipes (EC1) and a hand pump (HP11), yielded identical fingerprint patterns which differed from E-II in lacking a 1.18-kb band but which amplified three additional bands of 3.5, 2.8, and 1.64 kb in size; this profile was designated E-III. However, a majority of strains, consisting of four strains from Eichhornia crassipes (EC2, EC3, EC7, and EC8) and two strains from the water of the River Ganga (GW3 and GW4), represented ERIC-PCR fingerprint profile E-IV, which amplified 16 bands, including five additional bands of 2.2, 1.13, 0.48, 0.42, and 0.31 kb in size.
FIG. 2.
(A) DNA fingerprints of environmental isolates of V. cholerae O139 strains generated by ERIC-PCR amplification. Fingerprint I, produced by one strain (WO4), is shown in lane 1; fingerprint II, produced by another strain (EC6) is shown in lane 2; fingerprint III, produced by two strains (EC1 and HP11) is shown in lanes 3 and 4; fingerprint IV, produced by six strains (EC8, GW3, GW4, EC2, EC3, and EC7) is shown in lanes 5 through 10. A lambda DNA digested with EcoRI and HindIII shown in lanes M was used as molecular weight marker. (B) Digitized ERIC-PCR profiles obtained from genomic DNA of environmental V. cholerae O139 isolates. The dendrogram was constructed by using the Bionumerics fingerprint analyst software (Applied Maths) with simple match similarity matrix, and data were clustered by the unweighted pair group method with arithmetic means.
It can be seen from the dendrogram that strains isolated from environmental sources form four different clusters (Fig. 2B). The observed difference in banding patterns within the O139 serogroup suggested divergence in genomic organization and can be hypothesized to have arisen from genetic assortment taking place in the environment over time. The results of the ERIC-PCR fingerprint profiles supported the results obtained by ribotyping that multiple clones of V. cholerae O139 were present in the aquatic environment. These observations are in agreement with those of others who demonstrated heterogeneity in O139 V. cholerae strains (10, 17, 20, 29).
Analysis of virulence and regulatory genes.
All of the V. cholerae O139 strains were positive for the ctxA, zot, ace, tcpA, ompU, and toxR genes (Table 2). The amplified tcpA gene in V. cholerae O139 strains was characteristic of the El Tor biotype tcpA gene. These strains were also positive for the tcpI and acfB genes. However, all were negative by PCR for the st gene. Colony blot hybridization reveals that all the environmental strains carried the ctxB gene (data not shown).
TABLE 2.
Antibiotic resistance, virulence profiles, and genomic patterns of Vibrio cholerae O139 used in this study
| Strain no. | Source | Antibiotic resistance patterna | Presence of virulence and regulatory genesb
|
Ribotype | ERIC profile | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ctxA | zot | ace | tcpA | ompU | acfB | tcpI | toxR | |||||
| MO45 | Diarrheal | A, Co, F, S | + | + | + | + | + | + | + | + | B-II | E-IV |
| EC1 | E. crassipes | Co, F, S | + | + | + | + | + | + | + | + | B-II | E-III |
| EC2 | E. crassipes | Co, F, S, Cf | + | + | + | + | + | + | + | + | B-II | E-IV |
| EC3 | E. crassipes | Co, F, S | + | + | + | + | + | + | + | + | B-II | E-IV |
| EC6 | E. crassipes | Co, F, S | + | + | + | + | + | + | + | + | B-II | E-II |
| EC7 | E. crassipes | Co, F, S | + | + | + | + | + | + | + | + | B-II | E-IV |
| EC8 | E. crassipes | A, Co, F, S, Ce | + | + | + | + | + | + | + | + | B-II | E-IV |
| HP11 | Hand pump | Co, F, S | + | + | + | + | + | + | + | + | B-II | E-III |
| GW3 | River water | A, Co, F, N, S | + | + | + | + | + | + | + | + | B-I | E-IV |
| GW4 | River water | A, Co, F, S | + | + | + | + | + | + | + | + | B-IV | E-IV |
| WO4 | Well water | Co, F, S | + | + | + | + | + | + | + | + | B-I | E-I |
A, ampicillin; Ce, cefotaxime; Cf, ciprofloxacin; Co, co-trimoxazole; F, furazolidone; N, neomycin; S, streptomycin.
None of the strains were positive for the st gene.
V. cholerae O1 and O139 strains isolated from the aquatic environment have been reported to be nontoxigenic, i.e., lacking the capability to produce CT (10, 15). However, we have demonstrated that environmental isolates of V. cholerae O1 and O139, like clinical isolates, possessed ctxA, zot, ace, tcpA, and ompU genes (27). Results obtained in this study also indicate that environmental V. cholerae O139 strains retained the core of the CTX genetic element, TCP pathogenicity islands, and the ompU and acfB genes, even in the aquatic environment.
Production of CT.
All of the O139 strains isolated from the environment produced CT, and the level of CT production varied between 20 and 400 pg/ml among the different strains. These strains had ability to adhere, colonize, and produce CT in the small intestine, indicating that environmental strains had all of the characteristics of epidemic cholera.
Antibiotic resistance.
The V. cholerae O139 Bengal strains, which emerged during 1992 and 1993, were sensitive to tetracycline and showed resistance to sulfamethoxazole, trimethoprim, streptomycin, and pteridine (19, 21). Waldor and coworkers (32) reported the presence of a 62-kb self-transmissible transposon-like element (SXT element) encoding resistance to sulfamethoxazole, trimethoprim, and streptomycin in V. cholerae O139 strains isolated from the epidemic. All the environmental O139 isolates included in this study were resistant to trimethoprim, sulfamethoxazole, streptomycin, furazolidone, and pteridine (Table 2). Three strains showed additional resistance to ampicillin, and one strain each was resistant to cefotaxime, ciprofloxacin, and neomycin (Table 2).
All the V. cholerae O139 strains were positive by PCR for the sxt gene and amplified portions of the SXT genetic element consisting of 946, 1,035, 1,151, and 1,240 bp, depending on the combinations of primers used (Table 1). Subsequent restriction analysis of the amplicon, generated by using sets of primers or with HindIII or MseI, produced sets of fragments whose sizes agreed with the expected sizes based on the published sequence of SXT (Table 1). All strains also showed positive results in a dot blot assay (data not shown). The results of this study thus indicate that environmental V. cholerae O139 strains, like clinical isolates, possessed the SXT genetic element encoding resistance for trimethoprim, sulfamethoxazole, streptomycin, and the vibriostatic agent pteridine.
Epidemiological significance of clonal diversity.
Faruque et al. (9, 10) reported the emergence in Bangladesh of new ribotypes of toxigenic V. cholerae O139 showing distinct ctxA genotypes and suggested that new clones possibly emerged from nontoxigenic progenitors. Since ribotype B-I and B-II V. cholerae O139 strains that caused an epidemic between 1992 and 1994 were present in the aquatic environment, these observations indicate that simultaneous changes may have occurred or are occurring in the rRNA operons and in other unidentified genes in the aquatic environment that might influence the prevalence and emergence of new clones of O139 strains by interacting with environmental factors. The emergence of a new pathogenic O139 serogroup in areas of endemicity allows it to be hypothesized that the change occurs in the survival capacity of the pathogenic clone for combating intestinal immunity or stresses in environmental habitats. In view of the fluctuations observed in the prevalence of V. cholerae O139 relative to that of V. cholerae O1 in human infection (9, 10) and the rapid genotypic and phenotypic changes, including changing patterns of antibiotics resistance, further ecological studies are required to explain the appearance and disappearance and the mobility of genetic elements encoding virulence properties as well as antibiotic resistance.
Acknowledgments
This research was supported by funds contributed to Rajiv Gandhi Centre for Biotechnology from the Department of Biotechnology, New Delhi, India. Junior research fellowships awarded by Council of Scientific and Industrial Research, New Delhi, India, to R. Bhanumathi and Sree Renjini Isac are gratefully acknowledged.
REFERENCES
- 1.Albert, M. J., A. K. Siddique, M. S. Islam, A. S. G. Faruque, M. Ansaruzzaman, S. M. Faruque, and R. B. Sack. 1993. Large outbreak of clinical cholera due to Vibrio cholerae non-O1 in Bangladesh. Lancet 341:704.. [DOI] [PubMed] [Google Scholar]
- 2.Barua, D. 1992. History of cholera, p. 1-36. In D. Barua and W. B. Greenough (ed.), Cholera. Plenum Publishing Co., New York, N.Y.
- 3.Bauer, A. W., W. M. M. Kirby, J. C. Sherris, and M. Truck. 1966. Antibiotic susceptibility testing by a standardized single disc diffusion method. Am. J. Clin. Pathol. 45:493-496. [PubMed] [Google Scholar]
- 4.Blake, P. A., R. W. Weaver, and D. G. Hollins. 1980. Diseases of human (other than cholera) caused by vibrios. Annu. Rev. Microbiol. 34:341-367. [DOI] [PubMed] [Google Scholar]
- 5.Brosius, J., A. Ullrich, M. A. Raker, A. Gray, T. J. Dull, R. R. Gutell, and H. F. Noller. 1981. Construction and fine mapping of recombinant plasmids containing the rrnB ribosomal RNA operon of E. coli. Plasmid 6:112-118. [DOI] [PubMed] [Google Scholar]
- 6.Chongsa-nguan, M., W. Chaicumpa, P. Moolasart, P. Vandhsingha, T. Shimada, H. Kurazono, and Y. Takeda. 1993. Vibrio cholerae O139 Bengal in Bangkok. Lancet 342:430-431. [DOI] [PubMed] [Google Scholar]
- 7.Colwell, R. R., J. Kaper, and S. W. Joseph. 1977. Vibrio cholerae, Vibrio parahaemolyticus, and other vibrios: occurrence and distribution in Chesapeake Bay. Science 198:394-396. [PubMed] [Google Scholar]
- 8.Faruque, S. M., Asadulghani, M. N. Saha, A. R. M. Abdul Alim, M. J. Albert, K. M. Nasirul Islam, and J. J. Mekalanos. 1998. Analysis of clinical and environmental strains of nontoxigenic Vibrio cholerae for susceptibility to CTXφ: molecular basis for origination of new strains with epidemic potential. Infect. Immun. 66:5819-5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faruque, S. M., M. N. Saha, Asadulghani, P. K. Bag, R. K. Bhadra, S. K. Bhattacharya, R. B. Sack, Y. Takeda, and G. B. Nair. 2000. Genomic diversity among Vibrio cholerae O139 strains isolated in Bangladesh and India between 1992 and 1998. FEMS Microbiol. Lett. 184:279-284. [DOI] [PubMed] [Google Scholar]
- 10.Faruque, S. M., M. N. Saha, Asadulghani, D. A. Sack, R. B. Sack, Y. Takeda, and G. B. Nair. 2000. The O139 serogroup of Vibrio cholerae comprises diverse clones of epidemic and nonepidemic strains derived from multiple V. cholerae O1 or non-O1 progenitors. J. Infect. Dis. 182:1161-1168. [DOI] [PubMed] [Google Scholar]
- 11.Fischer-Hoch, S. P., A. Khan, I. U. Huq, M. A. Khan, and E. D. Mintz. 1993. Vibrio cholerae O139 in Karachi, Pakistan. Lancet 342:1422-1423. [DOI] [PubMed] [Google Scholar]
- 12.Hochhut, B., and M. K. Waldor. 1999. Site specific integration of the conjugal Vibrio cholerae SXT element into prfC. Mol. Microbiol. 32:99-110. [DOI] [PubMed] [Google Scholar]
- 13.Huq, A., R. R. Colwell, M. A. R. Chowdhury, B. Xu, S. M. Moniruzzaman, M. S. Islam, M. Yunus, and M. J. Albert. 1995. Co-existence of Vibrio cholerae O1 and O139 Bengal in plankton in Bangladesh. Lancet 345:1245.. [DOI] [PubMed] [Google Scholar]
- 14.Islam, M. S., B. S. Drasar, and R. B. Sack. 1994. The aquatic flora and fauna as reservoir of Vibrio cholerae: a review. J. Diarrh. Dis. Res. 12:87-96. [PubMed] [Google Scholar]
- 15.Kaper, J. B., S. L. Moseley, and S. Falkow. 1981. Molecular characterization of environmental and nontoxigenic strains of Vibrio cholerae. Infect. Immun. 32:661-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaper, J. B., J. G. Morris, Jr., and M. Nishibuchi. 1988. DNA probes for pathogenic Vibrio species, p. 65-77. In F. C. Tenover (ed.), DNA probes for infectious disease. C R C Press, Inc., Boca Raton, Fla.
- 17.Kurazono, H., S. Yamasaki, Orn-Anong Ratchtrachenchai, G. B. Nair, and Y. Takeda. 1996. Analysis of Vibrio cholerae O139 Bengal isolated from different geographical area using micro restriction analysis. Microbiol. Immunol. 40:303-305. [DOI] [PubMed] [Google Scholar]
- 18.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 19.Mukhopadhyay, A. K., S. Garg, R. Mitra, A. Basu, D. Datta, S. K. Bhattacharya, T. Shimada, T. Takeda, Y. Takeda, and G. B. Nair. 1996. Temporal shifts in traits of Vibrio cholerae strains isolated from hospitalized patients in Calcutta: a 3-year (1993 to 1995) analysis. J. Clin. Microbiol. 34:2537-2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Popovic, T., P. I. Fields, Ø. Olsvik, J. G. Wells, G. M. Evins, D. N. Cameron, J. J. Farmer III, C. A. Bopp, K. Wachsmuth, R. B. Sack, M. J. Albert, G. B. Nair, T. Shimada, and J. C. Feeley. 1995. Molecular subtyping of toxigenic Vibrio cholerae O139 causing epidemic cholera in India and Bangladesh; 1992-93. J. Infect. Dis. 171:122-127. [DOI] [PubMed] [Google Scholar]
- 21.Ramamurthy, T., S. Garg, R. Sharma, S. K. Bhattacharya, T. Shimada, T. Takeda, Y. Takeda, and G. B. Nair. 1993. Emergence of a novel strain of Vibrio cholerae with epidemic potential in southern India. Lancet 341:703-704. [DOI] [PubMed] [Google Scholar]
- 22.Rivera, I. G., M. A. R. Chowdhury, A. Huq, D. Jacobs, M. T. Martins, and R. R. Colwell. 1995. Enterobacterial repetitive intergenic consensus sequence and the PCR to generate fingerprints of genomic DNAs from Vibrio cholerae O1, O139, and non-O1 strains. Appl. Environ. Microbiol. 61:2898-2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rivera, I. N. G., J. Chun, A. Huq, R. B. Sack, and R. R. Colwell. 2001. Genotype associated with virulence in environmental isolates of Vibrio cholerae. Appl. Environ. Microbiol. 67:2421-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanyal, S. C. 1986. Vibrio toxin, p. 207-225. In F. Dorner and J. Drews (ed.), Pharmacology of bacterial toxins: international encyclopedia of pharmacology and therapeutics, vol. 119. Pergamon Press, Oxford, United Kingdom.
- 25.Shimada, T., G. B. Nair, B. C. Deb, M. J. Albert, R. B. Sack, and Y. Takeda. 1993. Outbreak of Vibrio cholerae non-O1 in India and Bangladesh. Lancet 341:1347.. [DOI] [PubMed] [Google Scholar]
- 26.Shukla, B. N., D. V. Singh, and S. C. Sanyal. 1995. Attachment of non-culturable toxigenic Vibrio cholerae O1 and non-O1 and Aeromonas spp. to the aquatic arthropod Gerris spinolae and plants in the River Ganga, Varanasi. FEMS Immunol. Med. Microbiol. 12:113-120. [DOI] [PubMed] [Google Scholar]
- 27.Singh, D. V., M. H. Matte, G. R. Matte, S. Ziang, F. Sabeena, B. N. Shukla, S. C. Sanyal, A. Huq, and R. R. Colwell. 2001. Molecular analysis of Vibrio cholerae O1, O139, non-O1, and non-O139 strains: clonal relationships between clinical and environmental isolates. Appl. Environ. Microbiol. 67:910-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh, D. V., Sree Renjini Isac, and R. R. Colwell. 2002. Development of a hexaplex PCR assay for rapid detection of virulence and regulatory genes in Vibrio cholerae and Vibrio mimicus. J. Clin. Microbiol. 40:4321-4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Son, R., G. Rusul, L. Samuel, Yuherman, S. Senthil, A. Rasip, E. H. Nasreldin, and M. Nishibuchi. 1998. Characterization of Vibrio cholerae O139 Bengal isolated from water in Malaysia. J. Appl. Microbiol. 85:1073-1077. [DOI] [PubMed] [Google Scholar]
- 30.Svennerholm, A. M., and J. Holmgren. 1978. Identification of Escherichia coli heat-labile enterotoxin by means of a ganglioside immunosorbent assay (GM1-ELISA). Curr. Microbiol. 1:19-23. [Google Scholar]
- 31.Vicente, A. C. P., A. M. Coelho, and C. A. Salles. 1997. Detection of Vibrio cholerae and V. mimicus heat stable toxin gene sequence by PCR. J. Med. Microbiol. 46:398-402. [DOI] [PubMed] [Google Scholar]
- 32.Waldor, M. K., H. Tschape, and J. J. Mekalanos. 1996. A new type of conjugative transposon encodes resistance to sulfamethoxazole, trimethoprim, and streptomycin in Vibrio cholerae O139. J. Bacteriol. 178:4157-4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.World Health Organization. 1987. Manual for laboratory investigation of acute enteric infections. CDD/83.3. Program for control of diarrheal disease. World Health Organization, Geneva, Switzerland.