Abstract
Thermobifida fusca TM51, a thermophilic actinomycete isolated from composted horse manure, was found to produce a number of lignocellulose-degrading hydrolases, including endoglucanases, exoglucanases, endoxylanases, β-xylosidases, endomannanases, and β-mannosidases, when grown on cellulose or hemicellulose as carbon sources. β-Mannosidases (EC 3.2.1.25), although contributing to the hydrolysis of hemicellulose fractions, such as galacto-mannans, constitute a lesser-known group of the lytic enzyme systems due to their low representation in the proteins secreted by hemicellulolytic microorganisms. An expression library of T. fusca, prepared in Streptomyces lividans TK24, was screened for β-mannosidase activity to clone genes coding for mannosidases. One positive clone was identified, and a β-mannosidase-encoding gene (manB) was isolated. Sequence analysis of the deduced amino acid sequence of the putative ManB protein revealed substantial similarity to known mannosidases in family 2 of the glycosyl hydrolase enzymes. The calculated molecular mass of the predicted protein was 94 kDa, with an estimated pI of 4.87. S. lividans was used as heterologous expression host for the putative β-mannosidase gene of T. fusca. The purified gene product obtained from the culture filtrate of S. lividans was then subjected to more-detailed biochemical analysis. Temperature and pH optima of the recombinant enzyme were 53°C and 7.17, respectively. Substrate specificity tests revealed that the enzyme exerts only β-d-mannosidase activity. Its kinetic parameters, determined on para-nitrophenyl β-d-mannopyranoside (pNP-βM) substrate were as follows: Km = 180 μM and Vmax = 5.96 μmol min−1 mg−1; the inhibition constant for mannose was Ki = 5.5 mM. Glucono-lacton had no effect on the enzyme activity. A moderate trans-glycosidase activity was also observed when the enzyme was incubated in the presence of pNP-αM and pNP-βM; under these conditions mannosyl groups were transferred by the enzyme from pNP-βM to pNP-αM resulting in the synthesis of small amounts (1 to 2%) of disaccharides.
Thermobifida spp. are gram-positive, compost- and soil-inhabiting bacteria with broad degradative activity on plant cell wall constituents. Thermobifida fusca, the most extensively studied species of this genus, is the model organism of thermophilic, aerobic cellulolytic bacteria (48). While there are ample data on the cellulolytic system of T. fusca (8, 24, 30, 47, 48), the hemicellulolytic enzyme system of this species is still poorly characterized. Only one endoxylanase- and an endomannanase-encoding gene have been cloned to date (20, 22), although the hemicellulolytic enzyme system of T. fusca may be as complex as the well-characterized cellulase system of this organism; the latter consists of three endoglucanases, two exoglucanases, one processive endoglucanase, and a cellobiase enzyme (23, 43).
The complexity of cell wall-degrading enzyme systems is a consequence of the complex nature of plant cell wall. Hemicelluloses act as linkers between lignin and cellulose. The high percentage of hemicellulose fraction in the cell wall of higher plants makes this material the second most abundant biopolymer in nature. Besides xylan, mannan is the other major hemicellulose constituent. Galactomannan, found in large quantities in seeds of leguminous plants, is composed of a homogeneous backbone of β-1,4-linked mannose residues, whereas acetylated galactoglucomannan, a main constituent of softwoods, has a heterogeneous backbone of β-1,4-linked glucose and mannose residues (7). The complete conversion of galactomannan into galactose and mannose requires the activity of three types of enzymes, namely, endomannanases (EC 3.2.1.78), α-galactosidases (EC 3.2.1.22), and β-mannosidases (β-d-mannopyranoside hydrolase [EC 3.2.1.25]). Endomannanases catalyze the random hydrolysis of the β-1,4-mannosidic backbone of the main mannan chain, α-galactosidases cleave the terminal α-1,6-linked d-galactosyl residues, and β-mannosidases hydrolyze β-1,4-linked mannose residues from the nonreducing ends of various oligosaccharides (3).
β-Mannosidase-encoding genes have already been cloned from mammals (4, 10, 14), fungi (1, 46), and bacteria (9, 17, 45). Almost every β-mannosidase belongs to family 2 of glycosyl hydrolases (GHs) (16), except for the Pyrococcus sp. β-mannosidase, which has been assigned to family 1 (9).
Hemicellulases are industrially important enzymes; their use has been shown to promote pulp bleaching in paper industry. More recent investigations are focused on mannan-degrading enzymes (15). These enzymes are widely used in coffee bean fermentation to promote the hydrolysis of β-mannan-based oligosaccharides (37).
In general, mannosidases constitute only a small percentage of the proteins secreted by hemicellulose degrading organisms; their purification is therefore rather difficult. This problem could be solved by cloning and heterologous expression of mannosidase-encoding genes. Earlier we isolated a number of thermophilic, cellulolytic actinomycetes from a compost pile (27), and one of them, T. fusca TM51, was found to show an outstanding cellulolytic-hemicellulolytic activity.
The aim of this work was to obtain a better insight into the hemicellulose degrading enzyme system of T. fusca, the model organism of thermophilic, aerobic cellulose-degrading bacteria, as only a single endoxylanase encoding gene of this system has been cloned from this species to date. In the present paper we describe cloning, sequencing, and heterologous expression of manB, a β-mannosidase encoding gene from T. fusca TM51, and report the purification and biochemical characterization of the enzyme.
Glycosidases, which cleave by retention, such as family 2 GHs, hydrolyze their substrates trough a two-step mechanism: first, a covalent glycosyl-enzyme intermediate is produced, which is then hydrolyzed by a general base-catalyzed attack of water. However, if an appropriate sugar, capable of binding to the aglycone site, is present in sufficient concentrations, transglycosylation may occur (11). This process provides the basis of using glycosidases for enzymatic synthesis of oligosaccharides of physiological and medicinal relevance (41). The synthesis of such complex carbohydrates is difficult to manage by traditional chemical procedures. Another aim of this work was to demonstrate transglycosylase activity of the β-mannosidase enzyme produced by T. fusca TM51.
MATERIALS AND METHODS
Chemicals.
Restriction endonucleases were from Promega, Gibco-BRL (Burlington, Ontario, Canada). T4 DNA ligase, Hybond-N nylon membrane, and thiostrepton were obtained from Boehringer Mannheim (Laval, Quebec, Canada), Amersham International (Little Chalfont, England), and Fluka (Buchs, Switzerland), respectively. The different p-nitrophenyl (pNP) glycosides (including pNP β-d-mannopyranoside [pNP-βM], pNP-αM, pNP β-d-glucopyranoside [pNP-βGlc], pNP-αGlc, pNP β-d-galactopyranoside [pNP-βGal], pNP-αGal, pNP β-d-fucopyranoside [pNP-βFuc], pNP β-d-xylopyranoside [pNP-βXyl], and pNP α-l-arabinopyranoside [pNP-αAra]), 4-methylumbelliferyl-β-d-mannoside (MUβMan), isopropyl-β-d-thiogalactopyranoside (IPTG), birch wood xylan, and locust bean gum (LBG) were purchased from Sigma Chemical Co (St. Louis, Mo.). MN 300 cellulose powder was obtained from Macherey-Nagel (Düren, Germany).
Media and growth conditions.
Escherichia coli strains were grown in Luria-Bertani medium at 37°C. Streptomyces lividans strain TK24 was cultivated on basal medium (NaNO3, 1.0 g; KCl, 0.3 g; MgSO4 · H2O, 0.5 g; K2HPO4, 1.0 g; yeast extract, 0.5 g; peptone, 0.5 g; distilled water, 1 liter [pH of medium, 7.6]), containing 0.2% sucrose, at room temperature for protein production. Media were supplemented with 5 μg of thiostrepton ml−1. T. fusca TM51 and T. fusca ATCC 27730T were grown at 50°C in basal salt medium supplemented with different carbon sources. Liquid cultures were shaken at 220 rpm. Solid media contained 1.5% (wt/vol) agar-agar (Reanal, Budapest, Hungary).
Induction and localization of mannosidase in T. fusca.
Spore suspensions (106 ml−1) of T. fusca TM51 were used to inoculate 100 ml of basal medium containing 0.2% glucose, 1% MN 300 cellulose, 0.2% xylan, or 0.2% LBG as carbon sources. Cultures were incubated at 50°C for 7 days, and β-mannosidase activities were determined on MUβMan substrate by fluorimetry.
Cell fractionation.
Cells from cultures, grown on 40 ml of LBG containing basal medium at 50°C for 4 days, were harvested by centrifugation at 5,000 × g for 10 min at 4°C and washed twice with an equal volume of 0.15 M phosphate buffer. For separating soluble and cell wall-bound intracellular proteins, cells were disrupted by sonication (Virsonic 300; Virtis, Gardiner, N.Y.) for 6 min at 40 W. After centrifugation at 20,000 × g for 15 min at 4°C, the supernatant, containing the soluble intracellular protein fraction, was collected and used directly for enzyme assay. The pellet, composed of cell wall and membrane debris, was washed two times with water, centrifuged again, resuspended in the same volume of 0.15 M phosphate (pH 7.0) buffer, and tested for mannosidase activity.
Mannosidase activities of four subsamples—culture supernatant, sonicated whole-cell lysate, cell wall plus membrane fraction, and soluble protein fraction—were determined on MUβMan substrate by fluorimetry. The reaction mixture contained 1 ml of sample, 1 ml of phosphate buffer (0.1 M, pH 7), and 10 μl of MUβMan solution (2 mg ml−1). The reaction mixture was incubated at 50°C for 20 min. One enzyme unit was defined as the amount of enzyme that released 10 nmol of 4-methylumbelliferone per min.
Construction of a genomic library of T. fusca TM51.
Total DNA isolated from T. fusca TM51 was partially digested with Sau3AI. Restricted DNA was fractionated by agarose gel electrophoresis. The excised ∼15-kb fragments were purified with a Qiaex II gel extraction kit according to the manufacturer's recommendations (Qiagen, Santa Clarita, Calif.). The fragments were ligated into plasmid pIJ699 (26) according to the method of Blanco et al. (12). S. lividans TK24 protoplasts were transformed with the vector, and transformants were selected on R2-agar plates by the method of Hunter (21), using thiostrepton (100 μg ml−1) as a positive selection marker. The plates were incubated for 3 days at 30°C, and transformants were screened for β-mannosidase activity on MUβMan; positive clones fluoresced under UV light.
Cloning and sequencing of manB.
The 15-kb insert, carrying the whole manB gene, was subcloned in E. coli DH5α. pBluescript and pUC19 plasmids were used for subcloning. Standard methods were used for other DNA manipulations (39). DNA and protein sequences were analyzed by using the Genetics Computer Group program. The amino acid sequence and domain structure of ManB were determined by using Swiss-Prot, EMBL, NCBI, Pfam, and InterPro data banks (5, 6). A phylogenetic tree was constructed by the neighbor-joining method (38) and with the ARB phylogenetic package.
Southern blot analysis.
Total DNA from T. fusca TM51, digested with MluI, SacI, XhoI, and PstI and separated on a 1% agarose gel, was transferred onto a Hybond-N nylon membrane and probed with the labeled manB gene fragment. All hybridization procedures were performed as described by Sambrook et al. (39). The DNA probe was labeled using a random-primed labeling kit, following the manufacturer's recommendations.
Production and purification of enzyme.
A mannosidase-positive S. lividans clone was grown on sucrose containing basal medium supplemented with 5 μg of thiostrepton ml−1 for 72 h. Cells harvested by centrifugation (10 min at 10,000 × g), washed in 100 mM phosphate buffer (pH 7.0), and then resuspended in 50 mM Tris-HCl (pH 7.4) were disrupted by sonication (10 min at 40 W). The suspension was centrifuged for 20 min at 12,000 × g, and the supernatant, used as cell extract (CFE), was purified with fast-performance liquid chromatography (FPLC) (Pharmacia) by anion-exchange chromatography on a Mono-Q column (buffer A: 50 mM Tris-HCl, pH 7.4; buffer B: buffer A plus 1 M NaCl). Enzyme activities of the fractions were measured by fluorimetry using the MUβMan substrate. The enzyme was further purified from the active fractions by gel chromatography on an FPLC Superose-12 column (50 mM Tris-HCl, pH 7.4). The activity and purity of the enzyme were checked by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) zymography and by silver staining.
PAGE zymography.
SDS-polyacrylamide slab gels (0.75 mm thick) were prepared according to the method of O'Farrell (35) with 10% acrylamide separating gel. The silver staining procedure was used for the detection of proteins in polyacrylamide gels (50). For zymography, gels were renatured by washing twice with isopropanol containing (25%, vol/vol) phosphate buffer (0.1 M, pH 7.0) at room temperature. After two subsequent washing steps with phosphate buffer (0.1 M, pH 7.0), gels were stained for activity with 1 mM MUβMan solution. Enzyme activity was detected under UV light.
Enzyme assays.
The β-d-mannosidase activity was assayed by using 100 μl of 0.005 M pNP-βM in 0.1 M sodium phosphate buffer (pH 7.0) and an appropriate dilution of the enzyme solution in a 1-ml final volume of 0.1 M sodium phosphate buffer (pH 7.0) at 50°C for 10 min. The reaction was stopped by the addition of 2 ml of 0.2 M sodium borate buffer (pH 10.0), and the release of pNP was monitored at 400 nm. Blank solutions always contained the same components except for the enzyme solution. Enzyme activity was calculated as units per milliliter. One unit is defined as the amount of enzyme which liberates 1 μmol of pNP per min under the assay conditions.
Substrate specificity was determined by using different pNP glycosides (pNP-βM, pNP-αM, pNP-βGlc, pNP-αGlc, pNP-βGal, pNP-αGal, pNP-βFuc, pNP-βXyl, and pNP-αAra, respectively) according to the protocol described above.
Protein contents were determined according to the method of Lowry as modified by Hartree (19), using bovine serum albumin as the standard.
Characterization of the β-mannosidase.
Temperature and pH optima of ManB were determined according to the method of Kurakake and Komaki (29).
For kinetic investigations the reaction rate was determined at six different concentrations of pNP-βM (Km, 0.5 and 5). The Km and Vmax values were calculated by using the GraFit software program (Erithacus Software Ltd., Staines, United Kingdom). Inhibition experiments were carried out at three different substrate concentrations and six different inhibitor (mannose and glucono-lacton) concentrations (Ki, 0.5 to 5). The Ki value was determined from Dixon plots by using the GraFit software.
Transferase activity was examined under different reaction conditions in a 0.5-ml final reaction volume. The enzyme was incubated in 0.1 M sodium phosphate buffer (pH 7.0) with pNP-M at a concentration of 2.5 mM, at room temperature in the presence of methyl-, ethyl-, propyl-, and butyl-alcohol added at 10 and 20% (vol/vol) concentrations. β-Mannosidase was also incubated using 25 mM pNP-αM as acceptor and 25 mM pNP-βM as donor compounds in 0.1 M sodium phosphate buffer (pH 7.0), containing 0, 10, and 20% (vol/vol) dimethyl sulfoxide at room temperature. Negative controls were run without adding β-mannosidase. Samples taken after 1, 12, 24, and 48 h were analyzed by thin-layer chromatography and high-pressure liquid chromatography (HPLC). A Hewlett-Packard 1090 series II liquid chromatograph equipped with a refractive index detector, an automatic sampler, and a ChemStation was used to separate products of the reaction. The separation was performed on a Hypersil APS (aminopropylsilica) column (4.6 mm by 100 mm), converted into the sulfate form by the method of Kahle et al. (25). Acetonitrile-water mixture (9:1) was used as the eluant, and 20-μl samples were analyzed. Column temperature was controlled by a column oven. Products containing a chromophore group were determined by diode array detector at room temperature, at 300 nm. In other cases, a refractive index detector was used at 40°C.
Nucleotide sequence accession number.
The GenBank accession number for the nucleotide sequence of manB is AF489440.
RESULTS AND DISCUSSION
Induction and cellular localization of β-mannosidase in T. fusca.
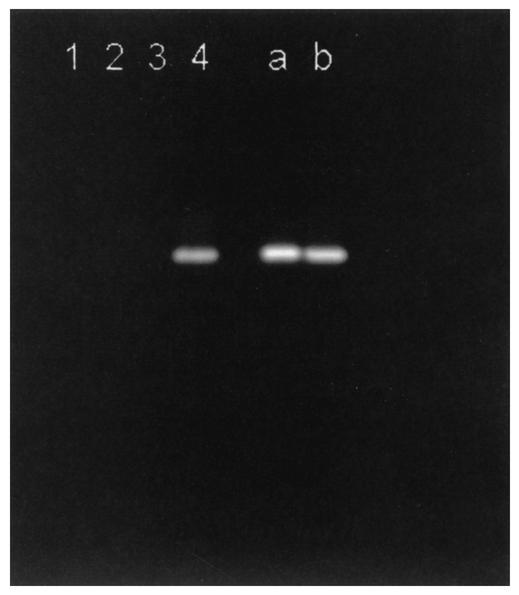
Cultures grown on LBG as the sole carbon source showed high mannosidase activity, whereas cultures grown on cellulose, xylan, or glucose produced no detectable mannosidase enzyme. Induction of β-mannosidase production by the mannan-containing carbon source is clearly shown in Fig. 1.
FIG. 1.
Induction of ManB on different carbon sources. Zymograms stained with MUβMan show that β-mannosidase of T. fusca TM51 is induced only by a mannan-containing substrate. Carbon sources were glucose (lane 1), MN300 cellulose (lane 2), birch wood xylan (lane3) and LBG (lane 4). The right part of the gel demonstrates the identity of mannanases produced by T. fusca ATCC 27730T (lane a) and the J9 transformant of S. lividans (lane b).
The cellular location of β-mannosidase activity was determined by comparing the activity of various cell fractions. The activity of cell extracts was about sevenfold higher than that found in the culture supernatant (15.2 versus 2.1 U ml−1). The activity of the cell wall-plus-membrane fraction was much lower (1.8 U ml−1) than that of the soluble protein fraction obtained from the disrupted cells (12.1 U ml−1). The low levels of enzyme activities in the culture supernatant and the cell wall fraction may originate from spontaneous cell lysis during culturing and the occurrence of a small number of nondisrupted hyphae, respectively. Apart from these contaminations ManB can be classified as a typical soluble, cytoplasmic enzyme. The intracellular location of ManB is explained by its role in mannan decomposition; i.e., this protein acts as a terminal component of the mannan-degrading enzyme system. The β-glucosidase enzyme of T. fusca is also intracellularly localized, as demonstrated by Spiridonov and Wilson (43). Thermobifidas and other compost-inhabiting actinomycetes extracellularly produce the large variety of cellulases, xylanases, and mannanases. On the other hand, their cellobiases, β-xylosidases, and β-mannosidases, which catalyze the final steps of polysaccharide decomposition, are often intracellularly located (45). This strategy helps to prevent the consumption of the released sugars by other compost-inhabiting microorganisms that are unable to utilize cellobiose, xylobiose, or mannobiose as they lack the appropriate enzyme and transport systems. T. fusca seems to have the appropriate transport system, as the bglABC operon, which has recently been described for this organism (43), encodes two types of sugar permeases, sharing substantial homology with CebG, a cellobiose-cellotriose transport protein known from Streptomyces reticuli (40). Xylobiose and mannobiose permeases are known from Aureobasidium pullulans (31), and a similar mechanism may support the mannobiose uptake in thermobifidas as well.
Cloning, sequencing, and analysis of mannosidase gene from T. fusca.
A genomic library of T. fusca was screened for mannosidase-positive clones by growing individually 2,000 S. lividans transformants on liquid Luria-Bertani medium. Supernatants of each culture were tested with MUβMan, and this yielded one positive clone, S. lividans J9.
A 15-kb DNA fragment responsible for mannosidase activity was subcloned into pUC19 in E. coli as host. Pilot DNA sequencing of the insert showed that this clone contained an 8-kb SalI fragment, harboring the entire mannosidase gene. The fragment was further subcloned in pBluescript. The ORF is composed of 2,523 nucleotides and codes for a protein of 840 amino acids (aa), with a calculated molecular mass of 94 kDa and a predicted pI of 4.87. A potential ribosome-binding site, AAGAG, was located 63 bp upstream from the start codon, GTG. An AACCGGTT motif was found at position −115; the same motif was present in two copies at the promoter region of the endomannanase-encoding gene of T. fusca (sequence data for the promoter region of the endomannanase of T. fusca YX was obtained from The Institute for Genomic Research website [http://www.tigr.org]).
Southern blot analysis clearly showed that manB is present in one copy in the T. fusca genome (data not presented).
Domain analysis of ManB.
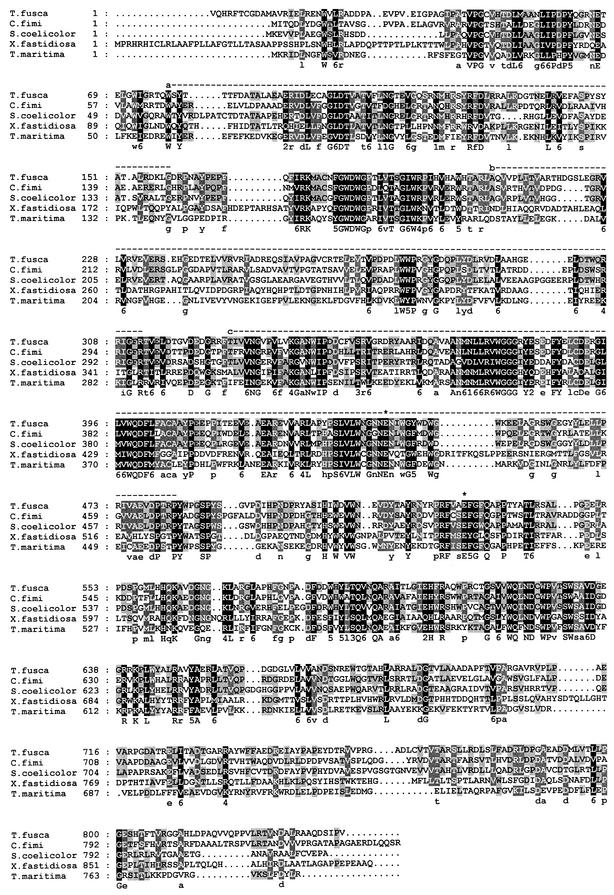
The N-terminal part of ManB contained no signal peptide sequence characteristic of extracellular enzymes. Computer-assisted homology analyses using the National Library of Medicine Retrieval System (http://www.ncbi.nlm.nih.gov) and the BLAST algorithm to scan GenBank and other databases indicated that ManB is a modular protein. It contains an N-terminal sugar binding domain (aa 79 to 157) with a jelly roll fold, an immunoglobulin-like β-sandwich domain (aa 208 to 312), and a TIM barrel domain (aa 329 to 483). At the C terminus of the protein there is a domain of unknown function (aa 671 to 816), which has also been identified in Man2A of Cellulomonas fimi.
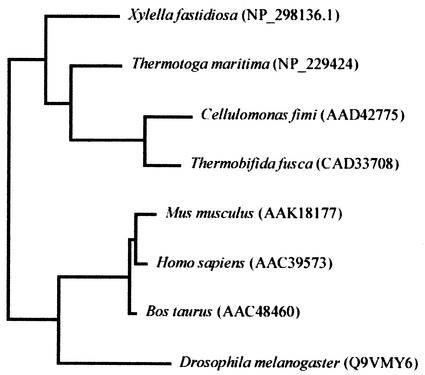
Two glutamate residues have been identified in the catalytic site of family 2 GHs; the identity of Glu 519 as a nucleophile partner was confirmed experimentally in the Man2A protein (44). In ManB the predicted nucleophile is Glu 530, whereas the acid base catalyst is Glu 443. ManB was found to show high similarities to β-mannosidases grouped into family 2 of the GHs (16). This family contains enzymes with β-galactosidase, β-mannosidase, and β-glucuronidase activities. Among the members of the GH family 2, ManB showed the highest overall similarity (53%) with Cellulomonas fimi Man2A and the putative β-mannosidases from Streptomyces coelicolor (54%) (Fig. 2). The similarity values were 34, 32, 31, 31, and 29% with β-mannosidases from Thermotoga maritima (36), Xylella fastidiosa (42), bovine (14), mouse (10), and human (4), respectively. A phylogenetic tree based on the full amino acid sequence of these enzymes was constructed by the neighbor-joining method (Fig. 3.). Among the GH family 2 enzymes, ManB clustered together in a phylogenetic group with β-mannosidases from eubacteria and was clearly separated from mannanases of mammalian origin. As can be seen from the topology of the tree, ManB is most closely related to Man2 of C. fimi. This is not surprising, as these two species are the closest relatives within this group of organisms and their plant cell wall-degrading enzyme systems share notable similarities.
FIG. 2.
Alignment of the amino acid sequences of β-mannosidases from T. fusca (CAD33708), C. fimi (AAD42775), X. fastidiosa (NP_298136.1), S. coelicolor (NP_630333), and T. maritima (NP_229424). Identical and similar residues are indicated on black and grey backgrounds, respectively. Dashed lines indicate a sugar binding domain (a), an immunoglobulin-like β-sandwich domain (b), and a TIM barrel domain (c). Asterisks indicate the putative acid base (443) and nucleophile (530) Glu residues of the catalytic center of ManB.
FIG. 3.
Phylogenetic relationships between ManB of T. fusca and a set of GH family 2 enzymes, including β-mannosidases of X. fastidiosa, T. maritima, C. fimi, Mus musculus, Homo sapiens, Bos taurus, and Drosophila melanogaster, respectively.
Production and purification of the ManB enzyme.
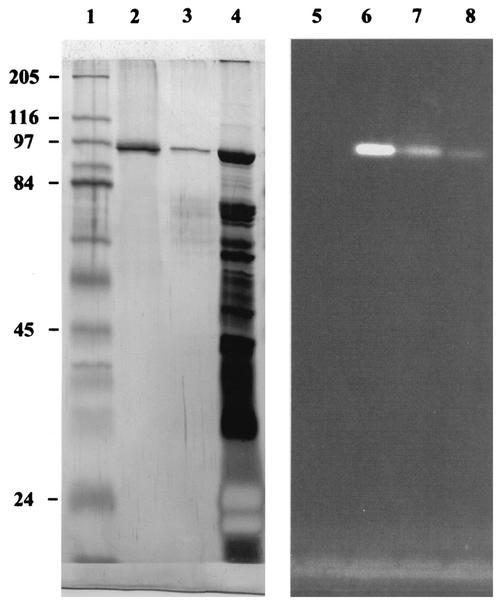
S. lividans clone TK24 harboring manB was grown in saccharose-containing medium. Due to the lack of enzyme secretion, no significant extracellular mannosidase activity was observed. The specific activity of CFE was in the range of 0.015 U mg−1. CFE was purified by anion-exchange chromatography on a Mono-Q column and was further purified by gel chromatography with FPLC on a Superose-12 column. The specific mannosidase activity of this purified extract increased to 5.6 U of protein mg−1. The mannosidase-positive band resolved by SDS-PAGE analysis was perfectly clear, confirming the high purity of the enzyme preparation (Fig. 4). The molecular mass of ManB as determined by SDS-PAGE was 94.5 kDa, approaching the deduced theoretical value of 94.0 kDa calculated from amino acid composition.
FIG. 4.
Purification of β-mannosidase (ManB) as demonstrated by SDS-PAGE. A silver-stained gel is shown on the left. Lanes: 1, broad-range marker (Sigma); 2, purified ManB eluted from Superose-12 column; 3, partially purified mannosidase eluted from Mono-Q column; 4, crude enzyme preparation. Zymograms demonstrating the activity of the same enzyme fractions using MUβMan as substrate are on the right. Lanes: 5, broad-range marker (Sigma); 6, purified ManB eluted from Superose-12 column; 7, partially purified mannosidase eluted from Mono-Q column; 8, crude enzyme preparation. Molecular masses are shown at right (in kilodaltons)
pH and temperature optima.
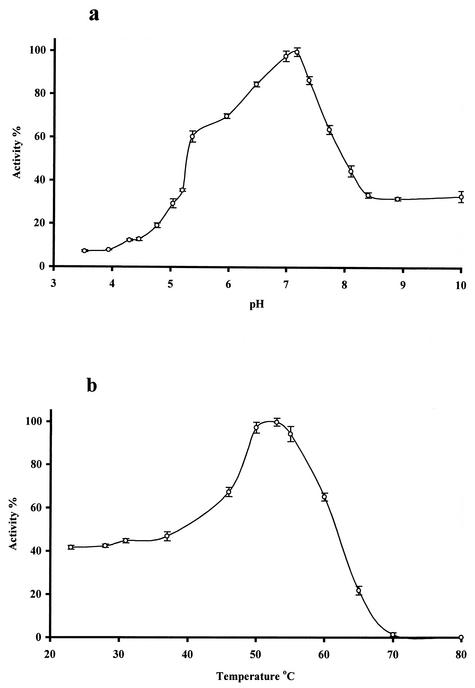
The pH and temperature optima of the enzyme produced in S. lividans were 7.17 and 53°C, respectively (Fig. 5). The thermal stability of ManB was relatively low, as 50% of the enzyme activity was destroyed after 10 min of incubation at 60°C and the half-life of its activity was 30 h at 40°C. However, this enzyme is still more heat tolerant than Man2A, a closely related β-mannosidase from the mesophilic organism C. fimi, which has a 27-h half-life at 37°C (45). The relative heat sensitivity of ManB, produced by a typical thermophilic organism, can be explained by its intracellular location. These data on pH and temperature optima are almost identical to those of BglC, a β-glucosidase enzyme (7.0 and 50°C), another intracellular member of the cellulolytic enzyme system of T. fusca (43).
FIG. 5.
Effect of pH and temperature on the activity of ManB. Hydrolysis of pNP-βM was measured at 53°C for 10 min in a pH range between 3.53 and 10.0 (a) and in 50 mM phosphate buffer, pH 7.17, between 23 and 80°C (b). Error bars show the standard deviation of the data.
Kinetic properties of the ManB protein.
The substrate specificity of ManB was investigated on nine different pNP glycosides (pNP-βM, pNP-αM, pNP-βGlc, pNP-αGlc, pNP-βGal, pNP-αGal, pNP-βFuc, pNP-βXyl, and pNP-αAra, respectively). The enzyme showed only β-d-mannosidase activity. Two other β-mannosidases from Thermotoga neapolitana (17) and Aspergillus niger (2, 18) have been reported to show similarly strict substrate specificity. In contrast, the β-mannosidase enzyme isolated from Pyrococcus furiosus exerted hydrolytic activity on pNP-βGal, pNP-βGlc, pNP-βXyl, and pNP-αGlc, besides the pNP-βM substrate (9). The kinetic parameters determined by using pNP-βM as substrate were as follows: Km = 180 (12.5) μM and Vmax = 5.96 (0.26) μmol min−1 mg−1, respectively (standard deviations shown in parentheses). The Km was similar to Km values of other β-d-mannosidases known from different organisms (Table 1.), but it is noteworthy that ManB has the second lowest Km value. The inhibition constant for mannose is Ki = 5.5 mM. Glucono-lacton had no effect on the enzyme activity.
TABLE 1.
Km values of β-d-mannosidases from different sources for pNP-βM as substrate
Glycosyl transferase activity of ManB.
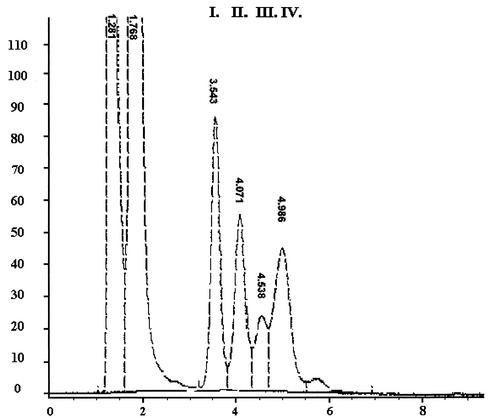
Due to its theoretical and practical importance we investigated the transglycosylase activity of ManB. When different alcohols and pNPMs were incubated in the presence of ManB the mannosyl moiety of the pNPMs was transferred to these alcohols and alkyl mannosides were formed. More interestingly, transglycosylase activity was also observed, when pNP-βM and pNP-βM were incubated with ManB, resulting in transfer of the mannosyl group from pNP-βM to pNP-αM. No reaction occurred in the absence of the enzyme. HPLC chromatograms clearly showed that disaccharides were synthesized: four different peaks were obtained, indicating that four disaccharides (forming 1,2, 1,3, 1,4 and 1,6 bonds) were produced in different ratios (Fig. 6.). The amounts of disaccharides were found to increase with duration of incubation and the amount of organic solvents, and the donor/acceptor ratios also affected the production. The percentage of disaccharides synthesized in this reaction ranged from 1 to 2% in all cases. The characteristic transglycosylation activity of ManB indicates that the enzyme cleaves the β-glycoside bond via retention, a mechanism constituting the prerequisite of the transglycosylation activity. This transglycosylation activity seems to be a common feature among β-mannosidases of microbial origin belonging to family 2 of GHs. Transmannosidase activity has also been reported for β-mannosidases from Cellulomonas (33) and Aspergillus species (3).
FIG. 6.
HPLC separation of the components obtained by transglycosylation reaction on an APS column. Samples were taken after 24 h of incubation. The reaction mixture contained 25 mM pNP-βM and pNP-αM as well as 20% dimethyl sulfoxide and β-mannosidase from T. fusca. Four different peaks (I to IV) indicate that four disaccharides were formed by 1,2, 1,3, 1,4, and 1,6 bonds, respectively.
Creating β-mannosidic bonds is still one of the most difficult issues in oligosaccharide chemistry. However, recent investigations on chemoenzymatic syntheses promise an efficient approach for solving this problem. The narrow substrate specificity and the extremely low Ki value of ManB make this protein an excellent candidate for chemoenzymatic utilization. Other mannosidases like the β-mannosidase from P. furiosus (9) and Man2A from C. fimi (34) have either a wider substrate specificity or a higher Ki for mannose. Due to these outstanding parameters ManB could be a promising subject of site-directed mutagenesis aiming at modifying its catalytic nucleophile to enhance the transmannosidase activity. According to recent reports the glycosynthase product yields have successfully been improved by this type of mutagenesis (32). Withers (49) produced a mutant Agrobacterium β-glucosidase (Abg) which converted 90% of the monosaccharides into oligosaccharides, demonstrating the efficiency of enzyme engineering in this field of research.
ManB is the first β-mannosidase enzyme characterized in detail in a representative of the thermophilic aerobic lignocellulose-degrading bacteria. The present work revealed one of the missing links in the lignocellulose-degrading system of these organisms. Proofs of the intracellular location of ManB are especially worthy of mentioning, as this finding points out that the compost-inhabiting bacteria and fungi follow basically different strategies for utilizing the final breakdown products of mannose containing polysaccharides. Bacteria, such as T. fusca or C. fimi, use intracellular enzymes for this purpose to prevent monosaccharide uptake by alien microbes, whereas fungi, such as Trichoderma, may rely on an extracellular mannosidase, as the strong antagonistic activity exerted by these organisms precludes the growth of other microbes in their microenvironment.
Acknowledgments
This work was supported by grants from OTKA (T 32407), FKFP (0315/99), and IUAP (P5/03).
We thank Gyöngyi Gyémánt for the HPLC measurements.
REFERENCES
- 1.Ademark, P., R. P. De Vries, P. Hägglund, H. Stålbrand, and J. Visser. 2001. Cloning and characterization of Aspergillus niger genes encoding an alpha-galactosidase and a beta-mannosidase involved in galactomannan degradation. Eur. J. Biochem. 268:2982-2990. [DOI] [PubMed] [Google Scholar]
- 2.Ademark, P., J. Lundqvist, P. Hägglund, M. Tenkanen, N. Torto, F. Tjerneld, and H. Stålbrand. 1999. Hydrolytic properties of a β-mannosidase purified from Aspergillus niger. J. Biotechnol. 75:281-289. [DOI] [PubMed] [Google Scholar]
- 3.Ademark, P., A. Varga, J. Medve, V. Harjunpaa, T. Drakenberg, F. Tjerneld, and H. Stålbrand. 1998. Softwood hemicellulose-degrading enzymes from Aspergillus niger: purification and properties of a beta-mannanase. J. Biotechnol. 63:199-210. [DOI] [PubMed] [Google Scholar]
- 4.Alkhayat, A. H., S. A. Kraemer, J. R. Leipprandt, M. Macek, W. J. Kleijer, and K. H. Friderici. 1998. Human beta-mannosidase cDNA characterization and first identification of a mutation associated with human beta-mannosidosis. Hum. Mol. Genet. 7:75-83. [DOI] [PubMed] [Google Scholar]
- 5.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Apweiler, R., T. K. Attwood, A. Bairoch, A. Bateman, E. Birney, M. Biswas, P. Bucher, L. Cerutti, F. Corpet, M. D. R. Croning, R. Durbin, L. Falquet, W. Fleischmann, J. Gouzy, H. Hermjakob, N. Hulo, I. Jonassen, D. Kahn, A. Kanapin, Y. Karavidopoulou, R. Lopez, B. Marx, N. J. Mulder, T. M. Oinn, M. Pagni, F. Servant, C. J. A. Sigrist, and E. M. Zdobnov. 2001. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 29:37-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bacic, A., P. J. Harris, and B. A. Stone. 1988. Structure and function of plant cell walls. p. 279-371. In J. Preiss (ed.), The biochemistry of plants: a comprehensive treatise, vol. 14. Academic Press, New York, N.Y.
- 8.Barr, B. K., Y. L. Hsieh, B. Ganem, and D. B. Wilson. 1996. Identification of two functionally different classes of exocellulases. Biochemistry 35:586-592. [DOI] [PubMed] [Google Scholar]
- 9.Bauer, M. W., E. J. Bylina, R. V. Swanson, and R. M. Kelly. 1996. Comparison of a beta-glucosidase and a beta-mannosidase from the hyperthermophilic archaeon Pyrococcus furiosus. Purification, characterization, gene cloning, and sequence analysis. J. Biol. Chem. 271:23749-23755. [DOI] [PubMed] [Google Scholar]
- 10.Beccari, T., L. Bibi, S. Stinchi, J. L. Stirling, and A. Orlacchio. 2001. Mouse beta-mannosidase: cDNA cloning, expression, and chromosomal localization. Biosci. Rep. 21:315-323. [DOI] [PubMed] [Google Scholar]
- 11.Blanchard, J. E., and S. G. Withers. 2001. Rapid screening of the aglycone specificity of glycosidases: applications to enzymatic synthesis of oligosaccharides. Chem. Biol. 8:627-633. [DOI] [PubMed] [Google Scholar]
- 12.Blanco, J., J. J. Coque, J. Velasco, and J. F. Martin. 1997. Cloning, expression in Streptomyces lividans and biochemical characterization of a thermostable endo-beta-1,4-xylanase of Thermomonospora alba ULJB1 with cellulose-binding ability. Appl. Microbiol. Biotechnol. 48:208-217. [DOI] [PubMed] [Google Scholar]
- 13.Bouquelet, S., G. Spik, and J. Montreuil. 1978. Properties of a β-D-mannosidase from Aspergillus niger. Biochim. Biophys. Acta 522:521-530. [DOI] [PubMed] [Google Scholar]
- 14.Chen, H., J. R. Leipprandt, C. E. Traviss, B. L. Sopher, M. Z. Jones, K. T. Cavanagh, and K. H. Friderici. 1995. Molecular cloning and characterization of bovine beta-mannosidase. J. Biol. Chem. 270:3841-3848. [DOI] [PubMed] [Google Scholar]
- 15.Clarke, J. H., J. E. Rixon, A. Ciruela, H. J. Gilbert, and G. P. Hazlewood. 1997. Family-10 and family-11 xylanases differ in their capacity to enhance the bleachability of hardwood and softwood paper pulps. Appl. Microbiol. Biotechnol. 48:177-183. [DOI] [PubMed] [Google Scholar]
- 16.Coutinho, P. M., and B. Henrissat. 1999. The modular structure of cellulases and other carbohydrate-active enzymes: an integrated database approach, p. 15-23. In K. Ohmiya, K. Hayashi, K. Sakka, Y. Kobayashi, S. Karita, and T. Kimura (ed.), Genetics, biochemistry and ecology of cellulose degradation. Uni Publishers, Tokyo, Japan.
- 17.Duffaud, G. D., C. M. McCutchen, P. Leduc, K. N. Parker, and R. M. Kelly. 1997. Purification and characterization of extremely thermostable beta-mannanase, beta-mannosidase, and alpha-galactosidase from the hyperthermophilic eubacterium Thermotoga neapolitana 5068. Appl. Environ. Microbiol. 63:169-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elbein, A. D., S. Adya, and Y. C. Lee. 1977. Purification and properties of a β-mannosidase from Aspergillus niger. J. Biol. Chem. 252:2026-2031. [PubMed] [Google Scholar]
- 19.Hartree, E. F. 1972. Determination of protein: a modification of the Lowry method that gives a linear photometric response. Anal. Biochem. 48:422-427. [DOI] [PubMed] [Google Scholar]
- 20.Hilge, M., S. M. Gloor, W. Rypniewski, O. Sauer, T. D. Heightman, W. Zimmermann, K. Winterhalter, and K. Piontek. 1998. High-resolution native and complex structures of thermostable beta-mannanase from Thermomonospora fusca—substrate specificity in glycosyl hydrolase family 5. Structure 6:1433-1444. [DOI] [PubMed] [Google Scholar]
- 21.Hunter, I. S. 1985. Gene cloning in Streptomyces, p. 19-44. In D. M. Glover (ed.), DNA cloning: a practical approach. IRL Press, Oxford, United Kingdom.
- 22.Irwin, D., E. D. Jung, and D. B. Wilson. 1994. Characterization and sequence of a Thermomonospora fusca xylanase. Appl. Environ. Microbiol. 60:763-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Irwin, D., D. H. Shin, S. Zhang, B. K. Barr, J. Sakon, P. A. Karplus, and D. B. Wilson. 1998. Roles of the catalytic domain and two cellulose binding domains of Thermomonospora fusca E4 in cellulose hydrolysis. J. Bacteriol. 180:1709-1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jung, E. D., G. Lao, D. Irwin, B. K. Barr, A. Benjamin, and D. B. Wilson. 1993. DNA sequences and expression in Streptomyces lividans of an exoglucanase gene and an endoglucanase gene from Thermomonospora fusca. Appl. Environ. Microbiol. 59:3032-3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kahle, V., and K. Tesaøik. 1980. Separation of saccharides and their anomers by high-performance liquid chromatography. J. Chromatogr. 191:121-128. [Google Scholar]
- 26.Kieser, D., and R. D. Melton. 1988. Plasmid pIJ699, a multicopy positive selection vector for Streptomyces. Gene 65:83-91. [DOI] [PubMed] [Google Scholar]
- 27.Kukolya, J., C. Dobolyi, and L. Hornok. 1997. Isolation and identification of thermophilic cellulolytic actinomycetes. Acta Phytopathol. Entomol. Hung. 32:97-107. [Google Scholar]
- 28.Kulminskaya, A. A., E. V. Eneiskaya, L. S. Isaeva-Ivanova, A. N. Savel'ev, I. A. Sidorenko, K. A. Shabalin, A. M. Golubev, and K. N. Neustroev. 1999. Enzymatic activity and β-galactomannan binding property of β-mannosidase from Trichoderma reesei. Enzyme Microb. Technol. 25:372-377. [Google Scholar]
- 29.Kurakake, M., and T. Komaki. 2001. Production of beta-mannanase and beta-mannosidase from Aspergillus awamori K4 and their properties. Curr. Microbiol. 42:377-380. [DOI] [PubMed] [Google Scholar]
- 30.Lao, G., G. S. Ghangas, E. D. Jung, and D. B. Wilson. 1991. DNA sequences of three beta-1,4-endoglucanase genes from Thermomonospora fusca. J. Bacteriol. 173:3397-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lubomir, K., and B. Peter. 1998. Disaccharides permeases: constituents of xylanolytic and mannanolytic systems of Aureobasidium pullulans. Biochim. Biophys. Acta 1425:560-566. [DOI] [PubMed] [Google Scholar]
- 32.Moracci, M., A. Trincone, and M. Rossi. 2001. Glycosynthases: new enzymes for oligosaccharide synthesis. J. Mol. Catal. B Enzyme 11:155-163. [Google Scholar]
- 33.Neustroev, K. N., A. S. Krylov, L. M. Firsov, O. L. Abroskina, and A. Y. Khorlin. 1991. Isolation and properties of β-mannosidase from Aspergillus awamori. Biokhimiya 56:1406-1412. [Google Scholar]
- 34.Nashiru, O., D. L. Zechel, D. Stoll, T. Mohammadzedeh, A. J. Warren, and S. G. Withers. 2001. β-Mannosynthase: synthesis of β-mannosides with a mutant β-mannosidase. Angew. Chem. Int. Ed. 40:417-420. [DOI] [PubMed] [Google Scholar]
- 35.O'Farrell, P. H. 1975. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 250:4007-4021. [PMC free article] [PubMed] [Google Scholar]
- 36.Parker, K. N., S. R. Chhabra, D. Lam, W. Callen, G. D. Duffaud, M. A. Snead, J. M. Short, E. J. Mathur, and R. M. Kelly. 2001. Galactomannanases Man2 and Man5 from Thermotoga species: growth physiology on galactomannans, gene sequence analysis, and biochemical properties of recombinant enzymes. Biotechnol. Bioeng. 75:322-333. [DOI] [PubMed] [Google Scholar]
- 37.Sachslehner, A., G. Foidl, N. Foidl, G. Gubitz, and D. Haltrich. 2000. Hydrolysis of isolated coffee mannan and coffee extract by mannanases of Sclerotium rolfsii. J. Biotechnol. 80:127-134. [DOI] [PubMed] [Google Scholar]
- 38.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 40.Schlosser, A., and H. Schrempf. 1996. A lipid-anchored binding protein is a component of an ATP-dependent cellobiose/cellotriose-transport system from the cellulose degrader Streptomyces reticuli. Eur. J. Biochem. 242:332-338. [DOI] [PubMed] [Google Scholar]
- 41.Scigelova, M., S. Singh, and D. H. G. Crout. 1999. Glycosidases a great synthetic tool. J. Mol. Catal. B Enzyme 6:483-494. [Google Scholar]
- 42.Simpson, A. J., F. C. Reinach, P. Arruda, F. A. Abreu, M. Acencio, R. Alvarenga, L. M. Alves, J. E. Araya, G. S. Baia, C. S. Baptista, M. H. Barros, E. D. Bonaccorsi, S. Bordin, J. M. Bove, M. R. Briones, M. R. Bueno, A. A. Camargo, L. E. Camargo, D. M. Carraro, H. Carrer, N. B. Colauto, C. Colombo, F. F. Costa, M. C. Costa, C. M. Costa-Neto, L. L. Coutinho, M. Cristofani, E. Dias-Neto, C. Docena, H. El-Dorry, A. P. Facincani, A. J. Ferreira, V. C. Ferreira, J. A. Ferro, J. S. Fraga, S. C. Franca, M. C. Franco, M. Frohme, L. R. Furlan, M. Garnier, G. H. Goldman, M. H. Goldman, S. L. Gomes, A. Gruber, P. L. Ho, J. D. Hoheisel, M. L. Junqueira, E. L. Kemper, J. P. Kitajima, C. L. Marino, et al. 2000. The genome sequence of the plant pathogen Xylella fastidiosa. Nature 406:151-157. [DOI] [PubMed] [Google Scholar]
- 43.Spiridonov, N. A., and D. B. Wilson. 2001. Cloning and biochemical characterization of BglC, a β-glucosidase from the cellulolytic actinomycete Thermobifida fusca. Curr. Microbiol. 42:295-301. [DOI] [PubMed] [Google Scholar]
- 44.Stoll, D., S. He, S. G. Withers, and R. A. Warren. 2000. Identification of Glu-519 as the catalytic nucleophile in beta-mannosidase 2A from Cellulomonas fimi. Biochem. J. 3:833-838. [PMC free article] [PubMed] [Google Scholar]
- 45.Stoll, D., H. Stalbrand, and R. A. Warren. 1999. Mannan-degrading enzymes from Cellulomonas fimi. Appl. Environ. Microbiol. 65:2598-2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takada, G., T. Kawaguchi, T. Kaga, J. Sumitani, and M. Arai. 1999. Cloning and sequencing of beta-mannosidase gene from Aspergillus aculeatus no. F-50. Biosci. Biotechnol. Biochem. 63:206-209. [DOI] [PubMed] [Google Scholar]
- 47.Trigo, C., and A. S. Ball. 1994. Production of extracellular enzymes during the solubilization of straw by Thermomonospora fusca BD25. Appl. Microbiol. Biotechnol. 41:366-372. [DOI] [PubMed] [Google Scholar]
- 48.Warren, R. A. J. 1996. Microbial hydrolysis of polysaccharides. Annu. Rev. Microbiol. 50:183-212. [DOI] [PubMed] [Google Scholar]
- 49.Withers, S. G. 2001. Mechanisms of glycosyl transferases and hydrolases. Carbohydr. Polym. 44:325-337. [Google Scholar]
- 50.Wray, W., T. Boulikas, V. P. Wray, and R. Hancock. 1981. Silver staining of proteins in polyacrylamide gels. Anal. Biochem. 118:197-203. [DOI] [PubMed] [Google Scholar]