Abstract
In a previous study we isolated the meta-cleavage enzyme gene, tesB, that encodes an enzyme that carries out a meta-cleavage reaction in the breakdown of testosterone by Comamonas testeroni TA441 (M. Horinouchi et al., Microbiology 147:3367-3375, 2001). Here we report the isolation of a gene, tesD, that encodes a hydrolase which acts on the product of the meta-cleavage reaction. We isolated tesD by using a Tn5 mutant of TA441 that showed limited growth on testosterone. TesD exhibited ca. 40% identity in amino acid sequence with BphDs, known hydrolases of biphenyl degradation in Pseudomonas spp. The TesD-disrupted mutant showed limited growth on testosterone, and the culture shows an intense yellow color. High-pressure liquid chromatography analysis of the culture of TesD-disrupted mutant incubated with testosterone detected five major intermediate compounds, one of which, showing yellow color under neutral conditions, was considered to be the product of the meta-cleavage reaction. The methylation product was analyzed and identified as methyl-4,5-9,10-diseco-3-methoxy-5,9,17-trioxoandrosta-1(10),2-dien-4-oate, indicating that the substrate of TesD in testosterone degradation is 4,5-9,10-diseco-3-hydroxy-5,9,17-trioxoandrosta-1(10),2-dien-4-oic acid. 4,5-9,10-Diseco-3-hydroxy-5,9,17-trioxoandrosta-1(10),2-dien-4-oic acid was transformed by Escherichia coli-expressed TesD. Downstream of tesD, we identified tesE, F, and G, which encode for enzymes that degrade one of the products of 4,5-9,10-diseco-3-hydroxy-5,9,17-trioxoandrosta-1(10),2-dien-4-oic acid converted by TesD.
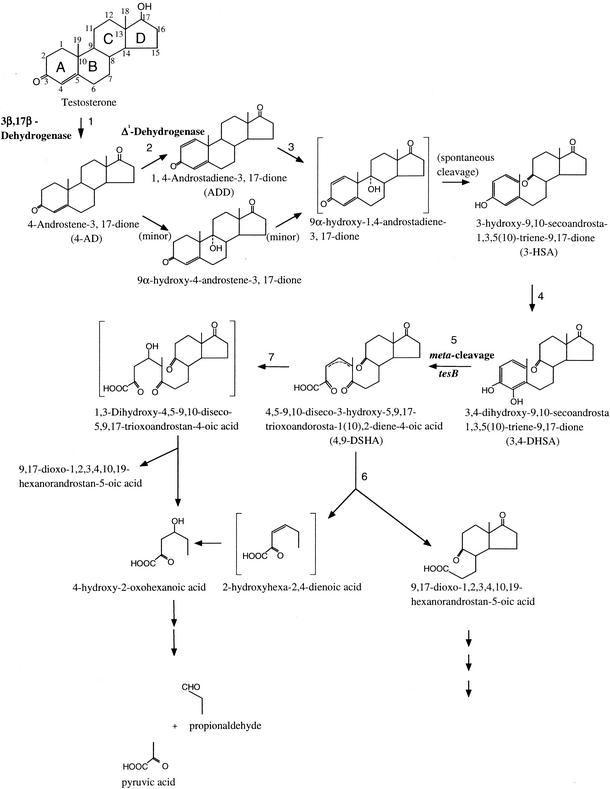
Via a complex metabolic pathway involving many steroid-inducible enzymes, Comamonas testosteroni is able to utilize certain C19 and C21 steroids, as well as a number of aromatic compounds as sole carbon and energy sources. The mechanism by which testosterone is degraded in Nocardia restrictus and C. testosteroni was proposed more than 30 years ago (Fig. 1) (4, 7, 16). The process is initiated by dehydrogenation of the 17β-hydroxyl group on testosterone to 4-androstene-3,17-dione (4AD) (reaction 1 in Fig. 1), which then undergoes Δ1-dehydrogenation to 1,4-androstadiene-3,17-dione (ADD) (reaction 2), followed by 9α-hydroxylation to produce 3-hydroxy-9,10-secoandrosta-1,3,5(10)-triene-9,17-dione (3-HSA) (reaction 3 and the following spontaneous cleavage) (16). The C-4 of 3-HSA is hydroxylated to yield 3,4-dihydroxy-9,10-secoandrosta-1,3,5(10)-triene-9,17-dione (3,4-DHSA) (reaction 4), which is cleaved between C-4 and C-5 via a meta-cleavage reaction (reaction 5) (7). The product of the meta-cleavage reaction of 3,4-DHSA, 4,5-9,10-diseco-3-hydroxy-5,9,17-trioxoandrosta-1(10),2-dien-4-oic acid (4,9-DSHA), is dark yellow in color and exists as a mixture of keto-enol forms in the culture (7). According to the speculation of Gibson et al. (7), 4,9-DSHA would be degraded in two ways: degraded into two compounds by hydrolysis (reaction 6) or hydroxylated at the 1-position before hydrolysis (reaction 7). 4,9-DSHA could not be isolated successfully from the reaction solution because it was extremely unstable (17). Therefore, the structure of 4,9-DSHA was predicted by analyzing the amorphous pyridine compound produced from 4,9-DSHA (17). For this metabolic pathway, the genes encoding 3β,17β-dehydrogenase (reaction 1) (1, 6) and Δ1-dehydrogenase (reaction 2) (5, 13) have been identified, but genes encoding enzymes for other reactions remain unknown.
FIG. 1.
Proposed pathway of testosterone degradation in N. restrictus and C. testosteroni (4, 7, 16). Names of the enzymes are given to those whose genes were isolated and charecterised. Compounds in brackets are speculation.
In a previous study, we isolated a gene encoding a meta-cleavage enzyme, tesB, which is indispensable for growth of C. testosteroni TA441 on testosterone and is induced during growth on testosterone (9). TesB probably catalyzes conversion of 3,4-DHSA to 4,9-DSHA (reaction 5 in Fig. 1). Downstream of tesB, three open reading frames (ORFs), ORF1 to ORF3, were identified—all of which are necessary for testosterone metabolism in TA441.
About 20 kb of the DNA region containing tesB was analyzed to isolate other genes for testosterone degradation of TA441, including the hydrolase gene for 4,9-DSHA, but none of the predicted ORFs showed significant homology with known hydrolase genes (unpublished data). In the present study we report the isolation of the hydrolase gene for 4,9-DSHA, tesD, by means of Tn5 mutants of TA441 showing limited growth on testosterone, and the analysis of the gene-disrupted mutant of tesD.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Bacterial strains and plasmids used in the present study are listed in Table 1. DNA manipulations were performed according to the standard method described previously (9). C. testosteroni TA441 and the mutant strains were grown at 30°C in Luria-Bertani (LB) medium, C medium (3), or LB+C medium (i.e., a mixture of equal volumes of LB and C media) with suitable carbon sources when necessary. Testosterone was added when necessary as a filtered dimethyl sulfoxide (DMSO) solution at a final concentration of 0.1% (wt/vol). A testosterone plate was made with C medium with the DMSO solution of testosterone. This plate is turbid due to the presence of testosterone, and when TA441 grows on the plate utilizing testosterone as a carbon source, a clear spot forms around the TA441.
TABLE 1.
C. testosteroni bacterial strains and plasmids
| Strain or plasmid | Characteristicsa | Source or reference |
|---|---|---|
| Bacterial strains | ||
| TA441 | Wild type, Tesb | 2 |
| TesB− | tesB::Kmr mutant of TA441 | This work |
| TesD− | tesD::Kmr mutant of TA441 | This work |
| TesE− | tesE::Kmr mutant of TA441 | This work |
| ORF11− | ORF11::Kmr mutant of TA441 | This work |
| ORF12− | ORF12::Kmr mutant of TA441 | This work |
| Plasmids | ||
| pUC19 | Apr; lacZ | 20 |
| pC29 | Apr; pSuperCosI with BamHI insert of TA441 | This work |
| pC26 | Apr; pSuperCosI with BamHI insert of TA441 | This work |
| pC29E3 | Apr; pUC19 with EcoRI insert from pC29 | This work |
| pC26ScH | Apr; pUC19 with SacI-HindIII insert from pC26 | This work |
| pORF11-Kmr | Apr; pC29E3 derivative with Kmr gene in NruI site of ORF11 | This work |
| pORF12-Kmr | Apr; pC29E3 derivative with Kmr gene in PstI site of ORF12 | This work |
| pTesD-Kmr | Apr; pC29E3 derivative with Kmr gene in NspI site of tesD | This work |
| pTesE-Kmr | Apr; pC29E3 derivative with Kmr gene in SmaI site of tesE | This work |
| pTesD | Apr; pC29E3 derivative coding tesD | This work |
| pTesE | Apr; pC29E3 derivative coding tesE | This work |
Apr, ampicillin resistance.
Grown on testosterone as the sole carbon source.
Tn5 mutagenesis.
pSUP5011 (18) was introduced into TA441 by electroporation. Tn5-inserted mutants were selected on an LB plate supplemented with kanamycin (400 μg/ml). The mutant colonies were transferred to a C medium plate supplemented with kanamycin (400 μg/ml) and testosterone (final concentreation, 0.1% [wt/vol], added as a 10% DMSO solution). Colonies showing a smaller clear zone around them on the testosterone plate with kanamycin were selected and grown in the liquid medium to confirm their lower ability to degrade testosterone. The mutants that grew as well as TA441 on C medium supplemented with p-hydroxybenzoate (positive control) but not with testosterone were selected. p-Hydroxybenzoate was used as a positive control, instead of commonly used carbon sources such as succinate, because the growth of TA441 was stable on p-hydroxybenzoate but unstable on succinate.
Cloning of TA441 DNA flanking the inserted Kmr gene of Tn5.
E. coli JM109 was transformed with pUC19 derivatives carrying a fragment of the total DNA of the Tn5 mutants digested with SalI, and kanamycin-resistant (Kmr) transformants were selected. Since Tn5 has a SalI site just next to the Kmr gene, the plasmid of the selected transformants contained the Kmr gene and the flanking DNA region originating from the Tn5 mutants of TA441. The cloned DNA fragment from the Tn5 mutant was sequenced to confirm that the potential testosterone degradation gene was cloned in the DNA region. Using a part of the DNA region as a probe, the cosmid library of the total DNA of TA441 was screened by colony hybridization. To construct the cosmid library, SuperCosI (Stratagene) was used according to the manufacturer's instructions. SuperCosI was introduced into E. coli by using the GigapackIII XL packaging kit (Stratagene). SuperCosI is maintained as a plasmid in transformed E. coli, and the selected E. coli colony was confirmed by Southern hybridization as carrying the plasmid containing the target gene fragment. Deletion plasmids for further study were constructed by inserting partial DNA of the SuperCosI derivatives into pUC19. Plasmids for the DNA sequence were constructed by using the genome priming system (New England Biolabs), and the plasmid solution for PCR was prepared by using the Wizard Plus Minipreps DNA purification system (Promega). DNA sequence determination and analysis were performed by using an ABI model 373A automated DNA sequencer and dye terminator sequencing protocols (Perkin-Elmer Japan, Chiba, Japan) as previously described (9).
Construction of plasmids and mutant strains for gene disruption experiments.
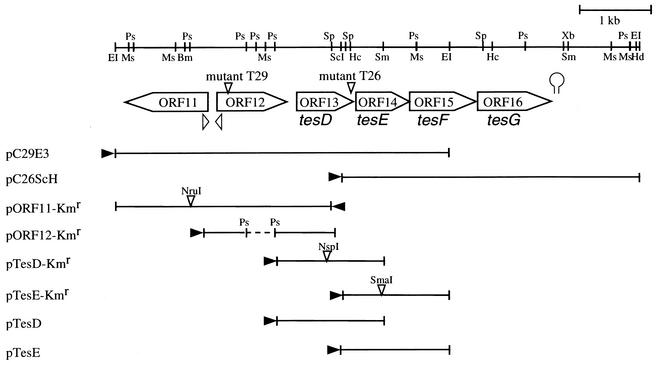
The tesD gene was disrupted by insertion of a Kmr gene into the NspI site in tesD. The resultant plasmid, pTesD-Kmr, was used for inactivation of the tesD gene in TA441 by homologous recombination. Insertion of the Kmr gene in tesD was confirmed by Southern hybridization. Gene disruption of tesE, ORF11, and ORF12 were performed in the same manner. The plasmids used for gene disruption are shown in Table 1, and the restriction site at which the Kmr gene was inserted is indicated in Fig. 2.
FIG. 2.
Partial restriction map of the 7.4-kb EcoRI-HindIII fragment, the insert in pC29E3 and pC26ScH that encodes genes for the testosterone-degrading enzymes of C. testosteroni TA441. The genes and putative ORFs are indicated by large open arrows. Open arrowheads below these ORFs indicate putative promoters, and the putative terminator locates just downstream of tesG. Deletion plasmids are indicated below the restriction map; the remaining gene segments (lines) are also shown. Plasmids pORF11-Kmr to pTesE-Kmr are gene-disrupted plasmids. Restriction sites where the Kmr gene was inserted are indicated by small open arrows above the lines. In pORF12-Kmr, the PstI fragment shown in the figure was lost. The closed arrowheads indicate the direction of transcription regulated by the lac promoter of the pUC19 vector. Abbreviations are as follows: Bm, BamHI; EI, EcoRI; Hc, HincII; Hd, HindIII; Ms, MstI; Ps, PstI; ScI, SacI; Sp, SphI; Sm, SmaI; Xb, XbaI.
Growth of TA441 and mutant strains on testosterone.
Growth was monitored as CFU because the culture to which testosterone was added as a DMSO solution was turbid, which prevented measurement of cell absorbance. The mutants were grown in C medium with 0.1% (wt/vol) testosterone, and growth was monitored by counting colonies that appeared on LB plates on which appropriate dilutions of the culture had been spread and incubated at 30°C as previously described (9).
HPLC analysis.
Twice the volume of methanol was added to the culture, which was then centrifuged and the supernatant directly injected into the high-pressure liquid chromatography (HPLC). For HPLC separations, a Waters 600 HPLC (Waters Corporation) was used with an Inertsil ODS-3 column (4.6 by 250 mm; GL Science, Tokyo, Japan), eluted with CH3CN-CH3OH-H2O-trifluoroacetic acid (TFA) (50:10:40:0.05; flow rate, 1 ml/min) at 40°C, and detected at 210 nm (for the data presented in Table 1). For the three-dimensional HPLC data, a Waters 2690 HPLC was utilized with an Inertsil ODS-3 column (4.6 by 250 mm for the data shown in Fig. 6 and 2.1 by 150 mm for the data shown in Fig. 7), and elution carried out by using a linear gradient from 20% solution A (CH3CN-CH3OH-TFA [95:5:0.05]) and 80% solution B (CH3CN-H2O-TFA [95:5:0.05]) to 65% solution A and 35% solution B over 10 min, then maintained for 3 min, and then changed again to 20% solution A for 5 min. The flow rate was 1 ml/min with the ODS-3 column with greater diameter and length (4.6 by 250 mm) and 0.21 ml/min for the narrower and shorter ODS-3 column (2.1 by 150 mm).
FIG. 6.
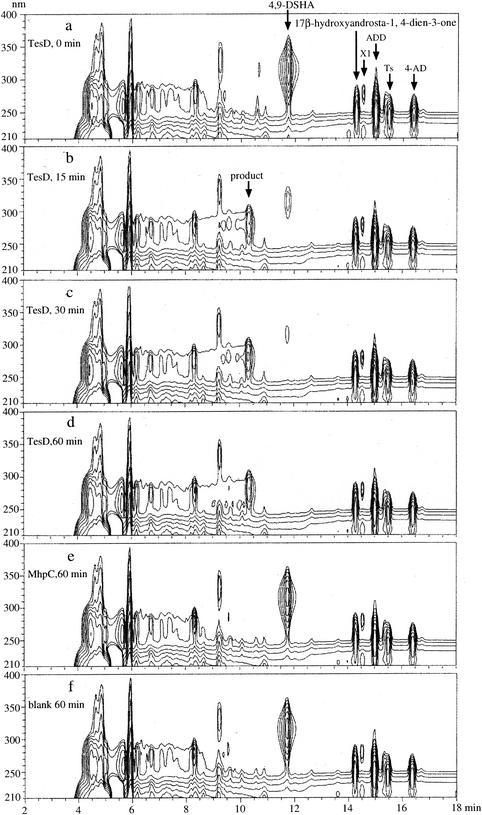
Conversion of 4,9-DSHA by TesD. Data are represented in a three-dimensional HPLC chart (the vertical axis indicates wavelength, the horizontal axis indicates the RT, and the UV absorbance of each compound is represented in contour). (a) HPLC chart of the culture of the TesD-disrupted mutant grown on the mixed LB+C medium with testosterone, which was used as the reaction solution. The charts show HPLC separations performed at 15 min (b), 30 min (c), and 60 min (d) after the addition of the cell extract of E. coli expressing TesD; 60 min after the addition of the cell extract of E. coli expressing MhpC (e); and without any enzymes (f). MhpC is the hydrolase for 2-hydroxy-6-keto-nona-2,4-diene-1,9-dioic acid, the product of meta-cleavage reaction in the 3-(3-hydroxyphenyl) propionic acid degradation pathway of TA441 (3), which was used as negative control. For the analysis, a Waters 2690 HPLC was used with an Inertsil ODS-3 column (4.6 by 250 mm; GL Science), and elution was carried out by using a linear gradient from 20% solution A (CH3CN-CH3OH-TFA [95:5:0.05]) and 80% solution B (CH3CN-H2O-TFA [95:5:0.05]) to 65% solution A and 35% solution B over 10 min, maintained for 3 min, and then changed to 20% solution A for 5 min. The flow rate was 1 ml/min.
FIG. 7.
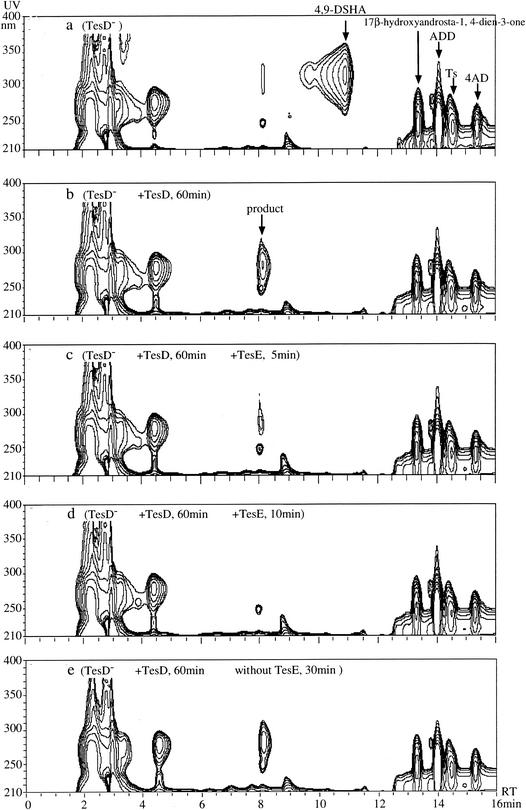
Conversion of the probable product of TesD by TesE presented as a three-dimensional HPLC chart. (a and b) HPLC chart of the culture of the TesD-disrupted mutant (a) and 60 min after treatment with TesD (b). The resultant solution was used as the reaction mixture for TesE. The charts show separations performed 0 min (b), 5 min (c), and 10 min (d) after the addition of the cell extract of E. coli expressing TesE and after 30 min without any enzymes (e). For the analysis, a Waters 2690 HPLC was used with an Inertsil ODS-3 column (2.1 by 150 mm; GL Science), and elution was carried out by using the same gradient stated in the legend of Fig. 6. The flow rate was 0.21 ml/min.
Isolation of the product of the meta-cleavage reaction.
For the isolation of the product of meta-cleavage reaction (the compound X5), a 400-ml culture of tesD-disrupted mutant incubated with 0.1% testosterone was twice extracted with the same volume of ethyl acetate. The aqueous layer was then acidified with HCl and extracted twice with the same volume of ethyl acetate. The ethyl acetate layer that was extracted under the acidic conditions was concentrated in vacuo, and the oily residue was dissolved in a small amount of methanol and loaded onto the Waters 600 HPLC (Nihon Waters, Tokyo, Japan) with an Inertsil ODS-3 column (20 by 250 mm; GL Science) and a solvent, with the composition CH3CN-CH3OH-H2O-TFA [50:10:40:0.05], at a flow rate of 1 ml/min at 40°C. X5 was detected at 316 nm. The fraction containing X5 was collected and studied further.
Isolation of the methyl esters of X5 and X6.
For the isolation of the methyl ester of the compound X5 (X5-Me), an 800-ml culture of TesD− incubated with 0.1% testosterone was extracted twice with the same volume of ethyl acetate. The aqueous layer was collected, acidified, and extracted twice with the same volume of ethyl acetate. The ethyl acetate layer that was extracted under the acidic conditions was concentrated in vacuo, and the oily residue was dissolved in ca. 5 ml of the solvent (methanol-benzene [3:2]) and treated with 3 ml of trimethylsilyldiazomethane solution (10% [wt/wt] trimethylsilyldiazomethane in hexane, purchased from Nacalai Tesque) for 1 h. The resultant solution was dried and concentrated in vacuo, and the residue was dissolved in 1 ml of methanol and loaded onto the Waters 600 HPLC with an Inertsil ODS-3 column (20 by 250 mm) and a solvent, with the composition CH3CN-CH3OH-H2O-TFA (50:10:40:0.05), and eluted at a flow rate of 8 ml/min at 40°C. X5-Me and X6-Me were detected at 300 nm. The fractions containing X5-Me and X6-Me were collected and dried.
Isolation of other intermediate compounds.
Steroid compounds without any ring cleavage were contained in the ethyl acetate layer extracted under neutral conditions. The ethyl acetate layer was dried, concentrated in vacuo, dissolved in a small amount of methanol, and loaded onto the Waters 600 HPLC as described above. These compounds were detected at 245 nm, and collected from the eluent.
General experimental procedures.
For gas chromotography-mass spectrometry, a GCMS-QP5050A mass spectrometer (Shimadzu, Tokyo, Japan) fitted with a HP-5MS column (0.25 mm [internal diameter] by 30 m 0.25 μm [film thickness]; Agilent Technologies) was used. Fast atom bombardment mass spectometry (FAB-MS) (positive-ion mode) were recorded on a JEOL JMS-SX 102 mass spectrometer by using a glycerin matrix. UV spectra were recorded with Ultrospec 3300 (Amersham Pharmacia Biotech). Nuclear magnetic resonance (NMR; 1D 1H and 13C, distortionless enhancement by polarization transfer [DEPT], pulsed field gradients [PFG]-DQF correlated spectroscopy [COSY], PFG-heteronuclear multiple quantum coherence [HMQC], and PFG-heteronuclear multiple bond coherence [HMBC]) spectra were taken on a JNM-ECP500 spectrometer (JEOL, Tokyo, Japan) and nuclear Overhauser effect (NOE) differential spectra were taken on a JNM-A400 spectrometer (JEOL) in CDCl3 solution with tetramethylsilane at 0 ppm and CDCL3 solvent at 77.0 ppm as an internal standard for 1H and 13C chemical shifts, respectively.
Conversion of the intermediate compounds by TesD and TesE.
The tesD-disrupted mutant incubated in LB medium for ca. 15 h was centrifuged, resuspended in a small amount of C-medium, inoculated into LB+C medium with 0.1% testosterone, and incubated at 30°C for ca. 15 h until the medium showed an intense yellow color. The culture medium was centrifuged, and the supernatant was transferred to a new test tube. This culture solution was used as the reaction solution containing 4,9-DSHA, the substrate of TesD, because 4,9-DSHA was not commercially available and was unstable after purification from the culture. E. coli JM109 harboring pTesD (Fig. 2) was cultured in LB medium containing 100 μg of ampicillin/ml and 200 μM IPTG (isopropyl-β-d-thiogalactopyranoside) at 30°C for ca. 15 h. After the incubation, 200 μM IPTG was added to the culture, and the culture was incubated for an additional 2 h. The cells were centrifuged, washed twice with C medium, resuspended in C medium, and disrupted by sonication. After centrifugation, the supernatant was used as a cell extract. The reaction was initiated by inoculation of the cell extract into the reaction solution and was carried out at 30°C. Samples were analyzed by HPLC at 0, 15, 30, and 60 min. MhpC, a hydrolase of TA441 for 3-(3-hydroxyphenyl) propionic acid degradation (3), was used as a negative control. The cell extract of E. coli expressing TesE was prepared in the same manner. The reaction solution after the conversion of 4,9-DSHA to the probable product by TesD was used as the reaction solution for TesE.
Northern analysis.
The total RNA of TA441 incubated in LB medium with testosterone or succinate (negative control) was purified every 2 h for 8 h. Northern analysis was carried out with the purified RNA by using ORF11 and ORF12 as probes.
Nucleotide sequence accession number.
The nucleotide sequence data reported here are listed in the DDBJ/EMBL/GenBank nucleotide sequence databases under accession number AB063482.
RESULTS
Isolation of TA441 mutants showing limited testosterone degradation.
In a previous study, we isolated and characterized the meta-cleavage enzyme gene, tesB, and three downstream ORFs (ORF1, ORF2, and ORF3), all of which are necessary for testosterone degradation in TA441 (9). Although the reaction of TesB was not confirmed because the proposed substrate, 3,4-DHSA (Fig. 1), was not available, TesB was considered to catalyze conversion of 3,4-DHSA to 4,9-DSHA (reaction 5 in Fig. 1) because a TesB-disrupted mutant showed little growth and accumulated 4AD, ADD, and a small amount of 3-HSA when grown with testosterone as the sole carbon source (9) (unpublished data). 4AD and ADD are intermediate compounds of testosterone degradation and are accumulated in most of the culture of gene-disrupted mutants incubated with testosterone (see Table 2). The functions of the three following ORFs were not clear. Since these genes were all necessary for testosterone degradation, we analyzed the gene region of ca. 20 kb containing tesB (unpublished data). In this 20-kb region, probable genes encoding expected enzymes, such as hydrolase for 4,9-DSHA, were not found. Thus, transposon mutagenesis was carried out in order to obtain Tn5 insertion mutants of TA441 with limited growth on testosterone. For the first screening of the target mutants, testosterone plates made turbid with testosterone were used. When TA441 grew on these plates, a clear zone formed around each growing colony due to the colony's utilization of testosterone. Of ca. 7,000 Kmr Tn5 mutants of TA441, ca. 30 showed a smaller clear zone on a testosterone plate than that formed by TA441. The growth of these mutants was examined on testosterone and p-hydroxybenzoate (carbon source used as positive control because TA441 shows unstable growth on commonly used carbon sources such as succinate) in liquid media, and mutants showing limited growth on testosterone but good growth on p-hydroxybenzoate were selected. Mutants which did not show good growth on p-hydroxybenzoate were not selected because they might contain a mutation in a vital gene. The selected mutants—T4, T9, T21, T26, T29, T42, T46, T47, T54, and T65—were studied further. The culture medium of T26 turned intensely yellow when it was incubated with testosterone. As the product of meta-cleavage reaction is well known for its intense yellow color, we considered that the yellow color of the T26 culture indicated the accumulation of 4,9-DSHA, the probable product of the meta-cleavage reaction in testosterone degradation of C. testosteroni (Fig. 1). Since the accumulation of the product of meta-cleavage reaction suggested that T26 had a mutation in the gene encoding the hydrolase for the product of meta-cleavage reaction, isolation of the gene region around the inserted Tn5 in T26 was carried out.
TABLE 2.
HPLC analysis of the culture media of the gene-disrupted mutants incubated with testosterone in C medium for 24 ha
| Compound | RT (min) | Relative peak area (%) of accumulated intermediate compounds with:
|
||||||
|---|---|---|---|---|---|---|---|---|
| TA441 | TesB− | ORF11− | ORF12− | TesD− | TesE− | TesD−c | ||
| X5 | 4.1 | NDb | ND | ND | ND | 100 | 45 | 156 |
| X1 | 6.5 | ND | 0.56 | 3.9 | 28 | 23 | 0.56 | 12.3 |
| ADD | 7.4 | ND | 3.3 | 31 | 13.9 | 23 | 0.56 | 2.8 |
| Tsd | 8.2 | ND | 20 | 2.8 | 3.9 | 3.9 | 2.2 | 0.56 |
| 4AD | 9.5 | ND | 10.6 | 2.8 | 8.4 | 2.8 | ND | 0.56 |
Data are presented with relative peak area (X5 in strain TesD− as 100%) of each compound detected at 210 nm.
ND, not detected.
Grown in LB+C medium with 0.1% testosterone.
Ts, testosterone.
Isolation and sequencing of the gene segment containing the hydrolase gene for the product of meta-cleavage reaction in testosterone degradation.
The total DNA of T26 was digested with SalI and ligated into pUC19, and E. coli was transformed with these plasmids. Since the Kmr gene of Tn5 has one SalI site just next to the terminal codon, the E. coli transformant carrying a plasmid containing Tn5 with the flanking gene region of T26 total DNA shows Kmr. Thus, we selected the transformant carrying the plasmid containing the T26 DNA in which Tn5 was inserted by using Kmr as a marker. A part of the cloned T26 DNA was sequenced to reveal that the sequenced region contained a putative ORF whose deduced amino acid sequence had homology with some hydrolases for the product of the meta-cleavage reaction in bacterial aromatic compound degradation.
Using a part of the cloned T26 DNA encoding the probable hydrolase as a probe, two plasmids, pCos26 and pCos29 carrying a part of TA441 DNA, were obtained by colony hybridization. These plasmids are SuperCosI (Stratagene) derivatives containing ca. 30 kb of TA441 DNA each. Together with a probe designed from T26 DNA, a probe of T29 also hybridized to pCos29. From pCos29, a pUC19 derivative plasmid, pC29E3, carrying a 4.7-kb EcoRI fragment containing the probable hydrolase gene, was constructed and sequenced. Analysis of the nucleotide sequence of the 4.7-kb EcoRI fragment revealed four ORFs (ORF11, ORF12, ORF13, and ORF14) (Fig. 2). ORF11 is encoded in the direction opposite to that of ORFs 12 to 14. The result of the sequence analysis showed that Tn5 was inserted into ORF13 in mutant strain T26 and into ORF12 in mutant strain T29. Since the cloned TA441 DNA ended soon after the end of ORF14 in pCos29, another pUC derivative plasmid pC26ScH, carrying a 4.2-kb SacI- HindIII fragment of TA441 DNA, was constructed from pCos26 (Fig. 2). In this region, two ORFs, ORF15 and ORF16, were identified just downstream of ORF14. A putative terminator was found just downstream of ORF16, and putative promoters were located in the gene regions upstream of ORF11 and 12 (Fig. 2). The deduced N-terminal amino acid sequences of ORF13 (MSQFSIPEGKYVDIGNVAGHPQRV), ORF14 (MSDKQFIETQGQRLYEALRSARTLAPLTD), and ORF16 (MSLKGKKITVHDMTLRDGMHP) were almost the same as the partial N-terminal sequences of TIP5 (MNIFSIPEGKYVDIGNVAG-PQRV), TIP8 (SDKQFIEQQGQRLYDALRTARTLAPLTD), and TIP4 (SLKEKKITVHDMTLRDGMHP) of C. testosteroni ATCC 11996, respectively (12). TIP5, TIP8, and TIP4 are testosterone-inducible proteins detected by two-dimensional gel electrophoresis analysis, and the partial N-terminal sequences are determined from the proteins purified from the gel (12). The deduced amino acid sequence of ORF13 showed the most similarity with the hydrolases involved in degradation of aromatic compounds. Since it showed the greatest identity, ca. 30 to 40%, with the hydrolases of biphenyl degradation in Pseudomonas, etc. (BphDs), ORF13 is named tesD. TesD showed greater homology with the BphDs than with the family of hydrolases involved in degradation of phenol derivatives but was not included in the group of BphDs in the phylogenetic tree. The deduced amino acid sequences of ORF14, ORF15, and ORF16 showed ca. 61, 83, and 85% identity with BphH, BphI, and BphJ of Pseudomonas sp. strain LB400, respectively, and were named tesE, tesF, and tesG, respectively. ORF11 and ORF12 were located upstream of tesD (Fig. 2). The deduced amino acid sequence of ORF11 showed homology with some hydroxylases, the maximum homology being ca. 33% with a hydroxylase of unknown substrate that is encoded by ORF11 in Rhodococcus erythropolis TA421 (10). ORF12 shows homology with the small component of two component hydroxylases, such as the nitrilotriacetate monooxygenase component B of Chelatobacter heintzii ATCC 29600 (ca. 30% identity) (19). ORF11 to tesG was not included in the gene region of ca. 20 kb containing tesB.
Growth of tesD, tesE, ORF11, and ORF12-disrupted mutants on testosterone.
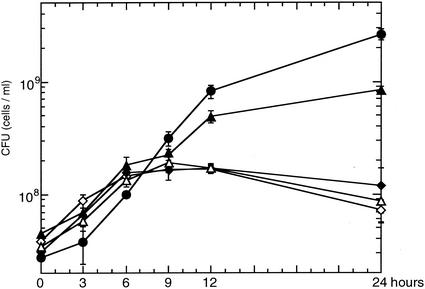
tesD, tesE, ORF11, and ORF12 in TA441 were individually disrupted with a Kmr gene by homologous recombination, and the resultant mutant strains were designated TesD−, TesE−, ORF11−, and ORF12−. Figure 3 shows comparison of the growth of each mutant strain and TA441 on testosterone as the sole carbon source. The growth of TesD−, ORF11−, and ORF12− on testosterone as the sole carbon source was limited, indicating that the associated genes are necessary for testosterone degradation in TA441. The growth of TesE− was lower than TA441 but far better than that of the other three mutants. The culture of TesD− showed a brilliant yellow color, and those of TA441 and TesE− showed a pale yellow color. TA441 showed the pale yellow color only ca. 9 to 12 h after the start of the incubation, and then the color disappeared gradually. The color implied temporary accumulation of the product of meta-cleavage reaction in testosterone degradation.
FIG. 3.
Growth of TA441 and gene disrupted mutants with testosterone as the sole carbon source. The genes were disrupted by insertion of a Kmr gene. Each strain was grown in 10 ml of C medium supplemented with 0.1% (wt/vol) testosterone (TA441, •; TesD-disrupted mutant, ▵; TesE-disrupted mutant, ▴; ORF11-disrupted mutant, ◊; ORF12-disrupted mutant, ⧫). The growth is represented by CFU. The data are an average of more than three experiments.
HPLC analysis of the culture media of the mutants.
The gene-disrupted mutant strains, TesB− (meta-cleavage enzyme gene, tesB, disrupted mutant described previously) (9), ORF11−, ORF12−, TesD−, TesE−, and TA441 were incubated in C medium with testosterone, and the culture media after 24 h were analyzed by HPLC. Table 2 shows the major compounds detected at 210 nm by HPLC in each culture. In the TesD− culture a compound, named X5, characteristically accumulated, indicating that this compound might be 4,9-DSHA that gives the yellow color to the TesD− culture. Before we isolated and purified compound X5, we examined several kinds of culture media in which TesD− accumulated more X5 than in C medium. These experiments showed that TesD− incubated in a mixed LB+C medium with testosterone accumulated more X5 than in C medium (Table 2). Thus, this medium was used for following studies when we needed to produce an accumulation of X5. The result also indicated that the induction of testosterone degradation genes in TA441 was not inhibited by any nutrients contained in LB medium.
Isolation and purification of the intermediate compounds in the culture of the TesD-disrupted mutant incubated with testosterone.
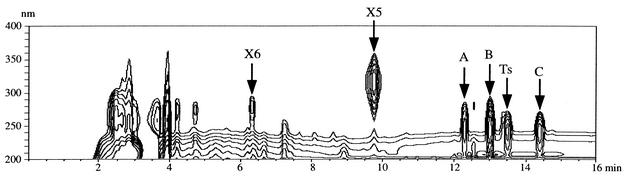
TesD. was grown in 400 ml of LB+C medium with 0.1% testosterone and incubated at 30°C for about 2 days until it showed an intense yellow color. The three-dimensional HPLC analysis of TesD− culture is shown in Fig. 4. Peaks were detected at retention time (RT) values of 6.4 min (compound X6), 9.7 min (compound X5), 12.3 min (compound A), 13.0 min (compound B), 13.4 min (testosterone), and 14.4 min (compound C). In the culture of TA441 incubated in the same manner, only a trace amount of testosterone was detected. Thus, the compounds X5, X6, A, B, and C accumulated by TesD− were considered to be the intermediate compounds in the testosterone degradation pathway.
FIG. 4.
Three-dimensional HPLC analysis of the culture of TesD-disrupted mutant incubated with testosterone. The vertical axis indicates wavelength, the horizontal axis indicates the RT, and the UV absorbance of each compound is represented in contour.
After the incubation, the culture was extracted twice with the same volume of ethyl acetate. Testosterone and compounds A, B, and C were extracted to the ethyl acetate fraction. The ethyl acetate fraction was dried, concentrated, and dissolved in a small amount of methanol. The compounds were separated by HPLC, and the fraction containing each compound was collected from the eluent. After extraction under neutral conditions, the aqueous fraction was acidified with HCl and then extracted twice with the same volume of ethyl acetate. Compounds X5 and X6 were extracted to this ethyl acetate fraction. The fraction was dried, and the residue was dissolved in 1 ml of methanol and loaded onto the HPLC column, and the fractions containing X5 and X6 were collected from the eluent. However, after the fraction containing X5 was dried, the residue became an oily material with a dark brown color, which was quite different from the first residue after the fraction that was extracted under acidic conditions was dried. HPLC analysis showed that the most of isolated X5 had broken down into unknown compounds, suggesting that purified X5 is unstable. Thus, the eluent from the HPLC containing X5 was prepared as described above, kept in the refrigerator, and subjected directly to FAB-MS and UV analyses. The X5 in the eluent gave a molecular ion at m/z 349.1661 (M+H)+, with FAB-MS analysis indicating its molecular formula to be C19H24O6. X5 showed an absorbance at 317 nm in acidic methanol solution (0.01 N HCl-MeOH). The absorbance shifted to 399 nm in alkaline methanol solution (0.01 N NaOH-MeOH). These typical UV/vis absorptions suggested the presence of a 2-hydroxy-6-oxo-2,4-dienoate structure as a chromophore. The fraction containing X6 was dried, and oily pale yellow residue was isolated. X6 was stable and successfully purified.
Isolation of the methyl ester of X5.
For the methyl ester of X5 (i.e., X5-Me), preexperiments showed that X5-Me was fairly stable and could be purified successfully. Thus, the isolation and purification of X5-Me was carried out. An 800-ml TesD− culture was prepared and extracted in the same manner as described previously. The acidic ethyl acetate fraction was concentrated in vacuo, and the residue was dissolved in a solvent mixture of 2 ml of methanol and 3 ml of benzene, followed by the treatment with trimethylsilyldiazomethane. The reaction solution was dried, and the brown residue was dissolved in 1 ml of methanol. The solution was centrifuged to remove the precipitate, and the supernatant was loaded onto the HPLC. The fraction containing X5-Me was collected and dried, and 22 mg of oily pale yellow material was obtained. The methyl ester of X6 (X6-Me) was also detected by HPLC and purified from the eluent.
Identification of X5-Me.
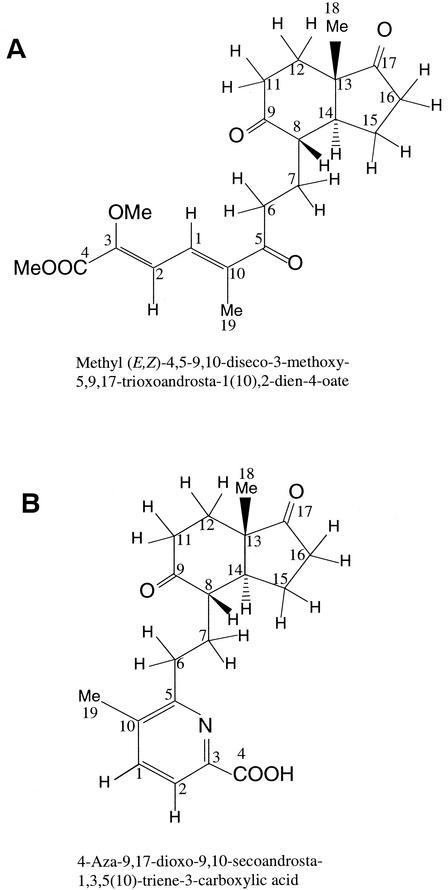
The molecular formula of X5-Me was determined to be C21H28O6 by high-resolution FAB-MS data [m/z 377.1929 (M+H)+]. In the UV spectrum, a maximum absorption was observed at 300 nm, which did not shift in either acidic (0.01 N HCl) or alkaline (0.01 N NaOH) conditions. The 13C NMR spectrum confirmed the presence of 21 carbons, including three characteristic ketones, one ester carbonyl, two sp2 quaternary carbons, two sp2 methine, and two methoxy carbons. The PFG-HMQC spectrum established all single-bond 1H-13C correlations, and PFG-DQFCOSY spectral data suggested the presence of C- and D-ring portions and two methylene units at C6 and C7 with three ketone groups at C5, C9, and C17 for the 9,10-secoandrostane skeleton. The positions of quaternary carbons including ketone carbonyl carbons were determined by analyses of 1H-13C long-range correlations of PFG-HMBC spectral data. PFG-HMBC data was also useful in analysis of the chromophore portion. Important long-range correlations were observed from olefinic methyl protons (Me-19) at 1.95 ppm to the C5 ketone carbonyl carbon at 202.2 ppm, the quaternary carbon C10 at 139.8 ppm, and the methine carbon C1 at 130.3 ppm. Two olefinic methine protons at 7.47 (H1) and 6.95 (H2) ppm were correlated in PFG-DQFCOSY spectrum, with a large spin coupling constant of 11.5 Hz. Two methoxy signals are observed at 3.86 and 3.85 ppm with long-range coupling to the quaternary sp2 carbon C3 at 149.4 ppm and the ester carbonyl carbon C4 at 163.9 ppm, respectively. The complete NMR assignments and long-range correlations for X5-Me are summarized in Table 3. The stereochemistry around the two double bonds was determined by NOE data. In the NOE differential spectrum, NOE were observed from H1 to the methyl protons of the enol ether methoxy group at 3.86 ppm, suggesting that the C2-C3 double bond had a Z configuration. The stereochemistry of C1-C10 double bond was determined to be an E configuration from NOE data between H2 and Me-19 and between H1 and Hs-6. Based on these spectral data the structure of X5-Me was determined to be methyl (1(10)E,2Z)-4,5-9,10-diseco-3-methoxy-5,9,17-trioxoandrosta-1(10),2-dien-4-oate (Fig. 5A).
TABLE 3.
NMR data for X5-Me and X6-Mea
| No. | X5-Me
|
X6-Me
|
|||
|---|---|---|---|---|---|
| 13C-NMR (δ [ppm]) | 1H-NMR (δ [ppm]), J (Hz)b | HMBC | 13C-NMR (δ [ppm]) | 1H-NMR (δ [ppm]), J (Hz)b | |
| 1 | 130.3 | 7.47 dq (11.5, 0.9) | C3, C5, C19 | 138.3 | 7.55 d (7.8) |
| 2 | 119.0 | 6.95 d (11.5) | C3, C4, C10 | 123.0 | 7.89 d (7.8) |
| 3 | 149.4 | 145.0 | |||
| 4 | 163.6 | 166.1 | |||
| 5 | 202.2 | 160.7 | |||
| 6 | 35.4 | 3.01 ddd (17.5, 6.9, 6.9) | C5, C7, C8 | 33.2 | 3.10 ddd (14.2, 8.8, 6.4) |
| 2.82 ddd (17.5, 7.3, 7.3) | C5, C7, C8 | 2.84 ddd (14.2, 9.3, 7.3) | |||
| 7 | 21.3 | 1.85 m | C5 | 24.8 | 1.93 m |
| 8 | 49.2 | 2.55 m | 49.4 | 2.69 ddd (11.7, 7.3, 3.9) | |
| 9 | 211.5 | 211.2 | |||
| 10 | 139.8 | 136.0 | |||
| 11 | 37.5 | 2.50 m | C9, C12, C13 | 37.5 | 2.50 ddd (15.6, 13.2, 6.4) |
| 2.44 m | C9, C12, C13 | 2.42 ddd (15.6, 6.4, 2.4) | |||
| 12 | 30.4 | 2.01 ddd (13.3, 6.0, 2.3) | 30.6 | 2.00 ddd (13.2, 6.4, 2.4) | |
| 1.68 ddd (13.3, 12.8, 6.4) | C18 | 1.68 ddd (13.2, 13.2, 6.4) | |||
| 13 | 47.6 | 47.6 | |||
| 14 | 49.6 | 1.84 m | 49.6 | 1.89 m | |
| 15 | 22.2 | 2.10 m | C17 | 22.3 | 2.10 m |
| 1.85 m | 1.85 m | ||||
| 16 | 36.1 | 2.59 dd (19.3, 7.8) | C17 | 36.2 | 2.58 dd (19.1, 8.3) |
| 2.24 ddd (19.3, 9.2, 9.2) | C17 | 2.23 ddd (19.1, 9.3, 8.8) | |||
| 17 | 218.7 | 218.2 | |||
| 18 | 13.5 | 1.18 s | C12, C13, C14, C17 | 13.5 | 1.17 s |
| 19 | 12.2 | 1.95 d (0.9) | C1, C5, C10 | 19.0 | 2.47 s |
| 3-OMe | 60.8 | 3.86 s | C3 | ||
| COOMe | 52.4 | 3.85 s | C4 | 52.6 | 3.97 s |
The molecular formula and weight of X5-Me and X6-Me are C12H28O6 and 376 and C20H25NO4 and 343, respectively. In HMBC data, carbons showing cross peaks with the proton are in the same row.
Abbreviations for NMR: s, singlet; d, doublet; q, quartet, m, multiplet.
FIG. 5.
(A) Structure of compound X5-Me. (B) Structure of compound X6.
Identification of X6 and the methyl ester of X6.
X6 was another compound isolated from the acidic fraction of the testosterone metabolites. The molecular formula of X6 was determined to be C19H23NO4. 1D and 2D NMR spectral data, together with UV spectral data, suggested that X6 was 4-aza-9,10-seco-9,17-dioxoandrosta-1,3,5(10)-triene-3-carboxylic acid(Fig. 5B). The methyl ester produced from direct methylation of X6 was shown to be identical to X6-Me isolated from the trimethylsilyldiazomethane-treated acidic fraction of the testosterone metabolites. Complete NMR assignments and long-range correlations for X6-Me are summarized in Table 3.
Identification of the compounds A, B, and C.
Three accumulated neutral metabolites—compounds A, B, and C—were identified as 4AD, ADD, and 17β-hydroxyandrosta-1,4-dien-17-one by comparison of the spectral data with data reported for these compounds (a brief summary of the data is shown in Table 4) (8, 11, 14). These compounds are intermediates in the early stage of testosterone degradation by C. testosteroni (cf. Fig. 1).
TABLE 4.
NMR data for 17β-hydroxyandrosta-1,4-dien-17-one, 4AD, and ADDa
| No. | 17β-hydroxyandrosta-1,4-dien-17-one
|
4AD
|
ADD
|
|||
|---|---|---|---|---|---|---|
| 13C-NMR (δ [ppm]) | 1H-NMR (δ [ppm]), J (Hz)a | 13C-NMR (δ [ppm]) | 1H-NMR (δ [ppm]), J (Hz)a | 13C-NMR (δ [ppm]) | 1H-NMR (δ [ppm]), J (Hz)a | |
| 1 | 157.2 | 7.14 d (10.1) | 35.7 | 2.05 ddd (13.3, 4.6, 3.2) | 155.3 | 7.06 d (10.2) |
| 1.70 m | ||||||
| 2 | 127.1 | 6.30 dd (10.1, 1.8) | 33.9 | 2.44 m | 127.7 | 6.25 dd (10.2, 2.0) |
| 2.34 m | ||||||
| 3 | 187.1 | 199.2 | 186.2 | |||
| 4 | 123.5 | 6.14 br.s | 124.1 | 5.76 s | 124.1 | 6.10 dd (2.0, 1.5) |
| 5 | 170.9 | 170.2 | 168.3 | |||
| 6 | 32.9 | 2.49 br.ddd (12.8, 12.8, 5.0) | 32.5 | 2.44 m | 32.5 | 2.52 dddd (13.7, 11.1, 5.4, 1.0) |
| 2.39 ddd (12.8, 4.1, 2.8) | 2.32 m | 2.44 ddd (13.7, 4.9, 2.9) | ||||
| 7 | 33.2 | 1.96 m | 30.8 | 1.98 m | 32.3 | 2.08 m |
| 1.03 m | 1.13 m | 1.13 m | ||||
| 8 | 35.5 | 1.68 m | 35.2 | 1.72 m | 35.1 | 1.83 m |
| 9 | 52.5 | 1.14 m | 53.9 | 1.00 ddd (12.4, 10.5, 4.1) | 52.3 | 1.12 m |
| 10 | 44.0 | 38.6 | 43.4 | |||
| 11 | 22.6 | 1.76 m | 20.3 | 1.72 m | 22.1 | 1.87 m |
| 1.70 m | 1.46 dddd (12.8, 12.8, 12.4, 4.1) | 1.70 m | ||||
| 12 | 36.2 | 1.88 m | 31.3 | 1.87 ddd (13.3, 4.1, 2.7) | 31.2 | 1.89 m |
| 1.10 m | 1.28 m | 1.30 m | ||||
| 13 | 43.1 | 47.5 | 47.7 | |||
| 14 | 50.0 | 0.96 m | 50.8 | 1.30 m | 50.4 | 1.28 m |
| 15 | 23.5 | 1.62 m | 21.7 | 1.98 m | 21.9 | 1.97 br.ddd (12.7, 8.8, 5.9) |
| 1.34 m | 1.58 dddd (12.4, 12.4, 9.2, 9.2) | 1.60 dddd (12.7, 12.7, 9.3, 8.8) | ||||
| 16 | 30.2 | 2.08 m | 35.7 | 2.48 dd (19.2, 9.2) | 35.6 | 2.48 dd (19.5, 8.8) |
| 1.48 m | 2.11 ddd (19.2, 9.2, 9.2) | 2.10 ddd (19.5, 9.3, 8.8) | ||||
| 17 | 81.5 | 3.66 dd (8.7, 8.3) | 220.2 | 219.9 | ||
| 18 | 11.1 | 0.83 s | 13.7 | 0.92 s | 13.8 | 0.95 s |
| 19 | 18.6 | 1.26 s | 17.4 | 1.22 s | 18.7 | 1.27 s |
br, broad. See Table 3, footnote a, for definitions of other terms used in this table.
Conversion of 4,9-DSHA by TesD.
TesD was expressed in E. coli, and its activity in degrading 4,9-DSHA was examined. The TesD-disrupted mutant TesD− was incubated in LB+C medium with testosterone, and the cells were removed by centrifugation. The yellow supernatant was collected and used as the reaction solution for TesD activity because purification of sufficient 4,9-DHSA was quite difficult and 4,9-DHSA is not commercially available. TesD or MhpC, the hydrolase of TA441 in 3-(3-hydroxyphenyl) propionic acid degradation (used as a negative control; the normal substrate of MhpC is 2-hydroxy-6-keto-nona-2,4-diene-1,9-dioic acid) (4), was expressed in E. coli. The cells were disrupted by sonication, and the resultant cell extract was added to the reaction solution. Figure 6a shows the three-dimensional HPLC chart of the reaction solution at the beginning, and Fig. 6b, c, and d show the HPLC runs at 15, 30, and 60 min, respectively, after the addition of TesD. Figure 6e and f are negative controls after 60 min, with MhpC and no enzymes added, respectively. In the TesD− culture, accumulation of 4,9-DHSA was detected at an RT of ca. 11.8 min. The 4,9-DHSA disappeared after the addition of TesD, and a probable product appeared at an RT of ca. 10.2 min (in Fig. 6b, c, and d). After 60 min, the reaction solution with TesD was no longer yellow. No degradation of 4,9-DHSA and accumulation of the probable product were observed in either negative controls with MhpC (Fig. 6e) or with no enzyme (Fig. 6f).
Conversion of the product from 4,9-DSHA by TesE.
The supernatant of the TesD− culture was treated with E. coli expressed TesD as mentioned in the previous section, and the resultant solution was used as the reaction solution for TesE. A cell extract of E. coli expressing TesE was prepared in the same manner as that used to prepare a cell extract of TesD. Figure 7a shows the three-dimensional HPLC chart of TesD− culture and Fig. 7b shows the culture after treatment with TesD in which 4,9-DSHA was converted to the probable product (RT differs a little from that in Fig. 6 as we used a column of a different size). Figure 7c and d show results at 5 and 10 min after the addition of TesE, respectively, in which the product disappeared. Little degradation was observed without TesE in 30 min (Fig. 7f, the negative control).
Induction of the testosterone degradation genes in TA441 during growth on testosterone.
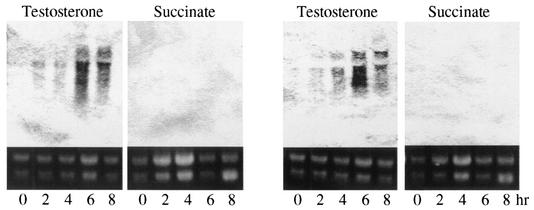
The total RNA of TA441 incubated in LB medium with testosterone or succinate (negative control) was purified every 2 h for 8 h and subjected to Northern analysis. LB medium, which did not inhibit the induction of testosterone degradation genes in TA441 (Table 2), was used to obtain almost the same volume of cells from cultures of both carbon sources. Northern analysis was carried out with the purified RNA by using ORF11 and ORF12 as probes. ORF11 and ORF12 were induced 2 h after the start of the incubation and showed the maximum induction at ca. 6 h (Fig. 8). No induction was observed in the culture incubated with succinate.
FIG. 8.
Induction of isolated testosterone degradation genes in TA441 incubated with testosterone. The total RNA was purified every 2 h and analyzed by Northern hybridization. Succinate was used as a negative control. The probes used were ORF12 (left panels) and ORF11 (right panels).
DISCUSSION
The meta-cleavage enzyme gene, tesB, reported previously, is necessary for testosterone degradation in C. testosteroni TA441 (9) and probably converts 3,4-DHSA to 4,9-DSHA (reaction 5 in Fig. 1). tesB is followed by three ORFs—ORF1, ORF2, and ORF3—all of which are necessary for testosterone degradation in TA441. In the present study, by using Tn5 mutants of TA441 with limited testosterone degradation ability, the hydrolase gene for 4,9-DSHA, tesD, was isolated with ORF11, 12, and tesEFG, all of which are involved in testosterone degradation. The tesD-disrupted mutant produced an accumulation of a characteristic yellow compound when it was incubated with testosterone. The product of meta-cleavage reaction in the testosterone degradation by N. restrictus was proposed to be 4,9-DSHA (7, 15, 17). In these earlier reports, the purification of 4,9-DSHA was not successful because 4,9-DSHA is quite unstable. They proposed the 4,9-DSHA structure based on the following two results: treatment of the probable 4,9-DSHA with diazomethane gave a mono methyl ester, which gave a characteristic singlet signal in 1H NMR spectrum, and treatment with ammonia gave 4-aza-9,17-dioxo-9,10-secoandrosta-1,3,5(10)-triene-3-carboxylic acid (4,9-DSHA-N) as its pyridine derivative. A TesD-disrupted mutant accumulated five major intermediates, designated X5, X6, A, B, and C. X5 showed an intense yellow color under neutral conditions. To clarify the structure and chemical properties of X5, we purified X5 (the eluent from the HPLC containing X5) for mass spectrometry and UV measurements and prepared the methylation derivative, which has the structure with a methyl enol ether on the C3-hydroxy group and a methyl ester on the C4 carboxyl group. The direct methylation product from X5 in the eluent and X5-Me isolated from culture filtrate after trimethylsilyldiazomethane treatment were completely identical (data not shown). X5-Me was determined to be methyl (1(10)E,2Z)-4,5-9,10-diseco-3-methoxy-5,9,17-trioxoandrosta-1(10),2-dien-4-oate. These data directly show that X5 is the yellow principle, i.e., 4,9-DSHA. The methyl enol at C3 and the stereochemistry of the two double bonds 1(10)E and 2Z indicate that the main tautomer of 4,9-DSHA is the free form of X5-Me. However, 4,9-DSHA presumably exists in the culture as the keto-enol tautomeric mixture with stereoisomerism around the double bonds because the mono methyl derivative prepared with diazomethane by Sih et al. in 1965 (17), which may be methyl 4,5-9,10-diseco-3-hydroxy-5,9,17-trioxoandrosta-1(10),2-diene-4-oate, is a complex mixture of keto-enol forms. Our report represents the first complete purification and identification of methyl 4,5-9,10-diseco-3-methoxy-5,9,17-trioxoandrosta-1(10),2-dien-4-oate. Moreover, our results include more direct evidence for the yellow principle of 4,9-DSHA than preparation of pyridine derivative by treatment with ammonia through multistep reactions. The analysis of X6, another compound accumulated by the TesD-disrupted mutant, by NMR and mass spectrometry revealed X6 to be 4,9-DSHA-N, which has the same partial structure as that of 4,9-DSHA except for the A-ring portion. 4,9-DSHA-N was reported by Sih et al. and Gibson et al. as the pyridine derivative of 4,9-DSHA isolated from the culture treated with ammonia (7, 15, 17). In the present study, we did not add ammonia for the purpose of producing the pyridine derivatives, but the culture media contained ammonium sulfate as a nitrogen source and the ammonium ion might produce 4,9-DSHA-N nonenzymatically. The other three compounds detected in the culture medium of the TesD-disrupted mutant, compounds A, B, and C, were identified as 17β-hydroxyandrosta-1,4-dien-3-one, 4AD, and ADD, respectively. 4AD is an intermediate compound produced from testosterone by initial 17β-dehydroxylation and ADD is produced by the following Δ1-dehydroxylation. 17β-Hydroxyandrosta-1,4-dien-3-one can be produced by Δ1-dehydroxylation of testosterone and will be one of the intermediates in the testosterone degradation. 17β-Hydroxyandrosta-1,4-dien-3-one corresponds to a previously reported “unknown compound” accumulated by the gene-disrupted mutant of tesB, ORF1, ORF2, and ORF3 and detected by thin-layer chromatography analysis (9). Accumulation of 4AD, ADD, and 17β-hydroxyandrosta-1,4-dien-3-one is often observed in the culture of gene-disrupted mutants of testosterone degradation genes (see Table 2). Gibson et al. proposed 1,3-dihydroxy-4,5-9,10-diseco-5,9,17-trioxoandrostan-4-oic acid as an intermediate in the testosterone degradation pathway by N. restrictus (7) (Fig. 1), but it was not identified in the culture of TesD-disrupted mutant in our current study.
4,9-DSHA was converted by E. coli expressed TesD into a probable product, which has yet to be identified. From these results, TesD is concluded to be the hydrolase for 4,9-DSHA in testosterone degradation by C. testosteroni TA441. Identification of the probable product of TesD is now under way, and at present, the product seems to be 2-hydroxyhexa-2,4-dienoic acid and/or its isomer(s). This compound was degraded by E. coli expressed TesE, which is encoded just next to TesD and has ca. 60% identity with BphH. BphH is the hydratase for conversion of 2-oxopent-4-enoic acid into 4-hydroxy-2-oxovaleric acid in biphenyl degradation by Pseudomonas etc. From these, TesE is considered to be a hydratase for 2-oxohex-4-enoic acid. The results of the growth experiments also support this proposed function of TesE; a TesE-disrupted mutant showed growth on testosterone but lower than that of TA441, probably because the mutant was not able to utilize 2-oxohex-4-enoic acid and only grew on the other of the two products of TesD, which was proposed as 9,17-dioxo-1,2,3,4,10,19-hexanorandrostan-5-oic acid by Gibson et al. in 1966. From these, we propose the testosterone degradation pathway of TA441 as presented in Fig. 9. Testosterone undergoes aromatization of A-ring and the resultant 3-HSA is hydroxylated to produce 3,4-DHSA, which is cleaved by meta-cleavage enzyme TesB. The product, 4,9-DSHA, which would exist as a keto-enol tautomeric mixture, is degraded into 9,17-dioxo-1,2,3,4,10,19-hexanorandrostan-5-oic acid and 2-hydroxyhexa-2,4-dienoic acid by TesD. 2-Hydroxyhexa-2,4-dienoic acid would be degraded by TesEFG.
FIG. 9.
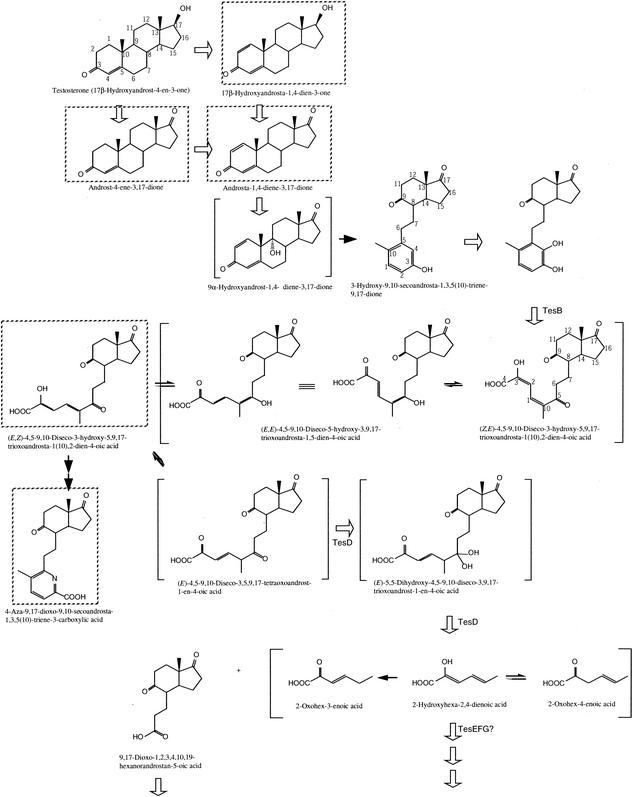
Proposed testosterone degradation pathway in C. testosteroni TA441. Boxed compounds are identified from the culture of TA441 and/or gene-disrupted mutant of TA441. Open arrows indicate the reactions that will be catalyzed by enzymes produced by TA441. Closed larger arrows indicate nonenzymatic reactions, and narrower arrows indicate that compounds on both sides of the arrow are tautomers.
TesB and TesD shared more identity (maximum ca. 40%) with BphCs and BphDs, respectively, the corresponding enzymes for biphenyl degradation. However, in the phylogenetic tree, neither of them was included in a group of Bphs. It is likely, therefore, that steroid degradation genes are a new family of aromatic compound degradation genes, but steroid degradation genes from other genera of bacteria need to be isolated to confirm this. ORF11 and ORF12, which encode probable hydroxylases and are located upstream of tesD, are also necessary and induced in TA441 grow on testosterone. But no suggestion is given about probable substrates by the similarities. The ORF12-disrupted mutant produced an accumulation of a significant amount of an unknown compound X1, whose identification study is under way. We are simultaneously identifying genes and intermediate compounds that are accumulated by the gene-disrupted mutants so as to clarify the whole degradation pathway of testosterone by C. testosteroni.
Acknowledgments
We thank Hideaki Kakeya (RIKEN) for useful discussions and Tamiko Chijimatsu (RIKEN) for technical assistance. We thank Y. Ichikawa and R. Nakazawa (Biodesign DNA Sequence Facility, RIKEN) for determination of the nucleotide sequence.
This work was partly supported by a grant from the Eco Molecular Sciences Research Program of RIKEN. M.H. was supported by a grant from the Special Postdoctoral Researchers Program of RIKEN.
REFERENCES
- 1.Abalain, J. H., S. Di Stefano, Y. Amet, E. Quemener, M. L. Abalain-Colloc, and H. H. Floch. 1993. Cloning, DNA sequencing, and expression of (3-17)β hydroxysteroid dehydrogenase from Pseudomonas testosteroni. J. Steroid Biochem. Mol. Biol. 44:133-139. [DOI] [PubMed] [Google Scholar]
- 2.Arai, H., S. Akahira, T. Ohishi, M. Maeda, and T. Kudo. 1998. Adaptation of Comamonas testosteroni TA441 to utilize phenol: organization and regulation of the genes involved in phenol degradation. Microbiology 144:2895-2903. [DOI] [PubMed]
- 3.Arai, H., T. Yamamoto, T. Ohishi, T. Shimizu, T. Nakata, and T. Kudo. 1999. Genetic organization and characteristics of the 3-(3-hydroxyphenyl)propionic acid degradation pathway of Comamonas testosteroni TA441. Microbiology 145:2813-2820. [DOI] [PubMed] [Google Scholar]
- 4.Coulter, A. W., and P. Talalay. 1968. Studies on the microbial degradation of steroid ring A. J. Biol. Chem. 243:3238-3247. [PubMed] [Google Scholar]
- 5.Florin, C., T. Kohler, M. Grandguillot, and P. Plesiat. 1996. Comamonas testosteroni 3-ketosteroid-delta 4(5 alpha)-dehydrogenase: gene and protein characterization. J. Bacteriol. 178:3322-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Genti-Raimondi, S., M. E. Tolmasky, L. C. Patrito, A. Flury, and L. A. Actis. 1991. Molecular cloning and expression of the beta-hydroxysteroid dehydrogenase gene from Pseudomonas testosteroni. Gene 105:43-49. [DOI] [PubMed] [Google Scholar]
- 7.Gibson, D. T., K. C. Wang, C. J. Sih, and H. Whitlock, Jr. 1966. Mechanisms of steroid oxidation by microorganisms. IX. On the mechanism of ring A cleavage in the degradation of 9,10-seco steroids by microorganisms. J. Biol. Chem. 241:551-559. [PubMed] [Google Scholar]
- 8.Hickey, J. P., I. S. Butler, and G. Pouskouleli. 1980. Carbon-13 NMR spectra of some representative hormonal steroids. J. Magn. Reson. 38:501-508. [Google Scholar]
- 9.Horinouchi, M., T. Yamamoto, K. Taguchi, H. Arai, and T. Kudo. 2001. Meta-cleavage enzyme gene tesB is necessary for testosterone degradation in Comamonas testosteroni TA441. Microbiology 147:3367-3375. [DOI] [PubMed] [Google Scholar]
- 10.Kosono, S., M. Maeda, F. Fuji, H. Arai, and T. Kudo. 1997. Three of the seven bphC genes of Rhodococcus erythropolis TA421, isolated from a termite ecosystem, are located on an indigenous plasmid associated with biphenyl degradation. Appl. Environ. Microbiol. 63:3282-3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mahato, S. B., S. Banerjee, and S. Podder. 1988. Oxidative side chain and ring fission of pregnanes by Arthrobacter simplex. Biochem. J. 255:769-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Möbus, E., M. Jahn, R. Schmid, D. Jahn, and E. Maser. 1997. Testosterone-regulated expression of enzymes involved in steroid and aromatic hydrocarbon catabolism in Comamonas testosteroni. J. Bacteriol. 179:5951-5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Plesiat, P., M. Grandguillot, S. Harayama, S. Vragar, and Y. Michel-Briand. 1991. Cloning, sequencing, and expression of the Pseudomonas testosteroni gene encoding 3-oxosteroid Δ1-dehydrogenase. J. Bacteriol. 173:7219-7227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reich, H. J., M. Jautelat, M. T. Messe, F. J. Weiert, and J. D. Roberts. 1969. Nuclear magnetic resonance spectra of steroids. J. Am. Chem. Soc. 91:7445-7454. [Google Scholar]
- 15.Sih, C. J., S. S. Lee, Y. K. Tsong, and K. C. Wang. 1965. 3,4-Dihydroxy-9,10-secoandrosta-1,3,5(10)-triene-9,17-dione: an intermediate in the microbiological degradation of ring A of androst-4-ene-3,17-dione. J. Am. Chem. Soc. 87:1385-1386. [PubMed] [Google Scholar]
- 16.Sih, C. J., S. S. Lee, Y. Y. Tsong, and K. C. Wang. 1966. Mechanisms of steroid oxidation by microorganisms: 3,4-dihydroxy-9,10-secoandrosta-1,3,5(10)-triene-9,17-dione, an intermediate in the microbiological degradation of ring A of androst-4-ene-3,17-dione. J. Biol. Chem. 241:540-550. [PubMed] [Google Scholar]
- 17.Sih, C. J., K. C. Wang, D. T. Gibson, and H. W. J. Whitlock. 1965. On the mechanism of ring A cleavage in the degradation of 9,10-seco steroids by microorganisms. J. Am. Chem. Soc. 87:1386-1387. [DOI] [PubMed] [Google Scholar]
- 18.Simon, R. 1984. High frequency mobilization of gram-negative bacterial replicons by the in vitro constructed Tn5-Mob transposon. Mol. Gen. Genet. 196:413-420. [DOI] [PubMed] [Google Scholar]
- 19.Uetz, T., R. Schneider, M. Snozzi, and T. Egli. 1992. Purification and characterization of a two-component monooxygenase that hydroxylates nitrilotriacetate from “Chelatobacter” strain ATCC 29600. J. Bacteriol. 174:1179-1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vieira, J., and J. Messing. 1987. Production of single-stranded plasmid DNA. Methods Enzymol. 153:3-11. [DOI] [PubMed] [Google Scholar]