Abstract
Cryptococcus neoformans is a pathogenic yeast that is currently divided into three varieties, five serotypes, and eight molecular types. The following report describes the use of PCR-restriction fragment length polymorphism (RFLP) analysis of the phospholipase B gene (PLB1) as a simple tool to differentiate between C. neoformans subgroups. A PLB1 fragment, 1,970 bp, was amplified and digested with either AvaI or HindIII. Both sets of profiles grouped the isolates into their respective varieties, but only the AvaI profiles allowed for the identification of the eight molecular types via the corresponding RFLP profiles A1 to A8. Digestion of the same fragments with HindIII resulted in RFLP profiles H1 to H5, which distinguished only between serotype A, AD, D, and B/C. Neither enzyme distinguished serotype B from serotype C. The serotype AD profile was a composite of the serotype A and D profiles. Further investigation showed that the serotype AD isolates used in this study are heterozygous, with one allele of PLB1 originating from a serotype A parent and the other from a serotype D parent.
The encapsulated, basidiomycetous yeast Cryptococcus neoformans is a human and veterinary pathogen that causes cryptococcosis. Three genetically, ecologically, and clinically different varieties exist. C. neoformans var. grubii (13) (serotype A), which has a worldwide distribution, is mainly associated with pigeon excreta, and C. neoformans var. neoformans (serotype D) is most commonly found in Europe and causes disease less frequently (1, 14). Both varieties cause disease predominantly in immunocompromised patients. C. neoformans var. gattii (serotypes B and C) is mainly found in tropical and subtropical climates, causing pulmonary cryptococcomas (24), affecting predominantly immunocompetent individuals, and being isolated from a variety of environmental sources (12, 16). In addition to these three varieties a hybrid serotype AD exists. Boekhout et al. and Diaz et al. (4, 10) have argued that C. neoformans var. gattii (serotypes B and C) is a separate species, proposing the name C. bacillisporus. Given these proposed changes and the ecological, epidemiological, and clinical differences within the C. neoformans complex, a clear and simple method of subgrouping isolates is desirable. Several molecular typing methods have been applied to C. neoformans isolates. These include multilocus enzyme electrophoresis (2), mating type locus PCR (6), sequencing (10, 34), karyotyping (23, 32), Southern hybridization with gene-specific probes (5, 28), random amplification of polymorphic DNA (7, 22), amplified fragment length polymorphism (AFLP) analysis (4), PCR-fingerprinting (8, 20, 21, 31), and a combination of PCR and restriction fragment length polymorphism (PCR-RFLP) analysis (27, 30). Most of these techniques distinguish individual isolates but had limitations in identifying serotypes and varieties at the same time.
PCR-fingerprinting using minisatellite (M13)- or microsatellite [(GACA)4, (GTG)5]-specific sequences as single primers in the PCR has grouped global isolates of C. neoformans into eight major molecular types based on previous publications (11, 20). These groupings have been confirmed by Cogliati et al. (8) and Viviani et al. (31), who have used a modified PCR-fingerprinting technique, with the primer (GACA)4, grouping Italian isolates of C. neoformans var. grubii and var. neoformans into four major molecular types, and by Boekhout et al. (4), using AFLP analysis to group C. neoformans isolates into six major AFLP groups. The comparable molecular types and genotypes identified in these independent studies are as follows: VNI = AFLP1 = Cogliati VN6 (serotype A, var. grubii); VNII = AFLP1A (serotype A, C. neoformans var. grubii); VNIII = AFLP3 = Cogliati VN3 and VN4 (serotype AD, hybrid between C. neoformans var. grubii and var. neoformans); VNIV = AFLP2 = Cogliati VN1 (serotype D, C. neoformans var. neoformans); VGI = AFLP4, VGII = AFLP6, VGIII = AFLP5 and VGIV (all corresponding to serotypes B and C, C. neoformans var. gattii) (4, 8, 11, 20, 31). PCR-fingerprinting and AFLP could not distinguish between the serotypes B and C, indicating that serotyping does not necessarily reflect the true genotype of the isolates.
The phospholipase B (PLB1) gene of C. neoformans was recently described to play an important role in virulence of C. neoformans var. grubii (9). Phospholipases are found in a wide range of organisms, including both pathogenic and nonpathogenic species of yeasts and moulds (3, 9, 17, 19). They are involved in the breakdown of phospholipids and may contribute to cell damage during cryptococcal infection. PLB1 was chosen in this study as a potential target for subtyping of C. neoformans isolates according to varieties, serotypes, and molecular types via PCR-RFLP analysis, since its entire sequence was either available from GenBank for serotypes A and D or had been generated in our own laboratory for serotypes B and C (15). The ploidy status of serotype AD isolates at the PLB1 locus was also investigated.
MATERIALS AND METHODS
Strains.
The 73 C. neoformans isolates used in this study are shown in Table 1. cells were cultured on Sabouraud's plates (SAB) (peptone, 10 g liter−1; glucose, 40 g liter−1; agar, 15 g liter−1 [pH 5.5]) at 30°C for 2 days before each experiment and were stored as 10% glycerol stocks at −70°C. Single colonies were picked and streaked onto a fresh plate before DNA was extracted. The serotype was determined using the IATRON Kit (IATRON, Tokyo, Japan) and the molecular type was determined by PCR-fingerprinting with the microsatellite specific primer M13 [5′ GAGGGTGGCGGTTCT 3′] (20).
TABLE 1.
Origin, serotype, molecular type, and PLB1 RFLP pattern of C. neoformans strains used in this study
| Strain | Origin | Serotype | Molecular typea | RFLP profile
|
|
|---|---|---|---|---|---|
| AvaI | HindIII | ||||
| Cryptococcus neoformans var. grubii | |||||
| BL-1 | Lung, Australia | A | VNI | A1 | H1 |
| CN042 | CSF, New Zealand | A | VNI | A1 | H1 |
| H99 | CSF, USA | A | VNI | A1 | H1 |
| LA190 = 76(a) | Pigeon droppings, Spain | A | VNI | A1 | H1 |
| LA193 = 6(a) | Pigeon droppings, Spain | A | VNI | A1 | H1 |
| LA194 = I-01 | CSF, Colombia | A | VNI | A1 | H1 |
| LA195 = I-21 | CSF, Colombia | A | VNI | A1 | H1 |
| LA199 = I-50 | CSF, Colombia | A | VNI | A1 | H1 |
| WM 148 | CSF, Australia | A | VNI | A1 | H1 |
| CN037 | CSF, New Zealand | A | VNII | A2 | H1 |
| CN038 | CSF, New Zealand | A | VNII | A2 | H1 |
| CN039 | CSF, New Zealand | A | VNII | A2 | H1 |
| CN041 | CSF, New Zealand | A | VNII | A2 | H1 |
| LA327 = MLP-1473 | CSF, Brazil | A | VNII | A2 | H1 |
| WM 626 | CSF, Australia | A | VNII | A2 | H1 |
| AD hybrid | |||||
| WM 328 = 88-1358 | No data | AD | VNIII | A3 | H2 |
| WM 329 = 90-7031 | No data | AD | VNIII | A3 | H2 |
| LA 184 = 2(c) | CSF, Spain | AD | VNIII | A3 | H2 |
| LA 185 = 3(c) | CSF, Spain | AD | VNIII | A3 | H2 |
| LA 192 = 4(a) | Pigeon droppings, Spain | AD | VNIII | A3 | H2 |
| LA 317 = 138027 | Blood, Argentina | AD | VNIII | A3 | H2 |
| RKI-A208 | Pigeon droppings, Germany | AD | VNIII | A3 | H2 |
| RKI-M1644/90 | Clinical, Germany | AD | VNIII | A3 | H2 |
| RKI-M364/98 | Clinical, Germany | AD | VNIII | A3 | H2 |
| WM 339 = RU-27809 | No data | AD | VNIII | A3 | H2 |
| TBS 28 | Clinical, India | AD | VNIII | A3 | H2 |
| TBS 54 | Clinical, India | AD | VNIII | A3 | H2 |
| WM 628 | CSF, Australia | AD | VNIII | A3 | H2 |
| Cryptococcus neoformans var. neoformans | |||||
| B3501 = ATCC 52817 | CSF, USA | D | VNIV | A4 | H3 |
| B-87455 | Blood, Australia | D | VNIV | A4 | H3 |
| CN3501 = ATCC 52817 | CSF, USA | D | VNIV | A4 | H3 |
| RKI-318 | Clinical, Germany | D | VNIV | A4 | H3 |
| RKI-1089/89 | Clinical, Germany | D | VNIV | A4 | H3 |
| RKI-M186/99 | Clinical, Germany | D | VNIV | A4 | H3 |
| TP0416a | Eucalyptus debris, Australia | D | VNIV | A4 | H3 |
| WM 629 | Blood, Australia | D | VNIV | A4 | H3 |
| Cryptococcus neoformans var. gattii | |||||
| LA174 = 48(a) | Goat, Spain | B | VGI | A5 | H4 |
| LA175 = 50(a) | Goat, Spain | B | VGI | A5 | H4 |
| LA176 = 52(a) | Goat, Spain | B | VGI | A5 | H4 |
| LA177 = 54(a) | Goat, Spain | B | VGI | A5 | H4 |
| LA178 = 56(a) | Goat, Spain | B | VGI | A5 | H4 |
| LA179 = 58(a0 | Goat, Spain | B | VGI | A5 | H4 |
| LA220 = I-628 | CSF, Colombia | B | VGI | A5 | H4 |
| SAVIM | No data, Papua New Guena | B | VGI | A5 | H4 |
| WM 276 = TSC/SC1 | Eucalyptus debris, Australia | B | VGI | A5 | H4 |
| WM 179 | CSF, Australia | B | VGI | A5 | H4 |
| WOWOPA | No data, Papua New Guena | B | VGI | A5 | H4 |
| LA196 = I-34 | CSF, Colombia | B | VGII | A6 | H4 |
| LA221 = I-638 | CSF, Colombia | B | VGII | A6 | H4 |
| LA222 = I-642 | CSF, Colombia | B | VGII | A6 | H4 |
| LA230 = I-357 | CSF, Colombia | B | VGII | A6 | H4 |
| LA231 = I-414 | CSF, Colombia | B | VGII | A6 | H4 |
| ATCC 32608 = NIH191 | CSF, USA | C | VGII | A6 | H4 |
| ARN1 | Eucalyptus debris, Australia | B | VGII | A6 | H4 |
| WM 178 | Lung, Australia | B | VGII | A6 | H4 |
| ATCC 34880 | CSF, USA | C | VGIII | A7 | H4 |
| CN043A | CSF, New Zealand | C | VGIII | A7 | H4 |
| CN0438 | CSF, New Zealand | C | VGIII | A7 | H4 |
| LA205 = I-78 | CSF, Colombia | C | VGIII | A7 | H4 |
| LA234 = I-559 | Almond tree, Colombia | C | VGIII | A7 | H4 |
| LA235 = I-737 | Almond tree, Colombia | C | VGIII | A7 | H4 |
| LA237 = I-871 | Almond tree, Colombia | C | VGIII | A7 | H4 |
| LA382 = BV19 | CSF, Venezuela | C | VGIII | A7 | H4 |
| UCLA380 | No data, USA | C | VGIII | A7 | H4 |
| UCLA385 | No data, USA | C | VGIII | A7 | H4 |
| WM 161 | Eucalyptus debris, USA | B | VGIII | A7 | H4 |
| WM 728 | Eucalyptus depris, USA | B | VGIII | A7 | H4 |
| B5242 | Clinical, India | B | VGIV | A8 | H5 |
| V869 | Urine, South Africa | C | VGIV | A8 | H5 |
| V709 | Urine, South Africa | C | VGIV | A8 | H5 |
| M27055 | Urine, South Africa | C | VGIV | A8 | H5 |
| MCS34 | CSF, Thailand | C | VGIV | A8 | H5 |
| WM 779 | Cheata, South Africa | C | VGIV | A8 | H5 |
Molecular type as determined by PCR-fingerprinting with the minisatellite specific primer M13 (21).
DNA extraction and PCR of the PLB1 gene fragment.
DNA was extracted from cryptococci using mechanical disruption with miniature pestles and detergents according to a previously described protocol (21). A pair of primers was designed to amplify a fragment of the PLB1 gene from each serotype. The sequence of the PLB1 gene of serotype A (strain H99; accession number AF223383) and serotype D (strain B-3501; accession number AF238241) were obtained from GenBank, and those of serotype B (strain WM276; accession number AJ238508) and serotype C (strain M27055; accession number AJ302038) were obtained from our own work (15). The four C. neoformans PLB1 genes, excluding introns, were aligned using the Jotun Hein function of the MegAlign program within the DNAStar (Madison, Wis.) software package, in order to identify regions within the gene that are conserved in all four serotypes. Two primers IDPLB1, 5′ TGA GCT TCA GGC GGA GAG AGG TTT GG 3′, and IDPLB1R, 5′ AGG CTG GGT GGT GTT GTC GTC ACC 3′, were designed from the serotype B sequence. The primer sequences were completely homologous to the four PLB1 sequences, except for the base shown in boldface type for the reverse primer. Serotypes A and D had a cytosine at that locus, while the serotype C sequence had a thymidine at that locus. The PCRs were carried out in duplicate, to eliminate PCR artifacts, in a Perkin-Elmer 480 thermocycler using the following parameters. For a 100-μl reaction mixture, 44 μl of distilled water was added to a tube containing 15 μl of genomic DNA (10 ng μl−1) along with 10 μl of GeneAmp PCR buffer (10×), 10 μl of deoxynucleoside triphosphates (2 nmol of each nucleotide μl−1), 8 μl of 50 mM magnesium acetate, 6 μl of IDPLB1 (10 ng μl−1), 6 μl of IDPLB1R (10 ng μl−1), and 1 μl of AmpliTaq DNA polymerase (5 U μl−1). A water blank was used as a negative control. The cycling conditions used were as follows: an initial denaturation step at 94°C for 3 min followed by 35 cycles of 94°C for 45 s, 62°C for 45 s, and 72°C for 1 min and the final extension step at 72°C for 7 min.
Restriction endonuclease digestion.
A restriction map was generated using the MapDraw program of the DNAStar package from all four PLB1 sequences. The restriction endonucleases, AvaI and HindIII, were predicted to yield specific RFLP profiles following digestion of the PCR products. For RFLP analysis 25 μl of the PCR products was digested with 3 μl of NEB buffer (10×, buffer used depended on enzyme), 1 μl of the endonuclease (20 U μl−1 for HindIII and 10 U μl−1 for AvaI), and distilled water to a final volume of 30 μl. Tubes were incubated at 37°C for at least 3 h before separation on a 3% agarose gel and visualization by staining in ethidium bromide and UV transillumination. The 1-kb GIBCO-BRL ladder (Life Technologies, Rockville, Md.) was used as a size marker.
Determination of ploidy of serotype AD isolates at the PLB1 locus.
Genomic DNA was extracted from the serotype AD isolate (WM329 = 90-7031) and was subjected to PCR with the primers IDPLB1 and IDPLB1R, as described above. The PCR product was run on a 1.5% agarose gel with a 1-kb GIBCO-BRL ladder as a size marker. The band, corresponding to the PLB1 fragment, was excised from the gel and purified using the Geneclean spin kit (Bio 101 Inc., Carlsbad, Calif.) according to the manufacturer's recommendations. The purified product was cloned into the PCR cloning vector, pGem-T Easy (Promega, Madison, Wis.) according to the manufacturer's instructions. White colonies were subjected to PCR directly, without prior DNA extraction, with IDPLB1 and IDPLB1R. The PCR products amplified from several clones were then subjected to digestion with AvaI as described above and separated on a 3% agarose gel.
RESULTS
PCR-RFLP profiles.
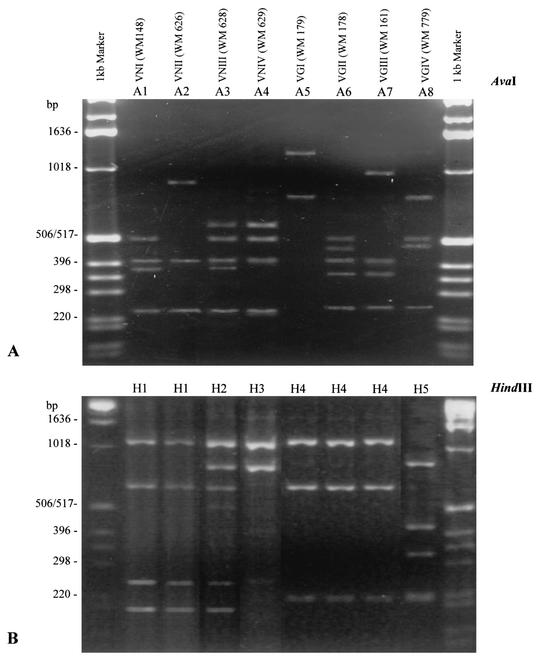
Amplification of the PLB1 fragment from the genomic DNA with the primers IDPLB1 and IDPLB1R generated a PCR product, approximately 1,970 bp in size, from all C. neoformans isolates investigated (results not shown). RFLP patterns generated by digestion with AvaI separated all isolates into eight groups (A1 through A8), which corresponded exactly with the 8 molecular types generated by PCR-fingerprinting (see Table 1 and Fig. 1A). RFLP groups A1 and A2 correspond to VNI and VNII (C. neoformans var. grubii), A3 to VNIII (the AD hybrid), and A4 to VNIV (C. neoformans var. neoformans). There was no correlation between serotype and RFLP profile among C. neoformans var. gattii isolates, with A5 to A8 corresponding to the molecular types VGI to VGIV (see right side of the top panel of Fig. 1).
FIG. 1.
AvaI (A) and HindIII (B) PCR-RFLP profiles of the PLB1 gene obtained from eight laboratory standard strains of C. neoformans, representing each of the eight major molecular types identified previously (20).
Digestion of the PCR products with the restriction endonuclease HindIII, produced a different set of profiles containing only five groups, H1 to H5 (see Table 1 and lower panel of Fig. 1). Three RFLP profiles, H1, H2, and H3, were generated by digestion of PCR products from C. neoformans var. grubii (serotype A, molecular types VNI and VNII), the serotype AD hybrid (molecular type VNIII), and C. neoformans var. neoformans (serotype D, molecular type VNIV), respectively. Two RFLP profiles were generated from isolates of C. neoformans var. gattii, H4 and H5, which corresponded to molecular types VGI to VGIV; neither was specific for serotype B or serotype C (see Table 1 and lower panel of Fig. 1).
Ploidy of serotype AD isolates at the PLB1 locus.
The RFLP profiles generated from serotype AD isolates, A3 and H2, consisted of a combination of bands from serotype A and serotype D profiles (Fig. 1, lanes 2 to 5, and Fig. 2, clones 1 to 6). More specifically, the RFLP profile A3 was found to be a mixture of A1 and A4. Similarly, the RFLP profile H2 is a composite of H1 and H3. The hybrid pattern obtained for serotype AD strains suggested that these isolates are heterozygous diploid strains, with a serotype A and a serotype D allele of the PLB1 gene. Given these observations, an investigation, aimed at confirming the ploidy of serotype AD strains at the PLB1 locus, was initiated.
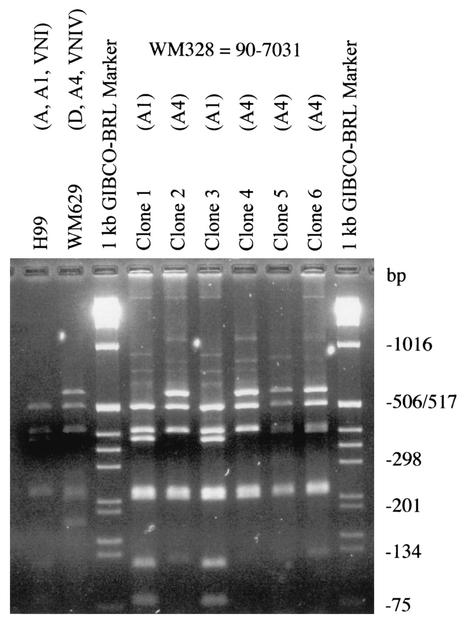
FIG. 2.
PCR-RFLP profiles of the PLB1 gene obtained from serotype AD clones of strain WM329 = 90-7031 (see hybrid RFLP profile in lane 3, Fig. 1A), with AvaI. Strain H99, serotype A, RFLP profile A1, molecular type VNI; strain WM629, serotype D, RFLP profile A4, molecular type VNIV; clones 1 to 6, clones of strain WM329 = 90-7031 (clones 1 and 3, RFLP profile A1; clones 2, 4, 5, and 6, RFLP profile A4).
The PLB1 fragment was amplified from the serotype AD strain WM329 = 90-7031 and cloned. Figure 2, shows the results obtained when the PLB1 fragment, amplified from six individual clones, was digested with AvaI. Two RFLP profiles were obtained, namely, A1 and A4, corresponding to the patterns obtained for serotype A (VNI) and serotype D (VNIV), respectively. The patterns obtained for the clones were comparable to those obtained for the serotype A strain (H99, RFLP profile A1) or to the D strain (WM629, RFLP profile A4) used as controls in Fig. 2 (see lanes 1 and 2). Clones 1 and 3, in lanes 4 and 6, contain the serotype A allele while clones 2, 4, 5, and 6 contain the serotype D allele of the PLB1 gene (Fig. 2).
DISCUSSION
The PCR-RFLP method was applied to C. neoformans isolates, with the aim of differentiating between varieties, serotypes, and molecular types. Amplification using the primers IDPLB1 and IDPLB1R resulted in a band of identical size for each isolate, regardless of variety, serotype, or molecular type. The RFLP profiles obtained with AvaI were more discriminating than those obtained with HindIII, yielding eight RFLP profiles, which correspond to the eight molecular types defined by PCR-fingerprinting (11, 20). This result is of great interest, given the fact that the PCR-RFLP technique described here highlights differences based on partial sequence of a single virulence gene, while PCR-fingerprinting, on the other hand, amplifies mostly noncoding DNA sequences (8) across the whole genome, although these observations may be coincidental, or more likely, reflect ongoing divergent evolution within the C. neoformans complex, into the current eight subgenotypes. This possibility is supported by the fact that PCR-RFLP of the URA5 housekeeping gene also distinguishes eight groups within the C. neoformans complex (S. Jackson and W. Meyer, personal communication [unpublished data]). In contrast, the digestion of PLB1 with the restriction enzyme HindIII differentiated isolates of C. neoformans into three varieties only. The distinction between serotypes A, D, and AD was achieved with both AvaI and HindIII, but neither distinguished serotype B from serotype C. This result shows again that serotype B and C isolates are much more closely related to each other than are serotypes A and D, as has already been suggested previously by Xu et al. (34).
The serotype AD profiles obtained with both AvaI and HindIII indicated that the serotype AD strains tested contained both the serotype A and D alleles of the PLB1 gene. Hybridization between serotype A and D genomes that result in the serotype AD profile, has been reported recently using random amplification of polymorphic DNA analysis (22), specific amplification and sequencing of PCR-fingerprinting bands (8; M. Cogliati, M. Allaria, A. M. Tortorano, G. Liberi, and M. A. Viviani, unpublished data [presented at the 4th International Conference on Cryptococcus and Cryptococcosis]), PCR-RFLP of the URA5 gene (29) and by sequencing of the laccase gene (33). Some isolates of serotype AD have been found to be diploid, and others have been found to be haploid (25, 26). Cloning of the PLB1 fragment followed by RFLP analysis of several clones demonstrated that the serotype AD strain used (WM329 = 90-7031) was indeed heterozygous diploid at that locus. Interestingly, the RFLP profile A3 obtained from the serotype AD strains indicated hybridization between RFLP profile A1 and A4. This suggests that serotype AD strains derive their serotype A PLB1 allele from the VNI genotype (RFLP profile A1) rather than VNII (RFLP profile A2). The results also suggest that VNI isolates may lend themselves better to hybridization with serotype D strains. The propensity for VNI isolates to hybridize may have implications for studies involving recombination and complementation. It must be noted, however, that the number of serotype AD isolates tested in this study was limited and that other diploid hybrid genotypes between serotypes A and D have been reported (W. Meyer and J. Heitman, personal communication). As a result, it cannot be irrefutably argued that VNI is dominant over VNII in its ability to hybridize to serotype D isolates. More serotype AD isolates need to be tested in order to confirm whether serotype D (VNIV) preferentially hybridizes to VNI rather than to VNII and to determine the extent of hybridization between molecular types. Notably, there were no hybrid profiles among the C. neoformans var. gattii isolates, which is in opposition to the findings reported previously using AFLP analysis (4).
It is reasonable to assume that diploid serotype AD strains probably contain two copies of each virulence gene, as has been shown for PLB1 in this report. It is surprising, therefore, that serotype AD is not encountered more commonly clinically, particularly in patients with AIDS, since the presence of the serotype A genome should make the serotype AD potentially as virulent as serotype A. The relatively low incidence of serotype AD isolates clinically may reflect the failure of antigen agglutination tests to adequately recognize this serotype, attributing the isolate to either serotype A or D. Alternatively, certain alleles may be dominant in diploid strains. In C. neoformans the virulence phenotype may be a recessive trait which is only expressed in haploid strains, whereas in diploid strains, the phenotype may or may not be expressed, depending on the alleles present in the particular strain. Lengeler et al. (18) have shown, using serotyping, that the serotype D defining gene was dominant over the serotype A defining gene in studies of inheritance. In an experiment looking at the serotype of segregants obtained from a diploid, self-fertile, laboratory-engineered strain of serotype AD, six out of eight isolates were serotype D and no serotype A isolates were recovered. Although these results point to the dominance of serotype D alleles over serotype A alleles in terms of inheritance, dominance may also be demonstrated in terms of gene expression and, therefore, virulence. Research in this area has not been undertaken in C. neoformans but may yield some interesting results.
In conclusion, PCR-RFLP of the PLB1 gene proved to be a simple, useful tool for differentiating between C. neoformans var. grubii, the AD hybrid, C. neoformans var. neoformans, and C. neoformans var. gattii. Significantly, the same eight molecular types or groups have now been defined by PCR-based methods of whole-genome sampling and RFLP of a virulence gene (PLB1), all of which will have evolved at different rates. This suggests that divergent evolution may be ongoing within the C. neoformans complex, leading to eight distinct subgenotypes, which are discordant with respect to serotype. Finally, some serotype AD strains contain two alleles of the PLB1 gene, one originating from serotype A (VNI) and the other from serotype D (VNIV).
Acknowledgments
We thank B. Currie (Menzies School of Research, Darwin, Australia), H. H. Crewe-Brown (University of the Witwatersrand, Johannesburg, South Africa), K. Tintelnot (Robert Koch Institute, Berlin, Germany), D. Howard (University of California, Los Angeles), A. Arechavala (Hospital de Infecciosas Francisco J. Muñiz, Buenos Aires, Argentina), E. Castañeda (Instituto Nacional de Salud, Bogotá, Colombia), J. Torres-Rodriquez (Institut Municipal d' Investigació Médica, Barcelona, Spain), M. S. C. Melhem (Adolfo Lutz Institute, Sao Paulo, Brazil), D. Parr (Auckland Children's Hospital, Auckland, New Zealand), J. R. Perfect (Duke University Medical Center, Durham, N.C.), Valerie Davis (South African Institute for Medical Research, Johannesburg), and D. Ellis (Woman's and Children's Hospital, Adelaide, Australia) for the contribution of cryptococcal strains used in this study.
This work was financially supported by the Harry Windsor Postgraduate Scholarship awarded by the Community Health and Tuberculosis Association (CHATA), Sydney, Australia; a Millennium Foundation (Westmead, Australia) Top-Up grant to N.G.L., and NH&MRC (Canberra, Australia) grant 9937187 to W.M. and T.C.S.
REFERENCES
- 1.Bennett, J. E., K. J. Kwon-Chung, and D. H. Howard. 1977. Epidemiological differences among serotypes of Cryptococcus neoformans. Am. J. Epidemiol. 105:582-586. [DOI] [PubMed] [Google Scholar]
- 2.Bertout, S., F. Renaud, D. Swinne, M. Mallie, and J.-M. Bastide. 1999. Genetic multilocus studies of different strains of Cryptococcus neoformans: taxonomy and genetic structure. J. Clin. Microbiol. 37:715-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birch, M., G. Robson, D. Law, and D. W. Denning. 1996. Evidence of multiple extracellular phospholipase activities of Aspergillus fumigatus. Infect. Immun. 63:751-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boekhout, T., B. Theelen, M. Diaz, J. W. Fell, W. C. J. Hop, E. C. A. Abeln, F. Dromer, and W. Meyer. 2001. Hybrid genotypes in the pathogenic yeast Cryptococcus neoformans. Microbiology 147:891-907. [DOI] [PubMed] [Google Scholar]
- 5.Casadevall, A., L. F. Freundlich, L. Marsh, and M. D. Scharff. 1992. Extensive allelic variation in Cryptococcus neoformans. J. Clin. Microbiol. 30:1080-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaturvedi, S., B. Rodeghier, J. Fan, M. McClelland, B. L. Wickes, and V. Chaturvedi. 2000. Direct PCR of Cryptococcus neoformans MATα and Mata pheromones to determine mating type, ploidy, and variety: a tool for epidemiological and molecular pathogenesis studies. J. Clin. Microbiol. 38:2007-2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, S. C. A., A. G. Brownlee, T. C. Sorrell, P. Ruma, D. H. Ellis, T. Pfeiffer, B. R. Speed, and G. Nimmo. 1996. Identification by random amplification of polymorphic DNA of a common molecular type of Cryptococcus neoformans var. neoformans in patients with AIDS or other immunosuppressive conditions. J. Infect. Dis. 173:754-758. [DOI] [PubMed] [Google Scholar]
- 8.Cogliati, M., M. Allaria, A. M. Tortorano, and M. A. Viviani. 2000. Genotyping Cryptococcus neoformans var. neoformans with specific primers designed from PCR-fingerprinting bands sequenced using a modified PCR-based strategy. Med. Mycol. 38:97-103. [DOI] [PubMed] [Google Scholar]
- 9.Cox, G. M., H. C. McDade, S. C. A. Chen, S. C. Tucker, M. Gottfredsson, L. C. Wright, T. C. Sorrell, S. D. Leidich, A. Casadevall, M. A. Ghannoum, and J. R. Perfect. 2001. Extracellular phospholipase activity is a virulence factor for Cryptococcus neoformans. Mol. Microbiol. 39:166-175. [DOI] [PubMed] [Google Scholar]
- 10.Diaz, M. R., T. Boekhout, B. Theelen, and J. W. Fell. 2000. Molecular sequence analyses of the intergenic spacer (IGS) associated with rDNA of the two varieties of the pathogenic yeast. Cryptococcus neoformans. Syst. Appl. Microbiol. 23:535-545. [DOI] [PubMed] [Google Scholar]
- 11.Ellis, D., D. Marriott, R. A. Hajjeh, D. Warnock, W. Meyer, and R. Barton. 2000. Epidemiology: surveillance of fungal infections. Med. Mycol. 38(Suppl. 1):173-182. [PubMed] [Google Scholar]
- 12.Ellis, D. H., and T. J. Pfeiffer. 1990. Natural habitat of Cryptococcus neoformans var. gattii. J. Clin. Microbiol. 28:1642-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franzot, S. P., I. F. Salkin, and A. Casadevall. 1999. Cryptococcus neoformans var. grubii: separate varietal status for Cryptococcus neoformans serotype A. J. Clin. Microbiol. 37:838-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwon-Chung, K. J., and J. E. Bennett. 1984. Epidemiologic differences between the two varieties of Cryptococcus neoformans. Am. J. Epidemiol. 120:123-130. [DOI] [PubMed] [Google Scholar]
- 15.Latouche, G. N., T. C. Sorrell, and W. Meyer. 2002. Isolation and characterisation of thepPhospholipase B gene of Cryptococcus neoformans var. gattii. FEMS Yeast Res. 2:551-561. [DOI] [PubMed]
- 16.Lazera, M. S., M. A. Salmito Cavalcanti, A. T. Londero, L. Trilles, M. M. Nishikawa, and B. Wanke. 2000. Possible primary ecological niche of Cryptococcus neoformans. Med. Mycol. 38:379-383. [DOI] [PubMed] [Google Scholar]
- 17.Lee, K. S., J. L. Patton, M. Fido, L. K. Hines, S. D. Kohlwein, F. Paltauf, S. A. Henry, and D. E. Levin. 1994. The Saccharomyces cerevisiae PLBI gene encodes a protein required for lysophospholipase and phospholipase B activity. J. Biol. Chem. 269:19725-19730. [PubMed] [Google Scholar]
- 18.Lengeler, K. B., G. M. Cox, and J. Heitman. 2001. Serotype AD strains of Cryptococcus neoformans are diploid or aneuploid and are heterozygous at the mating-type locus. Infect. Immun. 69:115-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masuda, N., N. Kitamura, and K. Saito. 1991. Primary structure of protein moiety of Penicillium notatum phospholipase B deduced from the cDNA. Eur. J. Biochem 202:783-787. [DOI] [PubMed]
- 20.Meyer, W., K. Marszewska, M. Amirmostofian, R. P. Igreja, C. Hardtke, K. Methling, M. A. Viviani, A. Chindamporn, S. Sukroongreung, M. A. John, D. H. Ellis, and T. C. Sorrell. 1999. Molecular typing of global isolates of Cryptococcus neoformans by polymerase chain reaction fingerprinting and randomly amplified polymorphic DNA: a pilot study to standardize techniques on which to base a detailed epidemiological survey. Electrophoresis 20:1790-1799. [DOI] [PubMed] [Google Scholar]
- 21.Meyer, W., and T. G. Mitchell. 1995. Polymerase chain reaction fingerprinting in fungi using single primers specific to minisatellites and simple repetitive DNA sequences: strain variation in Cryptococcus neoformans. Electrophoresis 16:1648-1656. [DOI] [PubMed] [Google Scholar]
- 22.Nakamura, Y. 2001. Molecular analyses of the serotype of Cryptococcus neoformans. Jpn. J. Med. Mycol. 42:69-74. [DOI] [PubMed] [Google Scholar]
- 23.Perfect, J. R., N. Ketabchi, G. M. Cox, C. W. Ingram, and C. L. Beiser. 1993. Karyotyping of Cryptococcus neoformans as an epidemiological tool. J. Clin. Microbiol. 31:3305-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sorrell, T. C. 2001. Cryptococcus neoformans var. gattii. Med. Mycol. 39:155-168. [PubMed] [Google Scholar]
- 25.Takeo, K., R. Tanaka, H. Taguchi, and K. Nishimura. 1993. Analysis of ploidy and sexual characteristics of natural isolates of Cryptococcus neoformans. Can. J. Microbiol. 39:958-963. [DOI] [PubMed] [Google Scholar]
- 26.Tanaka, R., K. Nishimura, and M. Miyaji. 1999. Ploidy of serotype AD strains of Cryptococcus neoformans. Jpn. J. Med. Mycol. 40:31-34. [DOI] [PubMed] [Google Scholar]
- 27.Varma, A., and K. J. Kwon Chung. 1989. Restriction fragment polymorphism in mitochondrial DNA of Cryptococcus neoformans. J. Gen. Microbiol. 135:3353-3362. [DOI] [PubMed] [Google Scholar]
- 28.Varma, A., and K. J. Kwon-Chung. 1992. DNA probe for strain typing of Cryptococcus neoformans. J. Clin. Microbiol. 30:2960-2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Velegraki, A., V. G. Kiosses, A. Kansouzidou, S. Smilakou, A. Mitroussia-Ziouva, and N. J. Legakis. 2001. Prospective use of RFLP analysis on amplified Cryptococcus neoformans URA5 gene sequences for rapid identification of varieties and serotypes in clinical samples. Med. Mycol. 39:409-471. [DOI] [PubMed] [Google Scholar]
- 30.Vilgalys, R., and M. Hester. 1990. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172:4238-4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Viviani, M. A., H. Wen, A. Roverselli, R. Caldarelli-Stephano, M. Cogliati, P. Ferrante, and A. M. Tortorano. 1997. Identification by polymerase chain reaction fingerprinting of Cryptococcus neoformans serotype AD. J. Med. Vet. Mycol. 35:355-360. [PubMed] [Google Scholar]
- 32.Wickes, B. L., T. D. E. Moore, and K. J. Kwon-Chung. 1994. Comparison of the electrophoretic karyotypes and chromosomal location of ten genes in the two varieties of Cryptococcus neoformans. Microbiology 140:543-550. [DOI] [PubMed] [Google Scholar]
- 33.Xu, J., G. Luo, R. J. Vilgalys, M. E. Brandt, and T. G. Mitchell. 2002. Multiple origins of hybrid strains of Cryptococcus neoformans with serotype AD. Microbiology 148:203-212. [DOI] [PubMed] [Google Scholar]
- 34.Xu, J., R. Vilgalys, and T. G. Mitchell. 2000. Multiple gene genealogies reveal recent dispersion and hybridisation in the human pathogenic fungus Cryptococcus neoformans. Mol. Ecol. 9:1471-1481. [DOI] [PubMed] [Google Scholar]