Abstract
Streptomyces coelicolor and Lemna minor were used as a model to study the modulation of bacterial gene expression during plant-streptomycete interactions. S. coelicolor was grown in minimal medium with and without L. minor fronds. Bacterial proteomes were analyzed by two-dimensional gel electrophoresis, and a comparison of the two culture conditions resulted in identification of 31 proteins that were induced or repressed by the presence of plant material. One-half of these proteins were identified by peptide mass fingerprinting by using matrix-assisted laser desorption ionization-time of flight mass spectrometry. The induced proteins were involved in energetic metabolism (glycolysis, pentose phosphate pathway, oxidative phosphorylation), protein synthesis, degradation of amino acids, alkenes, or cellulose, tellurite resistance, and growth under general physiological or oxidative stress conditions. The repressed proteins were proteins synthesized under starvation stress conditions. These results suggest that root exudates provide additional carbon sources to the bacteria and that physiological adaptations are required for efficient bacterial growth in the presence of plants.
Plant rhizospheres harbor diverse communities of microorganisms. It is generally assumed that rhizosphere microbes use compounds released by the plant roots as their major nutrient sources. This ability is the nutritional basis of rhizosphere competence (32). Streptomycetes are gram-positive bacteria that are frequently isolated from terrestrial plant rhizospheres (3, 15), but Wohl and McArthur (49) also showed that streptomycetes are associated with freshwater plants. Until now, few research efforts have been dedicated to the study of plant-streptomycete interactions, and most previous efforts have involved the common scab-inducing streptomycetes and their host plants (29). As several Streptomyces species have been shown to be effective biocontrol agents for plant diseases (10), studies on the interactions between saprophytic streptomycetes and plants need to be documented more.
We propose Streptomyces coelicolor and Lemna minor as an experimental model to study the modulation of bacterial gene expression during the interaction between a saprophytic streptomycete and a plant. Recent research in our laboratory has shown that L. minor, a small aquatic plant extensively used in bioremediation (38) and environmental studies (30), is colonized by various Streptomyces species (unpublished data). On the other hand, S. coelicolor is a common inhabitant of plant rhizospheres (11) that can be readily cocultivated in the presence of L. minor under sterile conditions. The fact that the S. coelicolor chromosome (6) has been fully sequenced is an important asset in proteomic studies.
Proteomics has become an integral part of gene expression analysis. The functional complement of genetic information can be analyzed by a combination of two-dimensional gel electrophoresis for the separation of complex mixtures of proteins and mass spectrometry for identification of proteins by tryptic peptide mass fingerprinting. Some bacterial proteins involved in different plant-bacterium interaction systems have been identified by using protein expression profiling (22, 34). In this study, we sought to identify modulated factors in the proteome of plasmid-free S. coelicolor M145 grown in the presence and in the absence of aseptic L. minor in a minimal culture medium. Cytoplasmic and secreted proteins of S. coelicolor were separated by two-dimensional electrophoresis. A comparison of the protein profiles obtained under the two culture conditions allowed identification of proteins that were induced or repressed by the presence of plant material. The results obtained might provide insight for elucidating the bacterial traits associated with rhizosphere competence.
MATERIALS AND METHODS
Culture media and growth conditions.
Aseptic L. minor, graciously provided by Gilles Grenier (Université de Sherbrooke), was propagated in 225 ml of modified Hoagland solution (4) at 20°C with a photon flux density of 180 μmol · m−2 · s−1 by using a 16-h photoperiod. S. coelicolor M145 was grown in 50 ml of J medium (25) for 24 h at 30°C. Bacteria were harvested by centrifugation (2,500 × g, 4°C), washed once in minimal MG medium (25), and resuspended in 25 ml of MG medium. One milliliter of the bacterial suspension was inoculated into two flasks containing 50 ml of MG medium, and the cultures were incubated for 24 h. Approximately 50 fronds of L. minor were then added to one of the cultures, and both cultures were incubated for an additional 24 h. The plants were removed from the induced culture with a sieve before proteins were isolated.
Protein isolation.
The S. coelicolor proteome from cultures with and without plants was obtained from two distinct fractions: (i) the soluble cytoplasmic proteins in the cell pellets and (ii) the secreted proteins present in the supernatants. For cytoplasmic protein extraction, bacteria were harvested by centrifugation, and the pellets were washed in sonication buffer (10 mM Tris-HCl, pH 7) and resuspended in 4 ml of sonication buffer. The suspensions were sonicated (Vibra-Cell; Sonics & Materials, Newtown, Conn.) four times during 1 min at 4°C and centrifuged to remove the cell debris. The protein concentrations in cytoplasmic and secreted fractions were measured by the method described by Bradford (7) by using the Bio-Rad protein assay reagent (Bio-Rad Laboratories, Hercules, Calif.). Proteins recovered from the cell pellets, as well as proteins from the supernatants, were precipitated with 5 volumes of acetone and resolubilized in rehydration buffer, which contained 8 M urea, 2% (wt/vol) 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 100 μl of IPG buffer (Amersham Pharmacia Biotech, Uppsala, Sweden) (pH 3 to 10), 0.3% (wt/vol) dithiothreitol, and 0.01% (wt/vol) bromophenol blue.
Two-dimensional gel electrophoresis.
Extracellular and soluble intracellular proteins were separated by two-dimensional gel electrophoresis. The first-dimension isoelectric focusing was performed in immobilized pH gradient (IPG) gel strips (pH 4 to 7 linear; 18 cm; Pharmacia, Peapack, N.J.). Reswelling of IPG gel strips was performed in a reswelling cassette (Pharmacia) overnight with rehydration buffer containing 500 μg of protein extract. An electrophoresis unit (LKB MultiPhor II; Pharmacia) equipped with a gradient power supply (EPS 3501XL; Pharmacia) was used to perform isoelectric focusing. Reswelled gels were loaded in a cooling bath and allowed to focus under a low-viscosity mineral oil (Dry Strip cover fluid; Pharmacia) at 20°C by using a four-stage ramped voltage program (2 min in a gradient from 0 to 200 V, 6 h at 200 V, 6 h in a gradient from 200 to 3,500 V, 36 h at 3,500 V). For the second-dimension sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE), IPG gels were equilibrated as described by Görg et al. (18). SDS-PAGE was performed as described by Laemmli (27) with a 12% polyacrylamide resolving gel and a 4% polyacrylamide stacking gel by using a Protean II unit (Bio-Rad Laboratories). The gels were electrophoresed at 45 mA/gel for 4 h. The molecular weights of the separated proteins were estimated by comparison with standards migrating alongside S. coelicolor proteins (SDS-PAGE standards, medium range; Bio-Rad Laboratories). Isoelectric points were estimated based on the linearity of the IPG strips.
Proteome analysis.
Two-dimensionally separated proteins were revealed with Coomassie brilliant blue R-250 (Sigma, St. Louis, Mo.) with a Hoeffer Processor Plus unit (Pharmacia) by using the method described by Wirth and Romano (48). Differential analysis of protein patterns was performed by using PhotoShop, version 5.0 (Adobe Systems, San Jose, Calif.), and Phoretix 2D Advanced, version 5.0 (NonLinear Dynamics, Durham, N.C.). Protein spots that were not detectable under one of the two conditions were excised from the gels.
In-gel digestion and tryptic peptide mass fingerprinting.
Excised protein spots were subjected to in-gel trypsin digestion. Briefly, excised spots were washed, reduced, S alkylated, and digested with trypsin (Promega, Madison, Wis.) as described by Williams et al. (47). Prior to analysis by mass spectrometry, the tryptic peptides were dissolved in 10 μl of 0.1% trifluoroacetic acid, desalted, and concentrated by using Zip Tip C18 (Millipore, Bedford, Mass.) as described by the manufacturer. A 1-μl aliquot of the desalted peptide extract was mixed with 1 μl of a saturated solution of α-cyano-4-hydroxycinnamic acid matrix (10 mg/ml) prepared in 0.1% trifluoroacetic acid-50% acetonitrile. The mixture was spotted onto the sample probe, and mass spectra of the tryptic peptide fragments were obtained by using a Voyager DE PRO matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometer (Applied Biosystems, Foster City, Calif.). Molecular masses obtained for the tryptic peptide profiles were used to search by peptide mass fingerprinting the National Center for Biotechnology Information (National Library of Medicine, Bethesda, Md.) databases by using the ProFound software (version 4.10.5; ProteoMetrics, Winnipeg, Canada). The defined search parameters were as follows: monoisotopic mass tolerance, 0.10 Da; singly protonated peptides (MH+); one missed cut allowed; cysteine as an S-carbamidomethyl derivative; and oxidation of methionine allowed.
RESULTS AND DISCUSSION
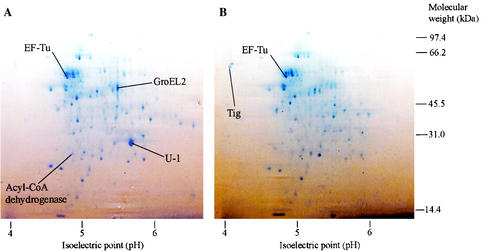
Approximately 400 cytoplasmic soluble protein spots per gel were revealed by Coomassie blue staining, while about 50 extracellular proteins were detected. In both cases, the majority of the visible proteins migrated to the acidic portion of the gel (between pH 4 and 5) during the first-dimension isoelectric focusing. The protein patterns were reproducible, and there were detectable visual intensity differences between the two culture conditions for close to 100 proteins. Protein spots that were undetectable under one of the two conditions in at least two independent experiments were selected for further analysis (Fig. 1). Thirty-two protein spots were excised from the gels and analyzed by MALDI-TOF mass spectrometry. Twenty-seven of these protein spots were from cytoplasmic fractions, and five spots were from the supernatants. Twenty-seven protein spots were induced in S. coelicolor by the presence of plant material, while four protein spots were repressed. The variation in protein spot intensities was not necessarily due to changes in transcriptional activity. Plant compounds could also affect bacterial translation, enzymatic modification of proteins, and protein degradation.
FIG. 1.
Two-dimensional gel electrophoresis of S. coelicolor cytoplasmic proteins grown in the presence (A) or in the absence (B) of L. minor fronds. The positions of some of the proteins identified in this study are indicated. U-1 is a protein spot that could not have been identified by MALDI-TOF mass spectroscopy.
Close to one-half of the proteins analyzed by MALDI-TOF mass spectrometry were identified with a high degree of confidence. The confidence level for the identification results was established by evaluating the probability that a protein identification hit in the National Center for Biotechnology Information databases corresponded to the protein being analyzed based on the ProFound probability score, the number of peptide mass matches, the percentage of sequence coverage, and the matched values for the isoelectric point and the molecular weight. High confidence levels for identification were not obtained for 15 good-quality mass spectra, probably because the corresponding proteins are not yet annotated in the database or because there was a mixture containing more than one protein in the excised spot. We could not exclude the possibility that some of the proteins were released by the plants in response to bacterial exposure, but no detectable amount of proteins was released by L. minor in the absence of S. coelicolor (data not shown).
Fifteen of the 16 high-confidence sequence matches were annotated in the S. coelicolor genome database, and the mass of the remaining sequence matched the predicted peptide mass derived from the actinomycete Streptomyces hygroscopicus. Table 1 shows a summary of the peptide mass fingerprinting results for the proteins identified. Bacterial proteins that were differentially expressed in the presence of plant material were associated with energy metabolism, carbon acquisition, and stress adaptations. The plant material that affected the proteome content was mainly composed of exudates since only a small part of the bacterial population was effectively attached to plant surfaces. Plant fronds were added to mid-log-phase bacterial cultures, but the amount of fronds used in this study was not sufficient to modify the bacterial growth curve.
TABLE 1.
Identification of proteins by peptide fingerprinting
| Protein designation | Localization | Level of expressiona | Mol wt (103) | Isoelectric point | Matching translated gene and corresponding proteinb | Protein accession no. | No. of matching peptides |
|---|---|---|---|---|---|---|---|
| Proteins involved in energetic metabolism or protein synthesis | |||||||
| NC-4 | Cytoplasmic | Constant | 50.1 | 5.4 | tuf1, protein synthesis elongation factor EF-Tu | 2021268A | 8 |
| NI-3 | Cytoplasmic | Induced | 95.5 | 5.3 | idh, isocitrate dehydrogenase | CAB88977 | 12 |
| NI-5 | Cytoplasmic | Induced | 58.9 | 5.2 | atpA, ATP synthase alpha chain | CAB94542 | 11 |
| NI-13 | Cytoplasmic | Induced | 35.0 | 6.1 | tsf, guanine-nucleotide exchange factor EF-Ts | 031213 | 8 |
| NI-14 | Cytoplasmic | Induced | 38.0 | 6.1 | fba, fructose biphosphate aldolase | Q9X8R6 | 8 |
| NI-22 | Cytoplasmic | Induced | 80.0 | 5.7 | tkt3, transketolase | T36007 | 6 |
| Proteins involved in the acquisition of carbon | |||||||
| NI-19 | Cytoplasmic | Induced | 31.0 | 5.7 | SC1G7.03, putative epoxide hydrolasec | CAC37878 | 3 |
| NI-20 | Cytoplasmic | Induced | 28.5 | 4.8 | fkb1, acyl-CoA dehydrogenase | AAF86388 | 8 |
| NI-21 | Cytoplasmic | Induced | 46.0 | 4.9 | SC4C6.23, putative transcription regulator (cellulose degradation)d | T35031 | 3 |
| Proteins involved in growth under stress conditions | |||||||
| NI-7 | Cytoplasmic | Induced | 55.0 | 4.8 | groEL2, stress-induced chaperonin | CAB93056 | 9 |
| NI-31 | Cytoplasmic | Induced | 15.0 | 4.9 | groES, stress-induced chaperonin | P40172 | 6 |
| NR-8 | Cytoplasmic | Repressed | 61.7 | 4.1 | SCC80.05c, putative cell division trigger factore | CAC09996 | 14 |
| XI-26 | Extracellular | Induced | 30.0 | 5.3 | sodF2, FeSOD | AAD33130 | 5 |
| XI-27 | Extracellular | Induced | 28.0 | 5.2 | sodF2, FeSOD | AAD33130 | 5 |
| XI-28 | Extracellular | Induced | 28.0 | 4.5 | SCC8A.26c, putative tellurite resistance proteinf | CAB92844 | 5 |
| XR-25 | Extracellular | Repressed | 13.5 | 5.0 | SC5C7.16, probable ATP-GTP binding proteing | T35223 | 4 |
Level of expression in the presence of plants.
Most proteins were associated with S. coelicolor; the only exception was NI-20, which was associated with S. hygroscopicus.
There was 29.6% identity in a 318-amino-acid overlap with Arabidopsis thaliana epoxide hydrolase.
There was 50% identity in a 304-amino-acid overlap with S. reticuli CebR and 50% identity in a 301-amino-acid overlap with T. fusca CelR.
There was 35.9% identity in a 412-amino-acid overlap with B. subtilis trigger factor.
There was 65.8% identity in a 190-amino-acid overlap with S. marcescens tellurium resistance protein.
There was 44.5% identity in a 137-amino-acid overlap with E. coli osmotically inducible protein C.
A major protein spot from the intracellular fraction showed a constant level of expression under both conditions and was identified as the elongation factor EF-Tu (protein NC-4 in Table 1). For several Streptomyces species, EF-Tu has been described as one of the most abundant cytoplasmic proteins in exponentially growing and late-stationary-phase cells (23, 28, 36). EF-Tu is actively involved in a kinetic proofreading mechanism of codon-anticodon pairing at the ribosome (45). Li et al. (28) described the use of EF-Tu as an internal calibration standard in two-dimensional electrophoretic studies. In our study, a fixed quantity of proteins was used for two-dimensional gel electrophoresis, but we could also rely on EF-Tu to confirm the efficiency of our protein assay procedure.
Three bacterial proteins induced by the presence of plant material are involved in the acquisition of carbon, suggesting that L. minor produces exudates that serve as nutriments for S. coelicolor. These proteins were a probable transcription regulator of a ceb operon or of a cellulolytic regulon (NI-21), a putative epoxide hydrolase (NI-19), and an acyl coenzyme A (acyl-CoA) dehydrogenase (NI-20). These proteins are involved in the degradation of cellulose, alkenes, and amino acids, respectively.
The transcription regulator identified exhibited 50% identity with both CebR of Streptomyces reticuli and CelR of Thermobifida fusca. CebR and CelR are related members of the LacI-GalR family (41, 46). These proteins control expression of transcripts from a ceb operon (41) and a cellulolytic regulon (44), respectively, which are required for cellobiose and cellotriose uptake. These carbohydrates are derived from degradation of cellulose present in plant residues. Ahmad and Baker (1) reported that extracellular cellulase activity contributed to the rhizosphere competence of the biocontrol fungus Trichoderma harzianum. Cellulase activity may also be an important determinant in rhizosphere colonization by streptomycetes which are known to secrete a wide range of hydrolytic enzymes (31).
The second identified protein of this group, the epoxide hydrolase, has been described as a protein that is essential for utilization of alkenes as carbon sources in prokaryotes (14). Alkenes are common compounds that exist in a variety of chemical forms and may be found in the root exudates of plants (35). Alkenes are also products released during the degradation of suberin, a nonhydrolyzable and insoluble compound found in the endoderm of plant roots (35). Several streptomycetes have been reported to be able to degrade complex lipidic polymers, such as cutin and suberin (5, 16, 33). It has also been suggested that a suberin-degrading esterase from Streptomyces scabiei is involved both in plant colonization (32) and in pathogenesis (5). Esterase and epoxide hydrolase may thus be linked in a catabolic pathway involved in the acquisition of carbon and energy from suberin-derived compounds. Epoxide hydrolase activity has not yet been reported to be a rhizosphere colonization determinant; however, several Streptomyces species are distinguished from other rhizosphere inhabitants by their capacity to degrade complex polymers (31). The ability to derive nutrients from recalcitrant organic compounds, such as suberin, may provide an additional competitive edge to rhizospheric streptomycetes.
The third protein of this group is an acyl-CoA dehydrogenase known to be involved in degradation of ramified aliphatic amino acids, such as leucine, isoleucine, and valine (50). Amino acids are present in plant root exudates (43) and provide carbon and nitrogen to rhizosphere bacteria. Recent studies have shown that the rhizosphere environment induces the expression of Pseudomonas genes involved in amino acid transport and catabolism (13, 39). The ability to derive nutrients from amino acids may be an important property for rhizosphere competence in the genus Streptomyces.
The additional carbon sources provided by plant exudates could explain the increases in bacterial metabolic activities observed when L. minor was added to the culture medium. Indeed, five bacterial proteins induced in the presence of plant material reflected an increase in the metabolic state. One of these proteins, the guanine-nucleotide exchange factor EF-Ts (NI-13), associates with the EF-Tu-GDP complex and induces the change of GDP to GTP (20). Studies have shown that expression of EF-Ts is induced when the transcription of rRNA and ribosomal protein genes is elevated (8, 23). Identification of the following four other proteins also suggests that there are increases in metabolic activities: a fructose bisphosphate aldolase (NI-14) involved in glycolysis, a probable transketolase (NI-22) that could catalyze reactions in the pentose phosphate pathway, an isocitrate dehydrogenase (NI-3) involved in the oxidoreduction reactions of the citrate cycle, and an alpha protomer (NI-5) from the soluble F1 component of the membrane-bound ATP synthase which couples proton entry with ATP formation in the oxidative phosphorylation process. Induction of these proteins in the S. coelicolor proteome suggests that plant exudates provide a supply of nutriments in the form of additional carbon sources.
The functions of other bacterial proteins identified in this study were linked to adaptations to different stress signals. The nature of two (NR-8 and XR-25) of the six stress proteins identified reflected a diminution of nutritional stress in the culture medium supplemented with L. minor fronds. Indeed, both NR-8 and XR-25 were repressed in the presence of plant material. NR-8 was identified as a putative cell division trigger factor in S. coelicolor and exhibited 35.9% identity in a 412-amino-acid overlap with Tig, a cell division trigger factor from Bacillus subtilis, while XR-25 was associated with an ATP-GTP binding protein from S. coelicolor. The Tig factor is a peptidyl-prolyl cis-trans isomerase which is involved in the proline-limited folding of proteins. Gothel et al. (19) generated disruptions in the encoding gene and found that growth of the mutants in poor medium was strongly decelerated, indicating that the protein is essential for growth under starvation conditions. In our experimental model, addition of plants to the S. coelicolor culture medium appeared to render expression of this Tig factor not essential. The probable ATP-GTP binding protein, which was identified in the secreted fraction of S. coelicolor, exhibited 44.5% identity in a 137-amino-acid overlap with osmotically inducible protein C (OsmC) from Escherichia coli. In E. coli, transcription of osmC is induced at the onset of deceleration, and this occurs earlier in cultures in which the osmotic pressure is elevated (17). Addition of plant material to S. coelicolor medium delayed the entrance of S. coelicolor into the deceleration phase, and since OsmC and Tig are known to be nutritional stress factors, the repression of these proteins in the presence of plant material suggests that L. minor introduced nutrients available for uptake by S. coelicolor.
Three other stress proteins reflect adaptation to physiological stress. These three proteins were induced in the presence of L. minor in the culture medium and included a conserved hypothetical protein (XI-28) and two chaperone proteins, GroEL2 (NI-7) and GroES (NI-31). The groEL2 and groES transcripts in S. coelicolor were previously described as transcripts that are induced in response to undefined physiological stress signals (12) and may reflect bacterial adaptation to changes in the medium conditions or growth phase. The conserved hypothetical protein XI-28 exhibited 65.8% identity in a 190-amino-acid overlap with TerD from Serratia marcescens. TerD is a protein that is associated with resistance to tellurium salts, but the mechanism of resistance has not been determined (42). In E. coli, the response to tellurite is controlled by the soxRS system that is also involved in the regulated expression of the oxidative stress regulon (37). A probable SoxR-like transcription regulator (protein accession number T36798) is found in the annotated gene database of S. coelicolor. With this putative homologue, similar regulation of the oxidative stress regulon and the tellurite resistance genes in S. coelicolor may be proposed.
The remaining identified stress protein is an Fe-containing superoxide dismutase (FeSOD) (protein spots XI-26 and XI-27) encoded by sodF2 in S. coelicolor. Its expression was induced by the presence of plant material in the culture medium. FeSOD is a first-line antioxidant defense protein, and it eliminates the superoxide anions (O2−) generated as a by-product of aerobic respiration (9). The increased bacterial expression of FeSOD in the presence of L. minor suggests that oxidative stress is induced in the presence of plant compounds. Plant roots possess surface enzymes capable of producing the activated oxygen species O2− (2, 21). The essential nature of the bacterial response to oxidative stress during plant-microbe interactions is supported by the fact that Pseudomonas putida FeSOD mutants are impaired in rhizosphere colonization (26). Several FeSODs are found in the cytosol or extracellular space of prokaryotes (9, 24). In this case, FeSOD was present in the supernatant fraction. This localization has been described in the actinomycete Mycobacterium tuberculosis as an adaptation against extracellular host defense when macrophage-ingested bacteria must persist in a reactive oxygen environment (40). Hammad et al. (22) also described identification in the proteome of the actinomycete Frankia of an extracellular FeSOD induced by exudates from the symbiotic plant host. Given that superoxide radicals cannot cross membranes under physiological conditions, the extracellular localization of FeSOD in S. coelicolor cultures grown in presence of plant material is coherent with bacterial adaptation to extracellular oxidative stress.
The presence of plant material in the culture medium of S. coelicolor caused differential gene expression, as determined by analysis of the S. coelicolor proteome. The types of proteins identified in this study suggest that carbon and energy are acquired through degradation of compounds found in plant exudates and that bacteria adapt to physiological and oxidative stress. These traits might be essential for rhizosphere competence. Research is under way to further investigate the roles of the bacterial proteins identified in this work during plant-streptomycete interactions.
Acknowledgments
This work was supported by a grant from the Natural Sciences and Engineering Research Council of Canada (NSERC).
REFERENCES
- 1.Ahmad, J. S., and R. Baker. 1987. Rhizosphere competence of Trichoderma harzianum. Phytopathology 77:182-189. [Google Scholar]
- 2.Alberts, F. G., L. W. Bennett, and A. J. Anderson. 1986. Peroxidase associated with the root surface of Phaseolus vulgaris. Can. J. Bot. 64:573-578. [Google Scholar]
- 3.Barakate, M., Y. Ouhdouch, K. H. Oufdou, and C. Beaulieu. 2002. Characterization of rhizospheric soil Streptomyces from Moroccan habitats and their antimicrobial activities. World J. Microbiol. Biotechnol. 18:49-54. [Google Scholar]
- 4.Beaumont, G., R. Bastin, and H. P. Therrien. 1976. Effets physiologiques de l'atrazine sur Lemna minor L. I. Influence sur la croissance, la teneur en chlorophylle, en protéines et en azote soluble. Nat. Can. 103:527-533. [Google Scholar]
- 5.Beauséjour, J., C. Goyer, J. Vachon, and C. Beaulieu. 1999. Production of thaxtomin A by Streptomyces scabies in plant extract containing media. Can. J. Microbiol. 45:764-768. [Google Scholar]
- 6.Bentley, S. D., K. F. Chater, A.-M. Cerdeno-Tarraga, G. L. Challis, N. R. Thompson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C.-H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsh, M.-A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 7.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 8.Choe, L. H., W. Chen, and K. H. Lee. 1999. Proteome analysis of factor for inversion stimulation (Fis) overproduction in Escherichia coli. Electrophoresis 20:798-805. [DOI] [PubMed] [Google Scholar]
- 9.Chung, H.-J., E.-J. Kim, B. Suh, J.-H. Choi, and J.-H. Roe. 1999. Duplicate genes for Fe-containing superoxide dismutase in Streptomyces coelicolor A3(2). Gene 231:87-93. [DOI] [PubMed] [Google Scholar]
- 10.Doumbou, C. L., M. K. H. Salove, D. L. Crawford, and C. Beaulieu. 2001. Actinomycetes, promising tools to control plant diseases and to promote plant growth. Phytoprotection 82:85-102. [Google Scholar]
- 11.Doumbou, C. L., V. Akimov, M. Côté, P.-M. Charest, and C. Beaulieu. 2001. Taxonomic study on nonpathogenic streptomycetes isolated from common scab lesions on potato tubers. Syst. Appl. Microbiol. 24:451-456. [DOI] [PubMed] [Google Scholar]
- 12.Duchêne, A. M., C. J. Thompson, and P. Mazodier. 1994. Transcriptional analysis of groEL genes in Streptomyces coelicolor A3(2). Mol. Gen. Genet. 245:61-68. [DOI] [PubMed] [Google Scholar]
- 13.Espinosa-Urgel, M., and J.-L. Ramos. 2001. Expression of a Pseudomonas putida aminotransferase involved in lysine catabolism is induced in the rhizosphere. Appl. Environ. Microbiol. 67:5219-5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faber, K., M. Mischitz, and W. Kroutil. 1996. Microbial epoxide hydrolases. Acta Chem. Scand. 50:249-258. [Google Scholar]
- 15.Faucher, E., T. Savard, and C. Beaulieu. 1992. Characterization of actinomycetes isolated from common scab lesions on potato tubers. Can. J. Plant Pathol. 14:197-202. [Google Scholar]
- 16.Fett, W. F., H. C. Gerard, L. E. Jones, S. F. Osman, and R. A. Moreau. 1994. Production of cutin-degrading enzymes by plant pathogenic bacteria, p. 641-646. In M. Lamattre, S. Freigoun, K. Rudolph, and J. G. Swings (ed.), Plant pathogenic bacteria, 8th International Conference, Versailles, France, June 9-12, 1992. Les Colloques no. 66. INRA Edition, Paris, France.
- 17.Gordia, S., and C. Gutierrez. 1996. Growth-phase-dependent expression of the osmotically inducible gene osmC of Escherichia coli K-12. Mol. Microbiol. 19:729-736. [DOI] [PubMed] [Google Scholar]
- 18.Görg, A., W. Postel, and S. Günther. 1988. The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis 9:531-546. [DOI] [PubMed] [Google Scholar]
- 19.Gothel, S. F., C. Scholz, F. X. Schmid, and M. A. Marahiel. 1998. Cyclophilin and trigger factor from Bacillus subtilis catalyse in vitro protein folding and are necessary for viability under starvation conditions. Biochemistry 37:13392-13399. [DOI] [PubMed] [Google Scholar]
- 20.Gromadski, K. B., H. J. Weiden, and M. D. Rodniva. 2002. Kinetic mechanism of elongation factor Ts-catalyzed nucleotide exchange in elongation factor Tu. Biochemistry 41:162-169. [DOI] [PubMed] [Google Scholar]
- 21.Gross, G. G., C. Jange, and E. F. Elstner. 1977. Involvement of malate, monophenol, and the superoxide radical in hydrogen peroxide formation by isolated cell walls from horseradish (Armoracia lapathifolia Gilib). Planta 140:81-88. [DOI] [PubMed] [Google Scholar]
- 22.Hammad, Y., J. Maréchal, B. Cournoyer, P. Normand, and A.-M. Domenach. 2001. Modification of the protein expression pattern induced in the nitrogen-fixing actinomycete Frankia sp. strain ACN14a-tsr by root exudates of its symbiotic host Alnus glutinosa and cloning of the sodF gene. Can. J. Microbiol. 47:541-547. [DOI] [PubMed] [Google Scholar]
- 23.Hoogvliet, G., G. P. van Wezel, and B. Kraal. 1999. Evidence that a single EF-Ts suffices for the recycling of multiple and divergent EF-Tu species in Streptomyces coelicolor A3(2) and Streptomyces ramocissimus. Microbiology 145:2293-2301. [DOI] [PubMed] [Google Scholar]
- 24.Kang, S. K., Y. J. Jung, C. H. Kim, and C. Y. Song. 1998. Extracellular and cytosolic iron superoxide dismutase from Mycobacterium bovis BCG. Clin. Diagn. Lab. Immunol. 5:784-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, England.
- 26.Kim, Y. C., C. D. Miller, and A. J. Anderson. 2000. Superoxide dismutase activity in Pseudomonas putida affects utilization of sugars and growth on root surfaces. Appl. Environ. Microbiol. 66:1460-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 28.Li, X. M., J. Vohradsky, and J. Weiser. 1994. The use of protein synthesis elongation factor EF-Tu as internal calibration standard in two-dimensional electrophoretic studies of differentiation in Streptomyces. Electrophoresis 15:1198-1204. [DOI] [PubMed] [Google Scholar]
- 29.Locci, R. 1994. Actinomycetes as plant pathogens. Eur. J. Plant. Pathol. 100:179-200. [Google Scholar]
- 30.Lomagin, A. G., and L. V. Ul'yanova. 1993. A new test for water pollution using duckweed, Lemna minor L. Russ. Plant Physiol. 40:302-303. [Google Scholar]
- 31.Loria, R., R. A. Bukhalid, B. A. Fry, and R. R. King. 1997. Plant pathogenicity in the genus Streptomyces. Plant Dis. 81:836-846. [DOI] [PubMed] [Google Scholar]
- 32.Lugtenberg, B. J. J., L. V. Kravchenko, and M. Simons. 1999. Tomato seed and root exudates sugars: composition, utilization by Pseudomonas biocontrol strains and role in rhizosphere colonization. Environ. Microbiol. 1:439-446. [DOI] [PubMed] [Google Scholar]
- 33.McQueen, D. A., and J. L. Schottel. 1987. Purification and characterization of a novel extracellular esterase from pathogenic Streptomyces scabies that is inducible by zinc. J. Bacteriol. 169:1967-1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Natera, S. H. A., N. Guerreiro, and M. A. Djordjevic. 2000. Proteome analysis of differentially displayed proteins as a tool for the investigation of symbiosis. Mol. Plant Microbe Interact. 13:995-1009. [DOI] [PubMed] [Google Scholar]
- 35.Nierop, K. G. J. 1998. Origin of aliphatic compounds in a forest soil. Org. Geochem. 29:1009-1016. [Google Scholar]
- 36.Olsthoorn, L. N., L. J. Plooster, and B. Kraal. 2001. The variant tuf3 gene of Streptomyces coelicolor A3(2) encodes a real elongation factor Tu, as shown in a novel Streptomyces in vitro translation system. Eur. J. Biochem. 268:3807-3815. [DOI] [PubMed] [Google Scholar]
- 37.Orsaria, L., L. Paoletti, and H. C. Gramajo. 1998. Characterization of stationary-phase proteins in Streptomyces coelicolor A3(2). FEMS Microbiol. Lett. 162:275-281. [Google Scholar]
- 38.Rahmani, G. N. H., and P. K. Sternberg. 1999. Bioremoval of lead from water using Lemna minor. Bioresource Technol. 70:225-230. [Google Scholar]
- 39.Rainey, P. B. 1999. Adaptation of Pseudomonas fluorescens to plant rhizosphere. Environ. Microbiol. 1:243-257. [DOI] [PubMed] [Google Scholar]
- 40.Raman, S., T. Song, X. Puyang, S. Bardarov, W. R. Jacobs, Jr., and R. N. Husson. 2001. The alternative sigma factor SigH regulates major components of oxidative and heat stress responses in Mycobacterium tuberculosis. J. Bacteriol. 183:6119-6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schlösser, A., T. Aldekamp, and H. Schrempf. 2000. Binding characteristics of CebR, the regulator of the ceb operon required for cellobiose/cellotriose uptake in Streptomyces reticuli. FEMS Microbiol. 190:127-132. [DOI] [PubMed] [Google Scholar]
- 42.Silver, S., and L. Phung. 1996. Bacterial heavy metal resistance: new surprises. Annu. Rev. Microbiol. 50:753-789. [DOI] [PubMed] [Google Scholar]
- 43.Simons, M., H. P. Parmentier, L. A. de Wegler, C. A. Wijffelman, and B. J. J. Lugtenberg. 1997. Amino acid synthesis is necessary for tomato root colonization by Pseudomonas fluorescens strain WCS365. Mol. Plant Microbe Interact. 10:102-106. [Google Scholar]
- 44.Spiridonov, N. A., and D. B. Wilson. 1999. A celR mutation affecting transcription of cellulase genes in Thermobifida fusca. J. Bacteriol. 182:252-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thompson, R. C. 1988. EF-Tu provides an internal kinetic standard for translational accuracy. Trends Biochem. Sci. 13:91-93. [DOI] [PubMed] [Google Scholar]
- 46.Weickert, M. J., and S. Adhya. 1992. A family of bacterial regulators homologous to Gal and Lac repressors. J. Biol. Chem. 267:15869-15874. [PubMed] [Google Scholar]
- 47.Williams, K. R., M. LoPresti, and K. Stone. 1997. Internal protein sequencing of SDS-PAGE separated proteins: optimization of in-gel digest protocol, p. 79-90. In D. Marshak (ed.), Techniques in protein chemistry, vol. VIII. Academic Press, New York, N.Y.
- 48.Wirth, P. J., and A. Romano. 1995. Staining methods in gel electrophoresis, including the use of multiple detection methods. J. Chromatogr. A 698:123-143. [DOI] [PubMed] [Google Scholar]
- 49.Wohl, D. L., and J. V. McArthur. 1998. Actinomycete flora associated with submersed freshwater macrophytes. FEMS Microbiol. Ecol. 26:35-140. [Google Scholar]
- 50.Zhang, Y. X., C. D. Denoya, D. D. Skinner, R. W. Fedechko, H. A. I. McArthur, M. R. Morgenstern, R. A. Davies, S. Lobo, K. A. Reynolds, and R. Hutchinson. 1999. Genes encoding acyl-CoA dehydrogenase (AcdH) homologues from Streptomyces coelicolor and Streptomyces avermitilis provide insights into the metabolism of small branched-chain fatty acids and macrolide antibiotic production. Microbiology 145:2323-2334. [DOI] [PubMed] [Google Scholar]