Abstract
The examination of microorganisms in glacial ice cores allows the phylogenetic relationships of organisms frozen for thousands of years to be compared with those of current isolates. We developed a method for aseptically sampling a sediment-containing portion of a Greenland ice core that had remained at −9°C for over 100,000 years. Epifluorescence microscopy and flow cytometry results showed that the ice sample contained over 6 × 107 cells/ml. Anaerobic enrichment cultures inoculated with melted ice were grown and maintained at −2°C. Genomic DNA extracted from these enrichments was used for the PCR amplification of 16S rRNA genes with bacterial and archaeal primers and the preparation of clone libraries. Approximately 60 bacterial inserts were screened by restriction endonuclease analysis and grouped into 27 unique restriction fragment length polymorphism types, and 24 representative sequences were compared phylogenetically. Diverse sequences representing major phylogenetic groups including alpha, beta, and gamma Proteobacteria as well as relatives of the Thermus, Bacteroides, Eubacterium, and Clostridium groups were found. Sixteen clone sequences were closely related to those from known organisms, with four possibly representing new species. Seven sequences may reflect new genera and were most closely related to sequences obtained only by PCR amplification. One sequence was over 12% distant from its closest relative and may represent a novel order or family. These results show that phylogenetically diverse microorganisms have remained viable within the Greenland ice core for at least 100,000 years.
A majority of the Earth's biosphere is constantly at 5°C or lower (1). Examples of cold environments are the world's oceans, high-altitude and -latitude terrestrial areas, deep lakes, glaciers, and the continent of Antarctica. Habitats that are considered “extreme” by human standards are usually thought to be of limited phylogenetic diversity. Interest in the diversity and survival of microorganisms in frozen samples has increased with the postulation that ice-covered extraterrestrial environments may support life. The discovery of an icy cover on Jupiter's moon Europa has raised the possibility that life exists in cold environments beyond Earth (7, 15).
Earth environments which may be analogous to extraterrestrial cold environments include glacier ice cores and bodies of water located beneath glaciers (28, 34). One glacier-associated environment receiving recent attention is Lake Vostok, located more than 3,000 m beneath the East Antarctic ice sheet. Although drilling has not penetrated through the ice into the lake, ice cores have been obtained where lake water has frozen onto the bottom of the glacier (accretion ice). Studies of PCR-amplified 16S rRNA genes from DNA extracted from Lake Vostok accretion ice (29) showed that five of the seven sequences were related to members of the beta Proteobacteria, one was an alpha Proteobacterium, and another was an Actinomyces sp. Epifluorescence microscopy of DNA-stained samples showed between 2 × 102 and 2.3 × 103 cells per ml (19). Christner et al. (5) extended the Vostok study of accretion ice by obtaining four isolates following aerobic incubation at 25°C and finding six different 16S rRNA gene sequences amplified from DNA extracted from a melted sample. In addition, Christner et al. (6) initiated studies of microbial diversity in deep glacier ice from different locations and have reported results obtained from ice cores ranging from 5 to 20,000 years in age.
The detection, isolation, and characterization of microorganisms from glaciers are still relatively novel enterprises, and there are many reasons for examining additional ice samples (6, 34). It has been proposed that liquid veins that exist in ice along three-grain boundaries may provide habitats for psychrophilic organisms, where metabolism might continue (28). A new microscopic approach has been used to observe in situ bacterial inhabitants of micrometer-scale brine pockets in Arctic sea ice at temperatures as low as −15°C (8, 18). These results suggest that microorganisms may be more numerous and active in ice than had been previously thought. If microbial metabolism is occurring in glacial ice cores, the geological interpretation of results from mineral and gas analyses of previous cores could change. Thus, the study of microbial communities and their possible metabolic activities has important implications for understanding nutrient cycling and for defining microbial survival and persistence strategies. The development of more-complete pictures of microbial diversity in ice frozen for thousands of years will also provide insight into methods needed for recovering and cultivating these organisms.
In previous work, we have isolated psychrophiles from a variety of environments and examined their cold-active enzymes (3, 23, 32, 33, 38). To compare organisms from glaciers with other isolates, we initiated the characterization of the microbial diversity of a deep Greenland glacier ice core sample that was over 100,000 years old. In this work we have developed a simple, inexpensive method for obtaining samples aseptically from ice cores and present findings on the cultivation of anaerobic psychrophilic microbial communities from these samples. We extracted DNA from mixed cultures grown at −2°C and amplified by PCR, cloned, and sequenced the 16S rRNA genes obtained. The phylogenetic analysis of 24 of the bacterial sequences demonstrated a highly diverse community originating from the ice core sample with representatives of the alpha, beta, and gamma Proteobacteria, Deinococcus group, gram-negative and gram-positive anaerobes, and sporeforming and nonsporeforming organisms.
MATERIALS AND METHODS
Aseptic sampling of the ice core.
The ice core was from the Greenland Ice Sheet Project (GISP 2) and corresponded to a depth of 3042.67 to 3042.80 m. To aseptically sample the ice core, a portion was placed in a laminar-flow hood and the surface layers were removed by pulling a platinum wire heated by a current from a 6-V lantern battery (Eveready Battery Company, Inc., Cleveland, Ohio) along the edges. After the outer surface layers (approximately 2 to 3 mm) were removed, the newly exposed surface was treated with 95% ethanol as described previously (6), a sterile wire was placed about 1 cm from the end, and a new slice was removed. Once the wire was about one-third down from the top, the remaining layer fractured and exposed a sterile surface. Samples were then drilled from the newly exposed edge of the ice core with sterile keyhole bits attached to a sterile drill bit extender. Several keyhole bits (volume, 20 ml each) could be used in sequence to remove samples from increasing depths within the core or to obtain replicate samples in the same area. The subcored material (50 to 60 ml) was placed in a sterile flask, and the ice was melted in the cold (4°C) for 15 to 18 h to provide material for inoculating media. Control experiments in which sterile water was frozen and then sampled with the above procedure demonstrated that we did not introduce any contamination that could be detected by cultivation or PCR amplification.
Bacterial cell size and number estimation.
Staining of 1-ml samples from the melted ice with propidium iodide and SYTO9 was according to the instructions of the LIVE/DEAD BacLight bacterial viability kit (Molecular Probes, Eugene, Oreg.), and samples were examined under an Olympus BX-60 wide-field microscope with a 40× objective. The same samples were subjected to flow cytometry (XL-MCL; Beckman-Coulter, Miami Lakes, Fla.) in order to enumerate the total number of cells. Counted fluorescent beads were added to the sample in order to estimate the exact volume measured, and 10,000 nonbead events were counted.
Media and enrichment conditions.
Two media, MM1 and MM2, were used for growth of anaerobic microbial communities. Both media consisted of basal salts, trace elements, vitamin solution, and different substrate supplements to enrich for different groups of microorganisms at −2°C. MM1 contained the following constituents: 0.5 g of KH2PO4, 0.4 g of NaCl, 0.16 g of NH4Cl, 0.05 g of MgCl2 · 6H2O, 0.01 g of CaCl2 · 2H2O, 7.5 g of NaHCO3, and 0. 01 mg of resazurin per liter and 1% (vol/vol) each of vitamin and trace element solutions as described previously (35). The pH was adjusted to 7.0 with HCl. Na2S was used as a reducing agent at a 0.06% final concentration, and either sodium acetate or sodium formate was added as a substrate to a 0.1 M final concentration. Medium MM2, as described by Ferrara-Guerrero et al. (11), was designed for enrichment of fermentative bacteria (MM2.6.1) and for methanogens (MM2.6.3). The anaerobically prepared media (50 ml in 125 ml bottles) were overgassed with N2-CO2 or H2-CO2 (4:1), inoculated from the melted ice (1:200 and 1:1,000 dilutions), and incubated with appropriate controls at −2°C with shaking at 150 rpm. Medium MM1 supplemented with acetate or formate was inoculated and incubated aerobically as well.
Anaerobic community genomic DNA extraction.
Cultures in similar media were combined by filtration through 0.2-μm-pore-size Millipore filters for total genomic DNA extraction. Cells enriched in the MM1 medium containing either acetate or formate were combined, and cells enriched in the MM2.6.1 and MM2.6.3 media were combined. Different cell lysis protocols were used without success, including treatment with various concentrations of lysozyme and mutanolysin, alternating freeze-thaw cycles, lengthy incubations at elevated temperatures in the presence of detergents, boiling, mechanical disruption with a French press, ultrasound or microwave treatment, and exposure to bead beating (14, 16, 25, 39). Also unsuccessful was the procedure used for isolation of DNA from spores, including treatment with NaOH, boiling, and incubation with HCl (30). No DNA was obtained by using the Soil UltraClean kit (MO BIO).
The most comprehensive lysis, as indicated by microscopic examination of the cells, incorporated modifications of the method reported by Porteous et al. (27), in which the cells were subjected to bead beater disruption. Turbid anaerobic cultures (total combined volume, 750 ml) were initially filtered by using a 0.2-μm-pore-size filter and resuspended in 10 ml of 10 mM Tris, pH 8.0. The cell pellets obtained after centrifugation were maintained at −20°C. Pellets (40 to 80 mg) were resuspended in 500 μl of a solution containing 250 mM NaCl, 100 mM Na2-EDTA (pH 8.0), and 4% sodium dodecyl sulfate, added to 500 mg of sterile glass beads (250 mg each of 0.1- and 0.5-mm-diameter beads) and beaten at maximum speed for 5 min with a MiniBeadbeater-8 cell disrupter (Biospec Products, Inc.). The cell suspension was transferred to a new tube, proteinase K was added to a final concentration of 250 μg/ml, and the samples were incubated for 2 to 4 h at 68°C. After the suspension was cooled to room temperature, the insoluble material was removed by centrifugation for 2 min at 14,000 × g. The supernatant fraction (∼500 μl) was precipitated with absolute ethanol. The pellet was washed with 70% ethanol, dried, and resuspended in 10 mM Tris, pH 8. The enriched community genomic DNA was stored at 5°C.
16S rRNA gene amplification, cloning, and grouping.
The 16S rRNA genes were amplified from chromosomal DNA by PCR with Vent polymerase (New England Biolabs, Beverly, Mass.) and the universal bacterial primers 8F and 1492R or an archaeal primer set consisting of 21F and 958R or 765F and 1492R (26, 40). The products were ligated into the PCR-Script Amp vector (Stratagene, La Jolla, Calif.) by using the supplied protocols. Ligation products were transformed into Escherichia coli strain DH5α cells made chemically competent by using the Z-Competent kit (Zymo Research, Orange, Calif.). For initial screening, insert-containing transformants were grown in 1.5 ml of Luria-Bertani broth containing ampicillin (100 μg/ml) at 37°C overnight, and plasmid DNA was isolated by the boiling procedure (31). Plasmid DNA was treated with the restriction endonucleases PstI and NotI to determine which ones contained inserts of the appropriate size.
Candidate plasmids were then used for reamplification of 16S ribosomal DNA (rDNA) and amplification ribosomal DNA restriction analysis (ARDRA) was performed on the individual products with the 4-bp-cutting restriction enzymes MspI, RsaI, HaeIII, and AluI for 3 to 4 h at 37°C to group the amplified products by unique restriction patterns. Representatives of each distinct pattern were further studied by purification of the plasmid DNA with the Wizard SV-plus kit (Promega), followed by precipitation with 2 volumes of 2.5 M NaCl-20% polyethylene glycol 8000 at −20°C for 1 h. Plasmid DNA pellets were washed with 70% ethanol and resuspended in 100 μl of H2O (molecular grade; Eppendorf). One strand was fully sequenced at the Pennsylvania State University Nucleic Acid Facility on an ABI 370 sequencer using T7, T3, 704F, and 907R primers.
16S rRNA gene phylogenetic analysis.
The resulting single-stranded 16S rRNA gene sequences were matched with those from the Ribosomal Database Project (RDP) (24) and from a Blast search of the National Center for Biotechnology Information database (http://www.ncbi.nlm.nih.gov). Alignments were based on 1,599 nucleotide positions for 52 taxa. The rRNA gene sequences were aligned with sequences obtained from the RDP and GenBank by using the Clustal W program found in the BioEdit platform (version 5.0.6). GenBank accession numbers for each of the 16S rRNA gene sequences are as follows: Thermus scotoductus strain SE-1 (DSM 8553), AF032127; Thermus sp. strain NMX2 A.1, L09661; Bacteroides merdae ATCC 43184, X83954; Brevundimonas vesicularis LMG 11141, AJ227781; Methylobacterium fujisawaense DSM 5686, AJ250801; Bradyrhizobium genospecies B strain BDV5040, Z94812; Variovorax paradoxus IAM 12373, D30793; Pseudomonas beteli ATCC 19861, AB021406; Stenotrophomonas maltophilia LMG 11087, X95924; Ruminococcus obeum, X85101; Clostridium nexile DSM 1787, X73443; Clostridium clostridioforme ATCC 25537, M59089; Eubacterium rectale ATCC 33656, L34627; Butyrivibrio fibrisolvens NCDO 2397, X89979; Lachnospira pectinoschiza strain 150-1, L14675; Tissierella praeacuta ATCC 25539, X80833; Peptostreptococcus magnus ATCC 15794, D14149; Fusobacterium prausnitzii ATCC 27766, X85022; unidentified butyrate-producing bacterium L1-93, AJ270478; uncultured bacterium clone p-189-o5, AF371642; unidentified butyrate-producing bacterium A2-231, AJ270484; uncultured bacterium adhufec55, AF132275; beta proteobacterium A0647, AF236007; uncultured bacterium clone HuCA2, AJ408958; uncultured bacterium A54, AF052421; Peptostreptococcus genospecies 4, AB038360; uncultured bacterium clone p-2559-9F5, AF371715; uncultured eubacterium WD2115, AJ292627.
The BioEdit alignment was used in maximum-parsimony, maximum-likelihood, and distance analyses utilizing the PAUP package (D. L. Swofford, PAUP [phylogenetic analysis using parsimony], version 4 Ob10, Sinauer Associates, 2002). The sequence data were analyzed by using the maximum-parsimony method (heuristic search), the maximum-likelihood method (with a transition/transversion ratio of 2), and the distance method (neighbor-joining algorithm and Jukes-Cantor model), with 1,000 bootstrap replicates being performed for the last method. The initial distance analysis was performed by using the neighbor-joining algorithm and an uncorrected “p” distance measure with the PAUP program. To determine the effects of using various analysis methods on these results, we compared our results with distance trees generated by using the Jukes-Cantor, F81, F84, Kimura two-parameter, Kimura three-parameter, Tamura-Nei, Tajima-Nei, and HKY85 models with equal rates for variable sites. Unequal rates for variable sites were also examined for all eight models by using gamma parameters with shapes of 0.5, 1.0, 2.0, 3.0, and 5.0. Trees generated by all three methods were essentially congruent. A distance matrix was generated from the same alignment by the PAUP program using the Jukes-Cantor, F81, F84, Kimura two-parameter, Kimura three-parameter, Tamura-Nei, Tajima-Nei, and HKY85 models with equal rates for variable sites. The distance matrices resulting from each method were similar.
The CHECK-CHIMERA program of RDP was used to identify possible chimeric sequences in the clone library, and the number of rrn operons and genome sizes of close relatives were obtained from the rrndb database, release 2.3 (http://rrndb.cme.msu.edu).
Nucleotide sequence accession numbers.
GenBank accession numbers for the Greenland enrichment clone 16S rRNA gene sequences used in the alignment are as follows: 1-4, AY169411; 1-8, AY169412; 1-9, AY169413; 1-10, AY169414; 1-11, AY169415; 1-18, AY169416; 1-24, AY169417; 1-42, AY169418; 1-33, AY169419; 1-43, AY169420; 1-44, AY169421; 1-53, (AY169422; 1-54, AY169423; 1-56, AY169424; 1-68, AY169425; 1-71, AY169426; 1-79, AY169427; 1-82, AY169428; 1-84, AY169429; 1-88, AY169430; 1-94, AY169431; 1-95, AY169432; 28, AY169433; 15, AY169434.
RESULTS
Inoculation and cultivation.
The ice core sample came from the bottom portion of the GISP 2 ice core which had remained frozen at −9°C for over 100,000 years. This portion of the core was composed of “brown silty” ice and contains significant amounts of fine-grained sediment and rounded particles (12, 13). Microscopic examination showed many particles exceeding the size of bacterial cells. We also examined the melted ice core samples using the LIVE/DEAD BacLight kit and estimated the total number of cells by flow cytometry. Our results from two duplicate samples were similar, with 6.1 × 107 and 9.1 × 107 cells/ml and a ratio of live to dead cells of 5:1. Of special interest was the observation that the vast majority of the cells were small, with sizes 1 μm and below. Although these results suggested that a large microbial population exists within the ice core sample, the small sample available (about 50 ml) and the recalcitrance of most cells to lysis did not permit a direct large-scale DNA extraction and examination of diversity.
We inoculated different media to obtain communities in larger volumes that could be analyzed and to enrich for both aerobic and anaerobic microorganisms. Several aseptically removed ice core subcore samples were melted and used as inocula for aerobic MM1 and MM2 media and reduced, anaerobic MM1 and MM2 media. Turbidity was observed in the MM1 and MM2 aerobic cultures only after a few months of incubation. However, the anaerobic enrichments in MM1 and MM2 became lightly turbid after about 2 to 3 weeks of incubation at −2°C. The anaerobic turbid cultures were propagated by making dilutions (1:50) into new anaerobic media and reincubating at −2°C with shaking. New cultures became turbid within 4 to 10 days, and the communities could be maintained with periodic transfer for over 1 year. Microscopic examinations of the enrichment cultures showed numerous morphological shapes including extremely small cocci, large oblong rods, needle-shaped rods, filaments, and unevenly shaped cells. Many cells were motile, and some remained motile even after a few hours at room temperature, suggesting that they tolerated warmer temperatures and aerobic conditions. The viability of the cells was confirmed by epifluorescence microscopy using the BacLight staining. We also observed a high proportion of small cells among the viable population.
Development of DNA extraction protocol.
It was important that the lysis procedures disrupt cells generally recalcitrant to lysis in order to obtain a representative picture of the cultured community diversity. Microscopic examinations were used to evaluate the effectiveness of various lysis procedures. Unfortunately, many morphologically distinct cells remained intact, with some even remaining motile, following several standard methods, and the lysis procedure required optimization. A variety of protocols were used on relatively small cell pellets (30 to 50 mg wet weight) without success. High-molecular-weight DNA that yielded PCR amplification products was obtained by using a modified procedure of Porteous et al. (27) where the cells were treated with short microwave pulses. The genomic DNA yields were further increased by replacing the microwave treatment with bead beating. Two separate total-genomic-DNA preparations were extracted from combined cultures enriched in the MM2.6.1 and MM2.6.3 media. A third genomic preparation was obtained from a combined cell mass enriched in the MM1 medium containing acetate or formate.
Construction and screening of the 16S rDNA libraries.
After amplification of 16S rDNA genes with bacterial primers 8F and 1492R and cloning, approximately 60 inserts were screened by ARDRA and grouped into 27 restriction fragment length polymorphism (RFLP) patterns. Representative clones for each pattern were sequenced, and 24 were used for phylogenetic analysis. The 16S rDNA clones from DNA extracted from the MM1 medium containing formate or acetate had a larger diversity of RFLP types than those from the MM2.6.1 and MM2.6.3 media.
PCR products were seen when the archaeal primer sets (21F and 958R and 765F and 1492R) were used with DNA extracted from both types of media. Screening 600 clones revealed 78 with inserts with the appropriate size. However, the restriction digests revealed only a few unique ARDRA patterns. In addition, the sequence analysis of 10 of these inserts showed very low similarity with any known archaeal 16S rRNA gene sequence. Control experiments with DNA from a methanogen gave excellent PCR amplification and the correct insert size. Because it is not yet clear whether these sequences correspond to an unusual 16S rRNA gene sequence or are artifacts due to nonspecific primer binding, they have not been incorporated into the phylogenetic analyses.
Phylogenetic analysis of bacterial 16S rRNA gene sequences.
The analyses of the constructed bacterial 16S rDNA clone library used full-length or almost-full-length 16S rRNA gene sequences for the alignments and comparisons. The 16S rDNA sequences analyzed represented a wide diversity of both gram-positive and -negative organisms and were found in several of the major phylogenetic groups including Deinococcus, Bacteroides, Eubacterium, Clostridium, and Proteobacteria (Fig. 1). About one-half of the clones belonged to phylogenetic groups where the corresponding organisms are typically anaerobic and chemo-organotrophic, although some Clostridium species can be chemolithotrophic as well (4). Others were facultative anaerobes or aerobes that can also grow anaerobically in the presence of an alternative electron acceptor. Among the anaerobes were gram-positive sporeformers (Clostridium) and gram-positive nonsporeformers (Eubacterium, Ruminococcus, and Peptostreptococcus), gram-negative rods (Fusobacterium), and Firmicutes with unusual cell walls (Butyrivibrio and Lachnospira).
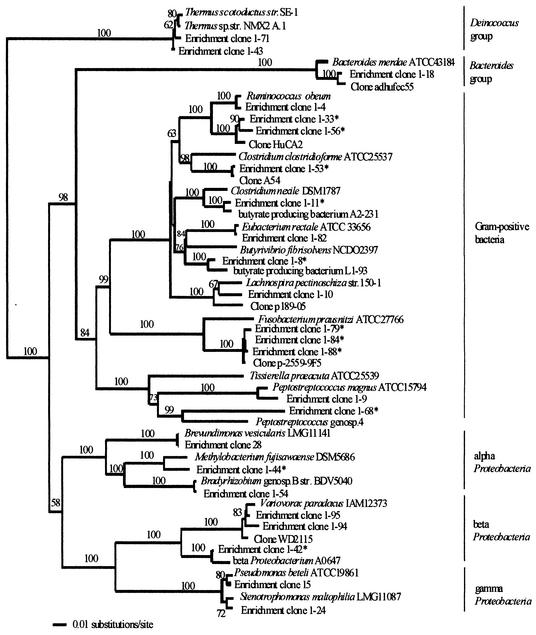
FIG. 1.
Phylogenetic relationships of the 24 rRNA gene sequences obtained from the community cultures and 28 closely related sequences, based on a distance analysis (neighbor-joining algorithm with Jukes-Cantor model; 1,000 bootstrap replicates performed). Accession numbers are provided in Materials and Methods. Asterisks, clones supposed to represent new species or genera. str., strain.
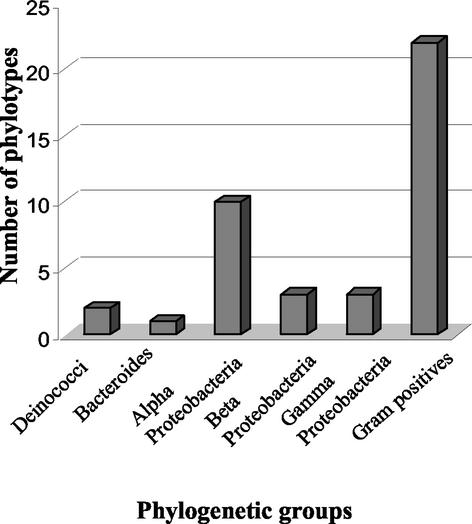
The ARDRA results were used to place the unsequenced 16S rDNA fragments into phylogenetic groups based on patterns identical to those for the sequenced fragments (Fig. 2). The most highly represented groups were the gram-positive bacteria and alpha Proteobacteria. Approximately equal numbers of gram-negative and gram-positive organisms are represented when the numbers from the separate gram-negative groups are combined.
FIG. 2.
Distribution of 16S rRNA gene clones according to ARDRA patterns and phylogenetic groups. Sequenced and nonsequenced fragments with identical ARDRA patterns were placed in the same group.
A Jukes-Cantor evolutionary-distance matrix was calculated (data not shown) in order to determine the degree of relatedness between the ice core enrichment clones and the sequences used in constructing the phylogenetic tree (Fig. 1). Over one-half (16 out of 24) of the enrichment clone sequences (1-71, 1-43, 1-18, 1-4, 1-33, 1-11, 1-82, 1-10, 1-9, 28, 1-44, 1-54, 1-95, 1-94, 15, and 1-24) were found to be closely related to sequences of isolated organisms already found in the GenBank and RDP databases. The similarity in sequence between 12 of these clones (1-71, 1-43, 1-18, 1-4, 1-82, 1-9, 28, 1-54, 1-95, 1-94, 15, and 1-24) and the most closely related isolates was such (<3% distance) that they should be considered strains of the known organisms. However, enrichment clone 1-10 was 3.4% different from Lachnospira pectinoschiza strain 150-1, enrichment clone 1-44 was 3.9% different from Methylobacterium fujisawaense DSM 5686, enrichment clone 1-33 was 4.6% different from Ruminococcus obeum, and enrichment clone 1-11 was 3.9% different from Clostridium nexile DSM 1787, suggesting that these clones could be from novel species.
Several enrichment clones were either distinct from any sequence in the databases (e.g., enrichment clone 1-68) or were closely related to sequences detected only by 16S rRNA gene amplification from various sources (enrichment clones 1-56, 1-53, 1-8, 1-79, 1-84, 1-88, and 1-42). The difference in sequence between the latter enrichment clones and characterized isolates was greater than 5%, suggesting that they are sufficiently different from known organisms to be representatives of new genera. Interestingly, enrichment clone 1-68 was 12, 16, and 18% different from Peptostreptococcus genospecies 4, Tissierella praeacuta ATCC 25539, and Peptostreptococcus magnus ATCC 15794, respectively. These large distances are greater than those found at the genus level.
Reliability of the clone libraries.
We examined our results to determine whether factors that are known to influence the relative proportions of taxa in a clone library could have biased our findings (36). The poor cell lysis observed in our study and the rigorous DNA extraction procedures used can lead both to underestimation of some members of the community and to DNA shearing, resulting in a large number of chimerical products. However, the CHECK-CHIMERA analysis did not show obvious evidence of artifacts for the sequenced 16S rRNA genes.
Another source of bias from PCR amplifications with DNA extracted from diverse organisms is that 16S rRNA genes that are more prevalent in a genome can predominate. Generally it is expected that species with small genomes and large numbers of rrn operons will prevail in a clone library. To determine whether such bias had occurred in our work, we examined the genome sizes and the numbers of rrn operons that had been reported (20) for the closest relatives of the cloned 16S rRNA genes we had sequenced (Table 1). Our results showed that we obtained clones related to Clostridium and Eubacterium, which have several rrn operons (5, 10, or more) (Table 1). However, other clones were related to organisms with small numbers of rrn operons and either small or large genome sizes. For example, Thermus scotoductus has a 1.74-Mb genome and two rrn operons and Brevundimonas vesicularis has a 4-Mb genome and two rrn operons. In addition, one of our clones was related to Bradyrhizobium japonicum, which has a single rrn operon contained in a very large genome (8.7 Mb). These results confirmed that our amplification reflected a range of rrn operons independent of their frequency and the genome size.
TABLE 1.
Numbers of rrn operons and genome sizes of species and phylogenetic groups related to the clones obtained from anaerobic enrichments
| Species or phylogenetic group | No. of rrn operons | Genome size (Mb) |
|---|---|---|
| Thermus thermophilus | 2 | 1.74 |
| Bradyrhizobium japonicum | 1 | 8.70 |
| Peptostreptococcus anaerobius | 5 | 1.3 |
| Peptostreptococcus asaccharolyticus | 3 | 2.0 |
| Pseudomonas aeruginosa | 4 | 6.26 |
| Pseudomonas fluorescens | 5 | 6.63 |
| Pseudomonas putida | 6 | 6.0 |
| Pseudomonas stutzeri | 4 | 4.37 |
| Bacteroides eggerthii | 4 | 4.40 |
| Bacteroides ovatus | 6 | 5.30 |
| Bacteroides uniformis | 5 | 6.67 |
| Bacteroides vulgatus | 4 | 4.80 |
| Fusobacterium nucleatum | 5 | 1.17 |
| Clostridium acetobutylicum | 11 | 3.94 |
| Clostridium beijerinckii | 12 | 6.70 |
| Clostridium saccharobutylicum | 12 | 5.34 |
| Clostridium perfringens | 10 | 3.60 |
| Clostridium paradoxum | 15 | NDb |
| Eubacterium and relatives | 10a | |
| Alpha Proteobacteria | 2.3a | |
| Beta Proteobacteria | 3.9a | |
| Gamma Proteobacteria | 5.6a |
Average number.
ND, not determined.
DISCUSSION
We report here the analysis of anaerobic microbial communities enriched from a sample taken from near the base of a Greenland glacier where the core contains a mixture of fine sediment and particles. Cells in this layer could have been trapped during the Greenland glaciation about 2 million years ago, could have been deposited at the top and packed down at temperatures as low as −30°C for thousands of years, or could have been mixed in from the underlying bedrock more recently. Independent of their derivation, cells within the sample have remained at −9°C or lower for over 100,000 years. Despite these extreme conditions, examination of a melted sample using epifluorescence microscopy and a BacLight staining kit to distinguish live and dead cells demonstrated that a very high proportion of the cells were viable. This is consistent with the growth of cells in our anaerobic media at a low temperature and our microscopic observation of numerous motile organisms with a variety of morphologies.
A unique aspect of examining enrichment cultures from an ice core sample that has been frozen for thousands of years is that one can ask whether the 16S rRNA gene sequences correspond to current organisms or if they represent distinct phylogenetic relationships. We considered the possibility that microorganisms deposited during or after the drilling process could contaminate ice core samples and interfere with the interpretation of the results. To avoid this problem, we took several precautions and developed an approach for removing the surface ice while maintaining cold temperatures so that any especially heat-sensitive psychrophiles would not be killed. Based on the results from the control experiments and the diversity of organisms reflected by the clone library, it is unlikely that the inner core samples we used were contaminated.
Our results show that 12 of the Greenland enrichment clone sequences are closely related to those of isolated organisms already found in the GenBank and RDP databases. The differences for four others (enrichment clones 1-10, 1-44, 1-33, and 1-11) are sufficient for the organisms to be in new species or genera depending on other physiological traits. Seven enrichment clone sequences (1-56, 1-53, 1-8, 1-79, 1-84, 1-88, and 1-42) were closely related to sequences detected only by 16S RNA gene amplification rather than to any known isolate. In these cases, the differences in sequence between these enrichment clones and the most closely related characterized sequences are greater than 5% and the clones could be representatives of new genera. One enrichment clone (1-68) was distinct from any sequence in either database, with a difference of 12% from the closest known relative, Peptostreptococcus genospecies 4. This large distance indicates that enrichment clone 1-68 may be a representative of a novel bacterial order or family.
Not only are several phylogenetic groups represented in our results, but also many physiologically distinct organisms are found within these groups. For example, within the alpha Proteobacteria, we find clones related to a N2-fixing species of Bradyrhizobium, the methylotrophic species of Methylobacterium, and the oligotrophic bacteria in the genus Brevundimonas. In other cases, the clones are closely related to gram-positive or Bacteroides organisms that have been associated with human feces (17, 37) or animal gastrointestinal tracts (22). These relationships suggest an enteric origin, such as animal feces, and their increased numbers could reflect enrichment during the anaerobic incubation. An alternative origin is suggested by the discovery of anaerobic fermentative bacteria (Clostridium, Eubacterium, or Bacteroides spp.) in mats and sediments of Antarctic lakes; their presence was connected to biodegradation of organic polymers (2, 9). A similar explanation could apply to our results because the sample came from deep ice possibly containing a significant amount of organic material based on data from similar basal ice core samples reported by other authors (34). Another indirect indication for a high organic-material loading of the ice comes from the high bacterial numbers found. Independent of their origins, these organisms apparently have persisted for thousands of years at low temperatures.
One surprising relationship was between clones 1-71 and 1-43 and the thermophile Thermus scotoductus, originally isolated from a geothermal heating system in southern Iceland (21). Although the ice core was drilled in Greenland, which is relatively close to Iceland, it is unlikely that the ice core contained these organisms in sufficient numbers to be detected after numerous transfers into new media. Thus, it is possible that the organisms yielding the sequences for clones 1-71 and 1-43 were able to grow at −2°C. The isolation of such psychrophilic organisms, which are phylogenetically related to Thermus species would be of great interest.
The enrichment communities from this Greenland ice core sample show a greater diversity than that found for a Lake Vostok core where seven unique sequences were found by direct DNA extraction and amplification and where epifluorescence microscopy suggested that the population was only a few hundred cells per milliliter (19, 29). The richer diversity found in this study is likely due to the large amount of sediment deposited in the sample compared to that in the translucent Vostok ice and to the enrichment process applied. Interestingly, no amplification product was obtained from Lake Vostok samples by using archaeal primers. Although we obtained fragments following amplification with archaeal primers, the similarity values obtained by comparing their sequences with known sequences were abnormally low (unpublished data). One explanation for these results is that no Archaea were present in the communities and that the fragments we obtained were PCR artifacts. An alternative explanation is that the fragments correspond to new, unusual archaeal sequences.
Even though we found gene sequences representing many different bacterial genera, no one approach can provide a complete unbiased picture of the organisms residing in a sample or their relative numbers. The inability to cultivate all species within a sample and the possibility that some organisms outgrow others are well accepted, but biases associated with molecular methods (10, 36) have received less attention. We examined the different methods we used in order to address these issues. For example, we found that many of the cells cultured and observed microscopically were resistant to lysis, which would result in the underestimation of diversity. Another factor is differences in genome size and the number, distribution, and microdiversity of the rrn operons. The quantity of the PCR amplification products of different 16S rRNA genes can be shifted by these genomic properties. Therefore, finding greater numbers of certain 16S rRNA gene fragments can reflect greater PCR amplification rather than a greater proportion of a particular cell. For example, the Clostridium and Eubacterium species have the greatest number of operons. In addition a larger number of Clostridium organisms might be expected since their spores might have survived in the ice core and the cells could have outgrown other organisms in our anaerobic enrichments. Indeed, we found sequences related to Clostridium species. Some of our clones are also related to organisms with a small number of rrn operons and either small or large genome sizes such as Thermus thermophilus and Bradyrhizobium japonicum. Our ability to find 16S rRNA gene sequences from organisms with a range of genome sizes and numbers of rrn operon shows that no single operon dominates the PCR amplification.
Since spore formation is a known survival mechanism, it is noteworthy that only two 16S rRNA gene sequences were related to the anaerobic sporeforming Clostridium species. This suggests that the cells used survival mechanisms other than spore formation. This is consistent with our observation that, although no growth occurred initially when aerobic cultures were inoculated, growth was observed after several months of incubation (unpublished data). Thus, even though the cells from the ice core are viable, special media and incubation conditions are required for their recovery and cultivation. These results are consistent with those of Christner et al. (6) and point to the need for understanding these processes. This is especially relevant to the elaboration of strategies for recovering microorganisms from extraterrestrial samples. Innovative incubation methods could help cultivate many of the organisms that have been found here and in other studies only by their cloned 16S rRNA gene sequence from ice cores and many other habitats. Our future work focuses on using the phylogenetic results obtained here and our increasing knowledge of cultivation conditions to obtain and characterize psychrophilic isolates from ice core samples.
Acknowledgments
We thank T. Sowers and members of our laboratory for helpful discussions and E. Kunze for help with the epifluorescence microscopy and flow cytometry work.
The research was supported by Department of Energy grant DE-FG02-93ER20117 and partial funding from the Penn State Astrobiology Center NASA-Ames Cooperative Agreement no. NCC2-1057 and NSF/IGERT DGE-9972759 for Biogeochemical Research Initiative for Education.
REFERENCES
- 1.Baross, J. A., and R. Y. Morita. 1978. Microbial life at low temperatures: ecological aspects, p. 9-71. In J. Kushner (ed.), Microbial life in extreme environments. Academic Press, New York, N.Y.
- 2.Brambilla, E., H. Hippe, A. Hagelstein, G. Tindall, and E. Stackebrandt. 2001. 16S rDNA diversity of cultured and uncultured prokaryotes of a mat sample from Lake Fryxell, McMurdo Dry Valleys, Antarctica. Extremophiles 5:23-33. [DOI] [PubMed] [Google Scholar]
- 3.Brenchley, J. E. 1996. Psychrophilic microorganisms and their cold-active enzymes. J. Ind. Microbiol. 17:432-437. [Google Scholar]
- 4.Cato, E. P., W. L. George, and S. M. Finegold. 1986. Genus Clostridium Prazmowski 1880, 23AL, p. 1141-1200. In P. H. A. Sneath, N. S. Mair, M. E. Sharpe, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 2. Williams & Wilkins, Baltimore, Md.
- 5.Christner, B. C., E. Mosley-Thompson, L. G. Thompson, and J. N. Reeve. 2001. Isolation of bacteria and 16S rDNAs from Lake Vostok accretion ice. Environ. Microbiol. 3:570-577. [DOI] [PubMed] [Google Scholar]
- 6.Christner, B. C., E. Mosley-Thompson, L. G. Thompson, B. Zagorodnov, K. Sandman, and J. N. Reeve. 2000. Recovery and identification of viable bacteria immured in glacial ice. Icarus 144:479-485. [Google Scholar]
- 7.Chyba, C. F. 2000. Energy for microbial life on Europa. Nature 403:381-382. [DOI] [PubMed] [Google Scholar]
- 8.Deming, J. W. 2002. Psychrophiles and polar regions. Curr. Opin. Microbiol. 5:301-309. [DOI] [PubMed] [Google Scholar]
- 9.Dube, S., L. Singh, and S. I. Alam. 2001. Proteolytic anaerobic bacteria from lake sediments of Antarctica. Enzyme Microb. Technol. 28:114-121. [DOI] [PubMed] [Google Scholar]
- 10.Farrelly, V. F., F. A. Rainey, and E. Stackebrandt. 1995. Effect of genome size and rrn gene copy number on PCR amplification of 16S rRNA genes from a mixture of bacterial species. Appl. Environ. Microbiol. 61:2798-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferrara-Guerrero, M. J., D. G. Marty, and A. Bianchi. 1993. Isolation and enumeration of anaerobic and microaerophilic bacteria in aquatic habitats, p. 9-19. In P. Kemp (ed.), Handbook of methods in microbial ecology. Lewis Publishers, Boca Raton, Fla.
- 12.Gow, A. J., and D. A. Meese. 1996. Nature of basal debris in the GISP2 and Byrd ice cores and its relevance to bed processes. Ann. Glaciol. 22:134-140. [Google Scholar]
- 13.Gow, A. J., D. A. Meese, R. B. Alley, and J. J. Fitzpatrick. 1997. Physical and structural properties of the Greenland Ice Sheet Project 2 ice core: a review. J. Geophys. Res. 102:26559-26575. [Google Scholar]
- 14.Gray, J. P., and R. P. Herwig. 1996. Phylogenetic analysis of the bacterial communities in marine sediments. Appl. Environ. Microbiol. 62:4049-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greenberg, R. 2002. Tides and the biosphere of Europa. Am. Sci. 90:48-55. [Google Scholar]
- 16.Hengstmann, U., K.-J. Chin, P. H. Janssen, and W. Liesack. 1999. Comparative phylogenetic assignment of environmental sequences of genes encoding 16S rRNA and numerically abundant culturable bacteria from an anoxic rice paddy soil. Appl. Environ. Microbiol. 65:5050-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hold, G. L., S. E. Pryde, V. J. Russell, E. Furrie, and H. J. Flint. 2002. Assessment of microbial diversity in human colonic samples by 16S rDNA sequence analysis. FEMS Microbiol. Ecol. 39:33-39. [DOI] [PubMed] [Google Scholar]
- 18.Junge, K., C. Krembs, J. Deming, A. Stierle, and H. Eicken. 2001. A microscopic approach to investigate bacteria under in situ conditions in sea-ice samples. Ann. Glaciol. 33:304-310. [Google Scholar]
- 19.Karl, D. M., D. F. Bird, K. Bjorkman, T. Houlihan, R. Shackelford, and L. Tupas. 1999. Microorganisms in the accreted ice of Lake Vostok, Antarctica. Science 286:2144-2147. [DOI] [PubMed] [Google Scholar]
- 20.Klappenbach, J. A., P. R. Saxman, J. T. Cole, and T. M. Schmidt. 2001. rrndb: the ribosomal RNA operon copy number database. Nucleic Acids Res. 29:181-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kristjansson, J. K., S. Hjorleifsdottir, T. V. Marteinsson, and G. A. Alfredsson. 1994. Thermus scotoductus, sp.nov., a pigment-producing thermophilic bacterium from hot tap water in Iceland and including Thermus sp. X-1. Syst. Appl. Microbiol. 17:44-50. [Google Scholar]
- 22.Leser, T. D., J. Z. Amenuvor, T. K. Jensen, R. H. Lindecrona, M. Boye, and K. Moller. 2002. Culture-independent analysis of gut bacteria: the pig gastrointestinal tract microbiota revisited. Appl. Environ. Microbiol. 68:673-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loveland-Curtze, J., P. P. Sheridan, K. R. Gutshall, and J. E. Brenchley. 1999. Biochemical and phylogenetic analyses of psychrophilic isolates belonging to the Arthrobacter subgroup and description of Arthrobacter psychrolactophilus, sp. nov. Arch. Microbiol. 177:355-363. [DOI] [PubMed] [Google Scholar]
- 24.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, P. R. Saxman, J. M. Stredwick, G. M. Garrity, B. Li, G. J. Olsen, S. Pramanik, T. M. Schmidt, and J. M. Tiedje. 2000. The RDP (Ribosome Database Project) continues. Nucleic Acids Res. 28:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meakin, S. A., J. H. E. Nash, W. D. Murray, K. J. Kennedy, and G. D. Sprott. 1991. A generally applicable technique for the extraction of restrictable DNA from methanogenic bacteria. J. Microbiol. Methods 14:119-126. [Google Scholar]
- 26.Pace, N. R., D. L. Stahl, D. J. Lane, and G. J. Olsen. 1986. The analysis of natural microbial populations by ribosomal RNA sequences. Adv. Microb. Ecol. 9:1-55. [Google Scholar]
- 27.Porteous, L. A., J. L. Armstrong, R. J. Seidler, and L. S. Watrud. 1994. An effective method to extract DNA from environmental samples for polymerase chain reaction amplification and DNA fingerprint analysis. Curr. Microbiol. 29:301-307. [DOI] [PubMed] [Google Scholar]
- 28.Price, P. B. 2000. A habitat for psychrophiles in deep Antarctic ice. Proc. Natl. Acad. Sci. USA 97:1247-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Priscu, J. C., E. E. Adams, W. B. Lyons, M. A. Voytek, D. W. Mogk, R. L. Brown, C. P. McKay, C. D. Takacs, K. A. Welch, C. F. Wolf, J. D. Kirshtein, and R. Avci. 1999. Geomicrobiology of subglacial ice above Lake Vostok, Antarctica. Science 286:2141-2144. [DOI] [PubMed] [Google Scholar]
- 30.Redeker, D., H. Thierfelder, C. Walker, and D. Werner. 1997. Restriction analysis of PCR-amplified internal transcribed spacers of ribosomal DNA as a tool for species identification in different genera of the order Glomales. Appl. Environ. Microbiol. 63:1756-1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Sheridan, P. P., and J. Brenchley. 2000. Characterization of a salt-tolerant family 42 β-galactosidase from a psychrophilic Antarctic Planococcus isolate. Appl. Environ. Microbiol. 66:2438-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sheridan, P. P., N. Panasik, J. M. Coombs, and J. E. Brenchley. 2000. Approaches for deciphering the structural basis of low temperature enzyme activity. Biochim. Biophys. Acta 1543:417-433. [DOI] [PubMed] [Google Scholar]
- 34.Skidmore, M. L., J. M. Foght, and M. J. Sharp. 2000. Microbial life beneath a high Arctic glacier. Appl. Environ. Microbiol. 66:3214-3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sowers, K. R., M. J. K. Nelson, and J. G. Ferry. 1984. Growth of acetotrophic, methane-producing bacteria in a pH auxostat. Curr. Microbiol. 11:227-230. [Google Scholar]
- 36.Stackebrandt, E., R. Pukall, G. Ulrichs, and H. Rheims. 1999. Analysis of 16S rDNA clone libraries: part of the big picture, p. 1-9. In B. M. Bell and C. R. P. Jonson-Green (ed.), Proceedings of the 8th International Symposium on Microbial Ecology. Microbial biosystems: new frontiers. Atlantic Canada Society for Microbial Ecology, Halifax, Nova Scotia, Canada.
- 37.Suau, A., R. Bonnet, M. Sutren, J. J. Gordon, G. R. Gibson, M. D. Collins, and J. Dore. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trimbur, D. E., K. R. Gutshall, P. Prema, and J. E. Brenchley. 1994. Characterization of a psychrotrophic Arthrobacter gene and its cold-active β-galactosidase. Appl. Environ. Microbiol. 60:4544-4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vetriani, C., H. W. Jannasch, B. J. MacGregor, D. A. Stahl, and A.-L. Reysenbach. 1999. Population structure and phylogenetic characterization of marine benthic Archaea in deep-sea sediments. Appl. Environ. Microbiol. 65:4375-4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]