Abstract
The bacterial strain MM-B16, which showed strong antifungal and antioomycete activity against some plant pathogens, was isolated from a mountain forest soil in Korea. Based on the physiological and biochemical characteristics and 16S ribosomal DNA sequence analysis, the bacterial strain MM-B16 was identical to Pseudomonas fluorescens. An antibiotic active against Colletotrichum orbiculare and Phytophthora capsici in vitro and in vivo was isolated from the culture filtrates of P. fluorescens strain MM-B16 using various chromatographic procedures. The molecular formula of the antibiotic was deduced to be C10H11NO2S (M+, m/z 209.0513) by analysis of electron impact mass spectral data. Based on the nuclear magnetic resonance and infrared spectral data, the antibiotic was confirmed to have the structure of a thiazoline derivative, aerugine [4-hydroxymethyl-2-(2-hydroxyphenyl)-2-thiazoline]. C. orbiculare, P. capsici, and Pythium ultimum were most sensitive to aerugine (MICs for these organisms were approximately 10 μg ml−1). However, no antimicrobial activity was found against yeasts and bacteria even at concentrations of more than 100 μg ml−1. Treatment with aerugine exhibited a significantly high protective activity against development of phytophthora disease on pepper and anthracnose on cucumber. However, the control efficacy of aerugine against the diseases was in general somewhat less than that of the commercial fungicides metalaxyl and chlorothalonil. This is the first study to isolate aerugine from P. fluorescens and demonstrate its in vitro and in vivo antifungal and antioomycete activities against C. orbiculare and P. capsici.
Chemical fungicides not only may pollute the atmosphere but also can be environmentally harmful, as the chemicals spread out in the air and accumulate in the soil (40). Furthermore, the repeated use of such chemicals has encouraged the development of chemical resistance in the target organisms (9). Despite the undesirable problems caused by the use of synthetic fungicides, fungicides will be increasingly applied in agriculture in the near future, provided that safer and ecologically friendly fungicides become available. The requirements for the fungicides to be practically used in fields are excellent potency against a variety of plant pathogens and safety, not only for humans, animals, and host plants but also for the ecosystems. These concerns highlight the need for selective fungicides with higher degradability in nature. Fungicides of microbial origin, which are synthesized biologically, have been demonstrated to be not only specifically effective on the target organisms but also inherently biodegradable (62). Microbial products are so complex in their chemical structures that one compound may have two or more totally different chemical moieties which can interact with different receptors (31). The complexity in their structures may make natural products chemically and biologically diverse. However, the use of microbial products as fungicides has limitations. One disadvantage is that they may suffer from the emergence of natural resistance in plant pathogens due to their highly specific mechanisms of action (62).
Since the first success in the use of streptomycin for the control of pear fire blight caused by Erwinia amylovora in the United States (62), many attempts have been made to find microbial products available only for plant disease control. Eventually, an epoch-making substance, named blasticidin S, active against rice blast disease, was discovered in Japan (55). Since a variety of antibiotics have been screened from microbial metabolites, some have been put into practical use in agriculture for the control of fungal plant diseases. For instance, kasugamycin (57), polyoxins (20), and validamycin A (21) and mildiomycin (12) have been commercially used for control of rice blast, rice sheath blight, and powdery mildew of rose, respectively. Strong in vitro antifungal activities have recently been discovered from the microbial metabolites. Gopalamicin (37), tubercidin (17), a manumycin-type antibiotic (16), oligomycin A (25), streptimidone (24), daunomycin (26), rhamnolipid B (23), and phenylacetic acid (18) were also found to have a potent antifungal or antioomycete activity for control of economically important plant diseases.
Some pseudomonads have been recognized as antagonists of plant fungal pathogens and antibiotic producers (44). This is probably due to the abundance of this diverse group of bacteria and their obvious importance in the soils (5). A single strain of Pseudomonas can produce several different antibiotics. A similar spectrum of antibiotic production has been described in different strains (5). For instance, Pseudomonas fluorescens strain Pf-5 has been demonstrated to synthesize different antibiotics such as 2,4-diacetylphloroglucinol, pyoluteorin, and hydrogen cyanide (42). The P. fluorescens strain CHA0 has also been known to produce pyrrolnitrin, pyoluteorin, and 2,4-diacetylphloroglucinol (47). Antifungal antibiotics produced by Pseudomonas spp. include pyrrolnitrin (3), phenazines (56), pyoluteorin (14), 2, 4-diacetylphloroglucinol (50), rhamnolipids (23), oomycin A (15), cepaciamide A (22), and ecomycins (36). In particular, the pseudomonad product pyrrolnitrin serves as a lead compound for new fungicides fenpiclonil and fludioxonil (43). Antibacterial antibiotics pseudomonic acid (8) and azomycin (52), antitumor antibiotics FR901463 (39) and cepafungins (51), and antiviral antibiotic karalicin (30) have also been isolated from Pseudomonas spp.
In the present study, we isolated the bacterial antagonist MM-B16 of plant pathogenic fungi and oomycetes from the soil of a mountain forest in Korea. The bacterial strain MM-B16 that showed significant antifungal and antioomycete activity against Colletotrichum orbiculare and Phytophthora capsici was identified to be P. fluorescens. An antibiotic was purely isolated from the culture filtrates of P. fluorescens strain MM-B16 using various purification procedures. The chemical structure of the antibiotic was determined and identified to be aerugine, based on spectroscopic and physicochemical analyses. The in vitro and in vivo antifungal and antioomycete activities of aerugine were also evaluated against some plant diseases. The isolation of an antifungal and antioomycete thiazoline derivative, aerugine, from P. fluorescens is reported for the first time in the present study.
MATERIALS AND METHODS
Isolation of antibiotic-producing bacterial strain MM-B16.
The bacterial strain MM-B16, which showed strong in vitro antifungal and antioomycete activity against some plant pathogens such as Alternaria mali, Colletotrichum gloeosporioides, Fusarium oxysporum f. sp. cucumerinum, Magnaporthe grisea, P. capsici, and Rhizoctonia solani, was isolated from the forest soil of the mountain Man-jul, Chun-an, Korea. The strain MM-B16 was selected for identification and production of antifungal and antioomycete antibiotic substances. The strain MM-B16 was cultured in nutrient agar at 28 ± 2°C. For long-term maintenance, strain MM-B16 was preserved in nutrient broth containing 15% (vol/vol) glycerol at −70°C.
Identification of bacterial strain MM-B16.
The physiological and biochemical characteristics of the antagonistic bacterial strain MM-B16 were examined using the methods of Lanyi (32), Palleroni (45), and Smibert and Krieg (54). The morphological features of the strain MM-B16, including shape, size, or type of flagella, were examined by transmission electron microscopy of the bacterial cells from exponentially growing cultures on tryptic soy agar (Difco) for 24 h at 28°C (2). The cells of strain MM-B16 were negatively stained with 1% (wt/vol) phosphotungstic acid and were examined after the grids were air dried. Identification of the strain MM-B16 was also conducted using the Biolog system (Biolog Inc., Hayward, Calif.), based on the 95 different carbon source utilization profiles of the strain MM-B16. For the determination of 16S ribosomal DNA (rDNA) sequences of the strain MM-B16, chromosomal DNA was isolated using a method of Pospiech and Neumann (46). The 16 rRNA genes were amplified using PCR with the universal primers fD1 (5′-AGAGT TTGAT CCTGG CTCAG-3′) and rP2 (5′-ACGGC TACCT TGTTA CGACTT-3′) (59). Thermal cycling was performed with the GeneAmp PCR system (model 2400; Perkin-Elmer). The thermal profile used was 35 cycles consisting of 1 min of denaturation at 95°C, 1 min of annealing at 58°C, and 2 min of extension at 72°C. A final extension step consisting of 8 min at 72°C was included. Amplified 16S rDNA was purified from agarose gel by using the method of Wu et al. (61). Purified rDNA was ligated into the pCR2.1-TOPO T vector (Invitrogen Co., Carlsbad, Calif.) according to the manufacturer's instructions. Ligated plasmid was then transformed into Escherichia coli TOP10 cell (Invitrogen Co.) by electroporation. The transformants were selected on the basis of the blue-white screening procedure (48). Plasmids containing 16S rDNA were isolated using the Wizard plus SV Minipreps DNA purification system (Promega Co., Madison, Wis.). These RNA genes were sequenced on an AB1310 DNA sequencer (Applied Biosystems) using a Big Dye Terminator cycle sequencing ready reaction kit (PE Applied Biosystems, Foster City, Calif.). The 16S rDNA sequence of the strain MM-B16 was aligned with reference sequences obtained from the databases of GenBank (National Center for Biotechnology Information, Bethesda, Md.) using a Clustal method (DNASTAR Inc., Madison, Wis.). A phylogenetic tree was constructed based on the percentage of difference in the genetic relationships between the strains.
Production and isolation of antibiotic.
The 5-ml inoculum of P. fluorescens strain MM-B16 precultured in nutrient broth for 24 h at 28°C was transferred into 500 ml of modified King's medium B (KB) (30 g of glycerol, 10 g of proteose peptone, 0.5 g of K2HPO4, 0.5 g of MgSO4 · 7H2O, 1 liter of H2O [pH 7.6]), which was optimal for production of antibiotic substances. The fermentation was carried out for 6 days at 28°C on a rotary shaker at 150 rpm. A total of 50 liters of cultures of P. fluorescens strain MM-B16 was obtained for further isolation of antibiotics. The cells of P. fluorescens strain MM-B16 were removed from the culture broth (50 liters) by centrifugation at 30,000 × g. The culture filtrates were combined and applied to a column packed with Diaion HP-20 (Mitsubishi Chemicals, Tokyo, Japan) (5 liters), a hydrophobic, polyaromatic adsorption resin. The column was washed three times with 5 liters of H2O and then eluted with stepwise gradients of aqueous methanol (0, 20, 40, 60, 80, and 100%, vol/vol). Each fraction (10 liters) of the eluates was concentrated, and the antifungal activity of each fraction against C. orbiculare, P. capsici, and R. solani was evaluated using the paper disk-agar diffusion method. The concentrated crude antibiotics (5 liters) in aqueous layers were extracted three times with an equal volume of ethyl acetate. The concentrated ethyl acetate layers were pooled and purified using C18 reversed-phase flash chromatography. The column packed with C18 resin (Lichroprep RP-18 [40 to 63 μm mesh]; Merck) was washed three times with 3 liters of H2O and then eluted with stepwise gradients of aqueous methanol (0, 20, 40, 60, 80, and 100%, vol/vol). Each fraction (3 liters) of the eluates was concentrated and bioassayed for antifungal activity. The fractions with antifungal activity obtained from C18 reversed-phase flash chromatography were subjected to preparative silica gel thin-layer chromatography (TLC) plates (60F254 [20 by 20 cm, 2 mm thick]; Merck). The TLC plates were developed with a solvent system of chloroform-methanol (90:10, vol/vol). Each band on the plates was scrapped and extracted with methanol. The methanol extracts were concentrated under reduced pressure, followed by evaluation of the antifungal activity. The antifungal substances obtained by preparative TLC were further purified by gel-filtration chromatography using Sephadex LH-20 resin (Pharmacia). The Sephadex LH-20 column (26 by 950 mm) was eluted with methanol at a flow rate of 0.15 ml min−1. The 2 ml of each fraction was harvested using a fraction collector (RediFrac; Pharmacia), and all the fractions were bioassayed for antifungal activity. The putative antibiotic substances purified by Sephadex LH-20 column chromatography were subjected to high-pressure liquid chromatography (HPLC) using a C18 reversed-phase column (SymmetryPrep C18 [7 μm thick; 7.8 by 300 mm]; Waters). The HPLC was performed on a Gilson (Middleton, Wis.) system with a linear gradient from 50% to 100% methanol at a flow rate of 1.5 ml min−1. The eluates of each peak were collected at 248 nm by a 118 UV/VIS detector (Gilson). Finally, 41.0 mg of the pure antibiotic substance was obtained from a single peak with the retention time of 21.88 min at 248 nm.
Structure elucidation of antibiotic.
Nuclear magnetic resonance (NMR) spectra were recorded on a Bruker (Billerica, Mass.) AMX 500 NMR spectrometer. The 1H NMR spectrum was measured in deuterated methanol. Chemical shifts are given in δ values (parts per million) referenced to the methyl group of solvent, with 3.30 ppm as an internal standard. The 13C NMR spectrum was recorded in deuterated methanol using the broad-band proton decoupling. The distortionless enhancement by polarization transfer (DEPT) spectrum was measured in deuterated methanol with a pulse width, θ, of 90° and 135°. Chemical shifts of the 13C NMR and DEPT spectra are given in δ values (ppm) referenced to the methyl group of solvent at 49.9 ppm in deuterated methanol. The 1H-1H correlation spectroscopy (COSY) spectrum was recorded in a phase-sensitive, double-quantum mode using the standard pulse sequence in deuterated methanol. Heteronuclear correlations via multiple bond connectivities (HMBC) and heteronuclear multiple quantum coherence (HMQC) spectroscopies were performed on a Bruker AMX 500 NMR spectrometer. The low-resolution mass spectra were recorded on a VG70-VSEQ mass spectrometer (VG Analytical, Manchester, United Kingdom) using the electron impact (EI) and fast atom bombardment (FAB) electron ionization method. The high-resolution EI and FAB mass spectra were recorded on a JEOL (Peabody, Mass.) JMS-HX110/110A tandem mass spectrometer. Fourier transform (FT) infrared (IR) spectroscopy was done on a Bio-Rad (Randolph, Mass.) instrument with the antibiotic embedded in a matrix of anhydrous KBr and pressed into a pellet. The UV absorption spectrum was measured with a model DU650 spectrophotometer (Beckman Instruments Inc., Fullerton, Calif.).
Detection of in vitro antimicrobial activity.
MICs of aerugine against fungi, oomycetes, yeasts, and bacteria were determined in a 24-well microtiter dish (Cell Wells; Corning Glass Works, Corning, N.Y.) using a modified method from Nair et al. (38). The inocula used in this test were a mycelial suspension of R. solani; spore suspensions (105 spores ml−1) of the other fungi and oomycetes, such as A. mali, Aspergillus niger, Botrytis cinerea, Cladosporium cucumerinum, C. orbiculare, Cylindrocarpon destructans, Didymella bryoniae, F. oxysporum f. sp. lycopersici, M. grisea, P. capsici, Pythium ultimum, and Sclerotinia sclerotiorum; and cell suspensions (104 CFU ml−1) of Candida albicans, Saccharomyces cerevisiae, Bacillus subtilis, Erwinia carotovora subsp. carotovora, Ralstonia solanacearum, and Xanthomonas campestris pv. vesicatoria. A 10-μl suspension was added to each well containing 1 ml of potato dextrose broth (Difco) supplemented with aerugine in the range of 0 to 100 μg ml−1. The lowest concentrations of aerugine that completely inhibited mycelial, yeast, and bacterial growth were considered to be MICs.
In vivo evaluation of antifungal and antioomycete activity.
The control efficacy of aerugine against P. capsici infection on pepper plants was tested in a growth room. Pepper plants were raised in a growth room at 28 ± 2°C with approximately 80 μmol of photons m−2 s−1 (white fluorescent lamps) for 16 h per day. The commercial fungicide metalaxyl was used to compare the antioomycete activity with aerugine. Aerugine and metalaxyl both dissolved in methanol were diluted to give concentrations of 1, 10, 50, 100, and 500 μg ml−1. Each of the chemical solutions was sprayed on the surface of a pepper plant at the first-branch stage 1 day before inoculation with P. capsici. The pepper plants treated with aerugine and metalaxyl were wounded by making 1-cm longitudinal slits on the stems 1 cm from the soil surface. The zoospore suspension was prepared as described previously (27). The sterile cotton dipped in zoospore suspension (105 zoospores ml−1) was placed on the wounded sites of stem. The inoculated sites were covered with plastic tape to maintain moisture. Disease severity on pepper plants was rated daily after inoculation based on a scale from 0 to 5 as follows: 0, no visible disease symptoms; 1, leaves slightly wilted with brownish lesions beginning to appear on stems; 2, 30 to 50% of entire plant diseased; 3, 50 to 70% of entire plant diseased; 4, 70 to 90% of entire plant diseased; or 5, plant dead. Data are the means of results for six plants per treatment. The protective efficacy of aerugine on cucumber (Cucumis sativus L. cv. Baekrokdadaki) plants against foliar necrotic anthracnose caused by C. orbiculare was evaluated in a growth room at 28 ± 2°C with approximately 80 μmol of photons m−2 s−1 (white fluorescent lamps) for 16 h per day. Aerugine and the commercial fungicide chlorothalonil dissolved in methanol and water, respectively, were diluted with 0.05% (vol/vol) Tween 20 solution to give concentrations of 10, 50, 100, and 500 μg ml−1. Each of the chemical solutions was sprayed on the primary and secondary leaves of cucumber plants 1 day before inoculation with C. orbiculare at the third-leaf stage. Conidial suspensions (106 spores ml−1) of 14-day-old C. orbiculare, cultured on potato dextrose agar at 28°C, in 0.05% (vol/vol) Tween 20 solution were sprayed on the leaves of cucumber plants. The inoculated plants were placed in a dew chamber to maintain approximately 100% humidity at 28 ± 1°C for 24 h and then transferred to the growth room for further incubation. Lesions on the primary or secondary leaves of cucumber plants were counted 6 days after inoculation. Data are the means of lesion numbers per square centimeter of leaf area of six plants.
Statistical analysis.
Statistical analyses were conducted with the Statistical Analysis System for personal computers (SAS Institute, Cary, N.C.). Fisher's protected least significant difference at P = 0.05 was applied to determine whether differences between treatments were significant.
Nucleotide sequence accession number.
The nucleotide sequence of 16S rDNA of strain MM-B16 has been deposited in the EMBL/GenBank database under the accession number AY196702.
RESULTS AND DISCUSSION
Identification of bacterial strain MM-B16.
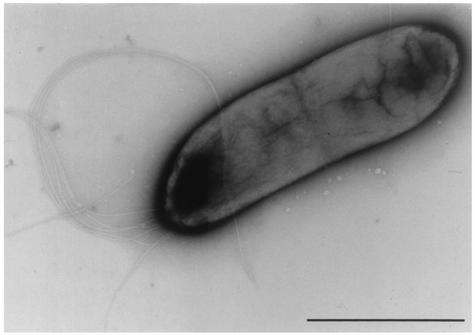
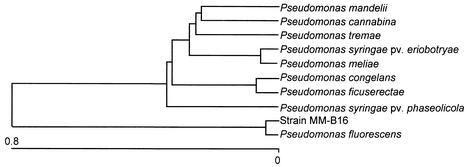
Under the transmission electron microscope, the cells of strain MM-B16 were ascertained to be rod shaped and have polar lophotrichous flagella (Fig. 1). The cell size of strain MM-B16 was found to be 0.4 to 0.6 μm by 1.7 to 2.1 μm. Strain MM-B16 was identified as an aerobic, motile, non-spore-forming, gram-negative bacterium. The strain MM-B16 produced a diffusible, fluorescent light green pigment on KB, but did not accumulate poly-β-hydroxybutyrate within the cells. The strain MM-B16 exhibited catalase, oxidase, urease, lecithinase, and arginine dihydrolase activity. Nitrate reduction to nitrite was not found, but oxidative response was detected in the oxidative/fermentative test. Indole, levan, or hydrogen sulfide was not produced. The reactions of methyl red and Voges-Proskauer were negative. The strain MM-B16 utilized citrate. Casein, gelatin, and Tween 80 were hydrolyzed. No hydrolysis of esculin and starch was observed. Strain MM-B16 grew optimally at pH 5 to 10 but was inhibited at pHs less than 5.0. Strain MM-B16 also grew at 4 to 37°C but not at temperatures above 40°C. Strain MM-B16 tolerated well up to 5% NaCl in the basal medium. The strain MM-B16 utilized N-acetyl-d-glucosamine, d-alanine, l-arabinose, l-arginine, d-fructose, d-glucose, meso-inositol, d-mannitol, d-mannose, trehalose, and l-valine but did not utilize adonitol, d-arabinose, d-cellobiose, d-galactose, lactose, maltose, l-rhamnose, d-sorbitol, sucrose, d,l-tartrate, and d-xylose. The similarity of strain MM-B16 with P. fluorescens in the 95 carbon source utilization profiles was found to be 0.923, which was the highest of the other microorganisms (data not shown), indicating that strain MM-B16 matched best with P. fluorescens. These results were obviously confirmed by the 16S rDNA sequence analysis of the strain MM-B16. The phylogenetic tree determined by the Clustal method showed that P. fluorescens was most closely related to the strain MM-B16 (Fig. 2). Among the 1,488 16S rDNA nucleotides of strain MM-B16 (GenBank accession number AY196702), 1,486 were identical to those of P. fluorescens ATCC 17386 (GenBank accession number AF094726), indicating that the sequence similarity of the 16S rDNA of the two bacteria was 99.9%. Based on the data of the physiological and biochemical characteristics and the 16S rDNA sequence analysis in the present study, the bacterial strain MM-B16 was identical to P. fluorescens.
FIG. 1.
Transmission electron micrograph of bacterial strain MM-B16 grown on tryptic soy agar for 24 h. Bar = 1 μm.
FIG. 2.
Phylogenetic location of bacterial strain MM-B16 based on 16S rDNA sequences. The phylogenetic tree was constructed based on the percent difference in the genetic relationships between the allied strains. The length of each pair of branches represents the distance between sequence pairs, while the units at the bottom of the tree indicate the number of substitution events.
Fluorescent pseudomonads have been known to produce a variety of metabolites, such as peptides, phenazines, pyocompounds, pyroles, and pterines (33), many of which are inhibitory to microorganisms and some of which are implicated in the biological control of plant diseases (5). Among the Pseudomonas spp., P. fluorescens has been well demonstrated to produce important antifungal metabolites, such as phenazine-1-carboxylic acid (56), phloroglucinol (50), pyoluteorin (14), pyrrolnitrin (3), and hydrogen cyanide (HCN) (58). In recent years, several antifungal antibiotics active against plant pathogens have been isolated from P. fluorescens and evaluated for efficacy of control of plant diseases. For instance, the cyclic peptide antibiotic tensin (41) exhibited strong antifungal activity against R. solani. The phenolic antibiotic 2-acetamidophenol (53) showed antifungal activity against take-all disease of wheat. The antifungal activity of sulfur-containing antibiotics N-mercapto-4-formylcarbostyril (6) against Fusarium sp. was also detected.
Production and purification of antibiotic.
Among the tested media having different carbon and nitrogen sources, the crude extracts from the culture filtrates of P. fluorescens strain MM-B16 in a modified KB exhibited the highest inhibitory effects on the mycelial growth of C. orbiculare, P. capsici, and R. solani. Carbon and nitrogen sources in culture media have been suggested to affect the types and levels of the antibiotics produced (5). In the present study, addition of the glycerol and proteose peptone, as carbon and nitrogen sources in modified KB, remarkably accelerated the production of antibiotic substances from the cultures of the strain MM-B16. Production of the antibiotic substances active against C. orbiculare, P. capsici, and R. solani gradually increased in modified KB 2 days after inoculation of P. fluorescens strain MM-B16 and reached a maximum level at 6 days, which indicates that the strain MM-B16 requires a relatively long period to produce antibiotic substances in modified KB compared to other media. The growth of strain MM-B16 reached a stationary phase 5 days after inoculation. Many secondary metabolites, including antibiotics, begin to be produced by microorganisms at the stationary phase when cell multiplication ceases (60). It seems likely that involvement of glycerol in modified KB may limit the vegetative growth of the strain MM-B16 but significantly accelerate the biosynthesis of antibiotic substances.
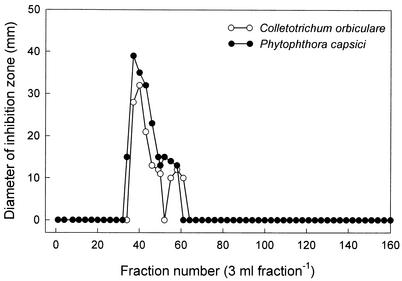
Individual antibiotics active against plant pathogenic fungi and oomycetes were purified from the culture filtrates (50 liters) of P. fluorescens strain MM-B16 using different chromatography procedures. The eluates of 60 and 80% methanol on a Diaion HP-20 column were most effective in inhibiting the plant pathogens C. orbiculare, P. capsici, and R. solani (data not shown). The fractions were combined and extracted with ethyl acetate. The concentrated ethyl acetate extracts were purified by a reversed-phase C18 flash column chromatography with stepwise gradients of methanol and water. The fractions eluted with 40 and 60% methanol that significantly reduced the mycelial growth of C. orbiculare, P. capsici, and R. solani were concentrated to a small volume and subjected to preparative silica gel TLC plates. The bands at the position of Rf 0.75 on the TLC plates exhibited strong antifungal activity against C. orbiculare and P. capsici. The antibiotics purified by preparative TLC were chromatographed on a Sephadex LH-20 column. Antifungal and antioomycete activities against C. orbiculare and P. capsici were found in fractions 34 to 61 (Fig. 3). These fractions were pooled and subjected to a preparative HPLC for further purification of antibiotic substances. An antibiotic which showed significant activities against C. orbiculare and P. capsici was finally obtained from a single peak at the retention time of 21.88 min at 248 nm. A total of 41.0 mg of the antibiotic was yielded from 50 liters of culture extracts of P. fluorescens strain MM-B16. The antibiotic, obtained as a pale brownish white powder, was soluble in most of the organic solvents, such as methanol, ethanol, chloroform, ethyl acetate, and dimethyl sulfoxide, but was poorly soluble in water. The maximum UV adsorption of the antibiotic was shown at 248 and 313 nm in methanol.
FIG. 3.
Sephadex LH-20 column chromatogram of active fractions from the eluates of preparative TLC. The Sephadex LH-20 column (26 by 950 mm) was eluted with methanol at a flow rate of 0.15 ml min−1. All fractions were bioassayed for the antifungal and antioomycete activity against C. orbiculare and P. capsici using the paper disk-agar diffusion method.
Structure elucidation of antibiotic.
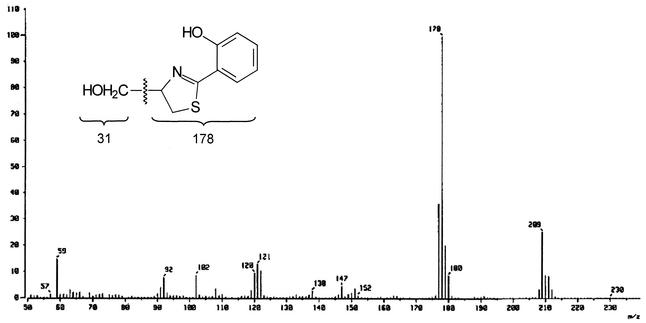
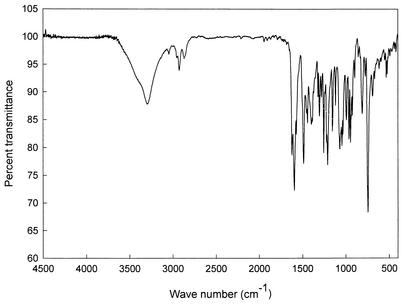
The structure of the antibiotic was determined by NMR, IR, and mass spectral analyses. The EI mass spectrum of the antibiotic confirmed its molecular weight to be 209 (Fig. 4). The major peak at m/z 178 in the mass spectrum reflects the molecular ion split out of a hydroxymethyl fragment. An ion with m/z 178 in the mass spectrum of the deuterium analogue of the antibiotic was shifted by one mass unit (data not shown). The molecular formula of the antibiotic was deduced as C10H11NO2S based on the analysis of high-resolution EI mass spectral data showing a molecular ion peak at m/z 209.0513 (M+) (calculated 209.0511), which is in accordance with the molecular composition (C10H11NO2S) (data not shown). The IR spectrum showed the following major absorption bands: 1,525 (C=C), 1,566, 1,606, 1,625, 3,010 (aromatic proton), and 3,335 (OH) cm−1 (Fig. 5). The structure of the antibiotic was further elucidated by 1H NMR and 13C NMR spectroscopy, DEPT experiments, and various two-dimension NMR spectral studies, including 1H-1H COSY, HMBC, HMQC, nuclear Overhauser effect spectroscopy (ROESY), and rotating frame nuclear Overhauser effect spectroscopy (NOESY). The proton and carbon counts from 1H and 13C NMR spectra and the DEPT experiment indicated that the 11 protons and 10 carbons are present in its structure, thus supporting the molecular formula of the antibiotic.
FIG. 4.
EI mass spectrum of the antibiotic aerugine.
FIG. 5.
FT-IR spectrum of the antibiotic aerugine.
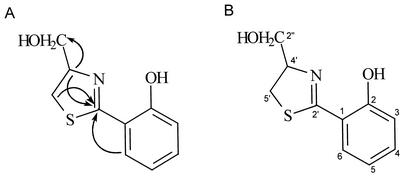
The 1H and 13C NMR spectral data of the antibiotic and their assignments are shown in Table 1. The 1H NMR spectrum indicated the presence of methine groups at δ 6.87, 6.92, and 7.41. From the 13C NMR spectrum, the presence of aromatic carbons was confirmed at positions δ 117.6, 117.8, 120.0, 131.6, and 134.1. The 13C NMR spectrum also indicated the presence of a hydroxyl carbon at δ 160.1. These results suggest that the structure of the antibiotic contained an aromatic group attached with a hydroxy moiety. The carbonyl-like moiety (C☰N) was detected at position δ 173.7 of the 13C NMR spectrum. The proton resonance at δ 3.29 and 3.45, corresponding to a methylene group, was attributed to a sulfur-bearing carbon atom, which was confirmed at position δ 33.8 of the 13C NMR spectrum. By analysis of the 13C NMR spectrum, the presence of a nitrogen-bearing carbon atom was confirmed at δ 79.4. The hydroxymethyl group was detected at positions δ 3.77 and 3.80 of the 1H NMR spectrum and δ 64.3 of the 13C NMR spectrum of the antibiotic. By the HMBC spectral analysis, the correlations of carbonyl-like carbon (δ 173.7, C-2′) with methylene protons (δ 3.29 and 3.45, H-5′) and a proton (δ 4.81, H-4′) attached with a carbon-bearing nitrogen were confirmed (Fig. 6A). These results indicate the presence of a thiazoline ring in the structure of the antibiotic. The HMBC spectrum also showed the correlations of the carbonyl-like carbon (δ 173.7, C-2′) with the aromatic proton (δ 7.41, H-6). The correlations of hydroxymethyl carbon (δ 64.3, C-2") with a proton (δ 4.81, H-4′) attached with a carbon-bearing nitrogen were ascertained by HMBC spectral analysis. These NMR and mass spectral results suggest that the antibiotic could be a thiazoline derivative containing a hydroxyphenyl and a hydroxymethyl group. In light of all the spectral data, the structure of the antibiotic was determined to be aerugine [4-hydroxymethyl-2-(2-hydroxyphenyl)-2-thiazoline] (molecular formula: C10H11NO2S) (Fig. 6B). The spectral data of the antibiotic were identical to those of aerugine presented by Zunnundzhanov et al. (64).
TABLE 1.
1H and 13C NMR spectral data of the antibiotic aerugine in CD3OD
| Carbon no. | 13C δa | 1H δb (mc, J in Hz) |
|---|---|---|
| 1 | 117.6 | |
| 2 | 160.1 | |
| 3 | 117.8 | 6.92 (br d, 1.0, 8.3) |
| 4 | 134.1 | 7.33 (dt, 1.6, 7.5, 8.3) |
| 5 | 120.0 | 6.87 (br t, 1.0, 7.5, 7.8) |
| 6 | 131.6 | 7.41 (dd, 1.6, 7.8) |
| 2′ | 173.7 | |
| 4′ | 79.4 | 4.81 (m) |
| 5′ | 33.8 | 3.29 (dd, 11.0, 7.8) |
| 3.45 (dd, 11.0, 8.8) | ||
| 2" | 64.3 | 3.77 (dd, 11.2, 6.0) |
| 3.80 (dd, 11.2, 5.2) |
125 MHz, chemical shift in parts per million.
500 MHz, chemical shift in parts per million.
Abbreviations of signal multiplicity: dd, doublet of doublets, dt, doublet of tiplets; brd, broad doublet; br t, broad triplet; m, multiplet.
FIG. 6.
(A). Correlations of partial structures of the antibiotic aerugine from HMBC spectra; (B) structure of the antibiotic aerugine isolated from P. fluorescens strain MM-B16.
Thiazolines have constituted a family of compounds known for their application in pharmaceutical and flavor chemistry (7). Many thiazoline derivatives have been chemically synthesized, while more than 30 thiazoline structures have been identified from natural sources such as bacteria (1) and actinomycetes (49). The thiazoline compounds have shown various biological activities such as antifungal activity (4), antibacterial activity (49), antitumor activity (34), antiviral activity (13) and anthelminthic activity (35). Recently, antimycoplasma activity (19) and antituberculosis activity (10) of the synthesized thiazoline derivatives have also been demonstrated. A variety of thiazoline compounds, such as curacin D (34), thiangazole (29), and watasemycins (49) have been isolated and characterized from the cyanobacterium Lyngbya majuscula, the myxobacterium Polyangium sp., and Streptomyces sp., respectively. Some thiazoline derivatives, β-methoxyacrylates melithiazols (1) produced by myxobacteria, and chemically synthesized quinazolones (11) have shown strong antifungal activity. In the present study, we isolated an antibiotic, identified as aerugine, 4-hydroxymethyl-2-(2-hydroxyphenyl)-2-thiazoline, from the culture filtrates of P. fluorescens strain MM-B16, which showed a high antifungal and antioomycete activity.
In vitro and in vivo antifungal and antioomycete activities of aerugine.
In most cases, antibiotics showing in vitro activity are generally active in vivo against plant diseases. The evaluation of in vitro antifungal activity of an antibiotic is the prerequisite for in vivo evaluation of its antifungal activity. In the present study, we ascertained not only in vitro antifungal and antioomycete activities of aerugine, against some plant pathogens, but also its in vivo antifungal and antioomycete activity in host plants. C. orbiculare, P. capsici, and P. ultimum were shown to be most sensitive to aerugine (the MICs for these organisms were approximately 10 μg ml−1) (Table 2). Aerugine also completely inhibited the mycelial growth of A. mali, B. cinerea, C. cucumerinum, C. destructans, D. bryoniae, and M. grisea, even at concentrations less than 50 μg ml−1. However, the growth of A. niger, F. oxysporum f. sp. lycopersici, R. solani, S. sclerotiorum, yeasts, and bacteria was not affected even at concentrations greater than 100 μg ml−1. These data indicated that aerugine was strongly inhibitory to plant pathogenic fungi and oomycetes, such as C. orbiculare, P. capsici, and P. ultimum, but did not exhibit antiyeast and antibacterial activities.
TABLE 2.
MICs of aerugine from P. fluorescens strain MM-B16 as determined by the microtiter broth dilution method
| Microorganismc | MIC (μg ml−1)a |
|---|---|
| Alternaria mali | 50b |
| Aspergillus niger | >100 |
| Botrytis cinerea | 30 |
| Cladosporium cucumerinum | 30 |
| Colletotrichum orbiculare | 10 |
| Cylindrocarpon destructans | 50 |
| Didymella bryoniae | 50 |
| Fusarium oxysporum f. sp. lycopersici | >100 |
| Magnaporthe grisea | 30 |
| Phytophthora capsici | 10 |
| Pythium ultimum | 10 |
| Rhizoctonia solani | >100 |
| Sclerotinia sclerotiorum | >100 |
| Candida albicans | >100 |
| Saccharomyces cerevisiae | >100 |
| Bacillus subtilis | >100 |
| Erwinia carotovora subsp. carotovora | >100 |
| Ralstonia solanacearum | >100 |
| Xanthomonas campestris pv. vesicatoria | >100 |
The lowest concentration of aerugine required for complete inhibition of microbial growth.
The value >100 represents growth of the microorganisms that was not completely inhibited by aerugine at a concentration of 100 μg ml−1.
Microorganisms include plant pathogenic fungi and oomycetes.
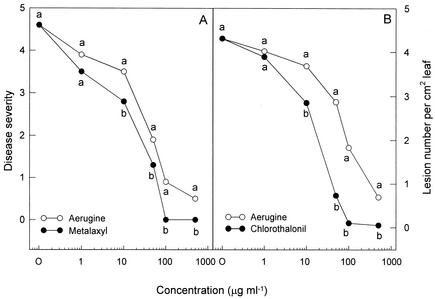
In vivo efficacy of aerugine and the commercial fungicide metalaxyl against phytophthora blight development in pepper plants is presented in Fig. 7A. As the concentration of the two compounds increased, phytophthora disease was gradually inhibited on the pepper plants at the first-branch stage. Treatments with aerugine and metalaxyl at 100 or 500 μg ml−1 exhibited a significantly high level of protective activity against P. capsici infection. The control efficacy against phytophthora blight was in general somewhat less than that of metalaxyl on pepper plants. Aerugine did not induce any phytotoxic symptoms on pepper plants even when they were treated with concentrations of 500 μg ml−1. These data suggest that aerugine from P. fluorescens may have a fungicidal potential against the oomycete pathogens. In vivo efficacy of aerugine and the commercial fungicide chlorothalonil for the control of anthracnose disease on cucumber plants was evaluated under greenhouse conditions. Treatments with the two compounds significantly reduced the C. orbiculare infection on cucumber leaves. Aerugine began to show protective activity against C. orbiculare infection at a concentration of 10 μg ml−1 (Fig. 7B). A few anthracnose lesions were produced on the cucumber leaves treated with aerugine at 500 μg ml−1. Chlorothalonil was more effective than aerugine in controlling the anthracnose disease on cucumber leaves. No disease symptoms were found on the cucumber leaves treated with chlorothalonil at 100 μg ml−1. Aerugine did not show any phytotoxicity on cucumber plants even when they were treated with concentrations of 500 μg ml−1.
FIG. 7.
In vivo efficacy of aerugine for control of plant diseases in host plants. (A) Effects of aerugine and metalaxyl on disease development in pepper plants inoculated with P. capsici at the first-branch stage. Disease severity is based on a scale from 0 to 5, where 0 indicates no visible symptoms and 5 indicates a dead plant. (B) Effects of aerugine and chlorothalonil on anthracnose development on cucumber leaves inoculated with C. orbiculare. Numbers of lesions on the leaves of cucumber plants were rated on day 6 after inoculation. Means at each concentration labeled with the same letter are not significantly different (P = 0.05) according to the least significant difference test.
Aerugine, a sulfur-containing thiazoline compound, the structure of which was established as 4-hydroxymethyl-2-(2-hydroxyphenyl)-2-thiazoline, was first discovered from culture filtrates of Pseudomonas aeruginosa strain 590 (64). More recently, aerugine has been synthesized by the oxidation of the siderophore pyochelin, which is structurally similar to aerugine (63). Carmi and Carmeli (4) isolated an analogous compound of aerugine, (+)-(S)-dihydroaeruginoic acid, from cultures of P. fluorescens strain PFM2, which exhibited its antimicrobial activity against some plant pathogenic fungi and bacteria. However, there is little information about the isolation and biological activities of aerugine and its analogues from other microorganisms. To our knowledge, this is the first study to isolate aerugine from P. fluorescens and demonstrate its in vitro and in vivo antifungal and antioomycete activities against C. orbiculare and P. capsici.
Taken together, all these results led us to conclude that aerugine produced by P. fluorescens strain MM-B16 has not only potent in vitro antifungal and antioomycete activities against some plant pathogens but also in vivo control efficacy against Phytophthora disease on pepper plants and anthracnose development on cucumber plants. However, further evaluation of the effectiveness of aerugine for disease control and the design of aerugine formulations and applications should be done in the field.
Acknowledgments
This research was financially supported by a grant (KRF-2001-041-G00017) from the Korea Research Foundation in 2001. J. Y. Lee is a recipient of a predoctoral fellowship (grant no. KRF-2000-908-G00002) from the Korea Research Foundation.
We thank Eun Jung Bang at the Korea Basic Science Institute for NMR spectroscopy.
REFERENCES
- 1.Bohlendorf, B., M. Herrmann, H. J. Hecht, F. Sasse, E. Forche, B. Kunze, H. Reichenbach, and G. Hofle. 1999. Antibiotics from gliding bacteria, 85 melithiazoles A-N: new antifungal β-methoxyacrylates from myxobacteria. Eur. J. Org. Chem. 1999:2601-2608. [Google Scholar]
- 2.Bozzola, J. J., and L. D. Russel. 1991. Electron microscopy: principles and techniques for biologists. Jones & Bartlett, Boston, Mass.
- 3.Burkhead, K. D., D. A. Schisler, and P. J. Slininger. 1994. Pyrrolnitrin production by biological control agent Pseudomonas cepacia B37w in culture and in colonized wounds of potatoes. Appl. Environ. Microbiol. 60:2031-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carmi, R., and S. Carmeli. 1994. (+)-(S)-Dihydroaeruginoic acid, an inhibitor of Septoria tritici and other phytopathogenic fungi and bacteria, produced by Pseudomonas fluorescens. J. Nat. Prod. 57:1200-1205. [DOI] [PubMed] [Google Scholar]
- 5.Dowling, D. N., and F. O'Gara. 1994. Metabolites of Pseudomonas involved in the biocontrol of plant disease. Trends Biotechnol. 12:133-141. [Google Scholar]
- 6.Fakhouri, W., F. Walker, B. Vogler, W. Armbruster, and H. Buchenauer. 2001. Isolation and identification of N-mercapto-4-formylcarbostyril, an antibiotic produced by Pseudomonas fluorescens. Phytochemistry 58:1297-1303. [DOI] [PubMed] [Google Scholar]
- 7.Fernandez, X., R. Fellous, and E. Dunach. 2000. Novel synthesis of 2-thiazolines. Tetrahedron Lett. 41:3381-3384. [Google Scholar]
- 8.Fuller, A. T., G. Mellows, M. Woolford, G. T. Banks, K. D. Barrow, and E. B. Chain. 1971. Pseudomonic acid: an antibiotic produced by Pseudomonas fluorescens. Nature 234:416-417. [DOI] [PubMed] [Google Scholar]
- 9.Goldman, G. H., C. Hayes, and G. E. Harman. 1994. Molecular and cellular biology of biocontrol by Trichoderma spp. Trends Biotechnol. 12:478-482. [DOI] [PubMed] [Google Scholar]
- 10.Gursoy, A., and N. Karali. 2000. 4-(3-Coumarinyl)-4-thiazolin-2-one-benzylidenehydrazones with antituberculosis activity. Arzneimittel-Forschung 50:167-172. [DOI] [PubMed] [Google Scholar]
- 11.Habib, M. S., and M. A. Khalil. 1984. Synthesis and antimicrobial activity of novel quinazolone derivatives. J. Pharm. Sci. 73:982-985. [DOI] [PubMed] [Google Scholar]
- 12.Harada, S., and T. Kishi. 1978. Isolation and characterization of mildiomycin, a new nucleoside antibiotic. J. Antibiot. 31:519-524. [DOI] [PubMed] [Google Scholar]
- 13.Harnden, M. R., S. Bailey, M. R. Boyd, J. B. Wilkinson, and N. D. Wright. 1979. Easily hydrolyzable, water-soluble derivatives of (+/-)-α-5-[1-(indol-3-yl)ethyl]-2-methylamino-δ-2-thiazoline-4-one, a novel antiviral compound. J. Med. Chem. 22:191-195. [DOI] [PubMed] [Google Scholar]
- 14.Howell, C. R., and R. D. Stipanovic. 1980. Suppression of Pythium ultimum-induced damping-off of cotton seedlings by Pseudomonas fluorescens and its antibiotic, pyoluteorin. Phytopathology 70:712-715. [Google Scholar]
- 15.Howie, W. J., and T. V. Suslow. 1991. Role of antibiotic biosynthesis in the inhibition of Pythium ultimum in the cotton spermosphere and rhizosphere by Pseudomonas fluorescens. Mol. Plant-Microbe Interact. 4:393-399. [Google Scholar]
- 16.Hwang, B. K., J. Y. Lee, B. S. Kim, and S. S. Moon. 1996. Isolation, structure elucidation, and antifungal activity of a manumycin-type antibiotic from Streptomyces flaveus. J. Agric. Food Chem. 44:3653-3657. [Google Scholar]
- 17.Hwang, B. K., S. J. Ahn, and S. S. Moon. 1994. Production, purification, and antifungal activity of the antibiotic nucleoside, tubercidin, produced by Streptomyces violaceoniger. Can. J. Bot. 72:480-485. [Google Scholar]
- 18.Hwang, B. K., S. W. Lim, B. S. Kim, J. Y. Lee, and S. S. Moon. 2001. Isolation and in vivo and in vitro antifugal activity of phenylacetic acid and sodium phenylacetate from Streptomyces humidus. Appl. Environ. Microbiol. 67:3739-3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ino, A., Y. Hasegawa, and A. Murabayashi. 1998. Total synthesis of the antimycoplasma antibiotic micacocidin. Tetrahedron Lett. 39:3509-3512. [Google Scholar]
- 20.Isono, K., J. Nagatsu, K. Kobinata, K. Sasaki, and S. Suzuki. 1965. Studies on polyoxins, antifungal antibiotics. Part I. Isolation and characterization of polyoxins A and B. Agric. Biol. Chem. 29:848-854. [Google Scholar]
- 21.Iwasa, T., E. Higashide, H. Yamamoto, and M. Shibata. 1970. Studies on validamycins, new antibiotics. II. Production and biological properties of validamycin A and B. J. Antibiot. 23:595-602. [DOI] [PubMed] [Google Scholar]
- 22.Jiao, Y., T. Yoshihara, S. Ishikuri, H. Uchino, and A. Ichihara. 1996. Structural identification of cepaciamide A, a novel fungitoxic compound from Pseudomonas cepacia D-202. Tetrahedron Lett. 37:1039-1042. [Google Scholar]
- 23.Kim, B. S., J. Y. Lee, and B. K. Hwang. 2000. In vivo control and in vitro antifungal activity of rhamnolipid B, a glycolipid antibiotic, against Phytophthora capsici and Colletotrichum orbiculare. Pest Manag. Sci. 56:1029-1035. [Google Scholar]
- 24.Kim, B. S., S. S. Moon, and B. K. Hwang. 1999. Isolation, antifungal activity, and structure elucidation of the glutarimide antibiotic, streptimidone, produced by Micromonospora coerulea. J. Agric. Food Chem. 47:3372-3380. [DOI] [PubMed] [Google Scholar]
- 25.Kim, B. S., S. S. Moon, and B. K. Hwang. 1999. Isolation, identification, and antifungal activity of a macrolide antibiotic, oligomycin A, produced by Streptomyces libani. Can. J. Bot. 77:850-858. [Google Scholar]
- 26.Kim, B. S., S. S. Moon, and B. K. Hwang. 2000. Structure elucidation and antifungal activity of an anthracycline antibiotic, daunomycin, produced by Actinomadura roseola. J. Agric. Food Chem. 48:1875-1881. [DOI] [PubMed] [Google Scholar]
- 27.Kim, Y. J., B. K. Hwang, and K. W. Park. 1989. Expression of age-related resistance in pepper plants infected with Phytophthora capsici. Plant Dis. 73:745-747. [Google Scholar]
- 28.Klingler, J. M., R. P. Stowe, D. C. Obenhuber, T. O. Groves, S. K. Mishra, and D. L. Pierson. 1992. Evaluation of the Biolog automated microbial identification system. Appl. Environ. Microbiol. 58:2089-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kunze, B., R. Jansen, L. Pridzun, E. Jurkiewicz, G. Hunsmann, G. Hofle, and H. Reichenbach. 1993. Thiangazole, a new thiazoline antibiotic from Polyangium sp. (myxobacteria): production, antimicrobial activity and mechanism of action. J. Antibiot. 46:1752-1755. [DOI] [PubMed] [Google Scholar]
- 30.Lampis, G., D. Deidda, C. Maullu, S. Petruzzelli, and R. Pompei. 1996. Karalicin, a new biologically active compound from Pseudomonas fluorescens/putida. I. Production, isolation, physico-chemical properties and structure elucidation. J. Antibiot. 49:260-262. [DOI] [PubMed] [Google Scholar]
- 31.Lange, L., and C. S. Lopez. 1996. Micro-organisms as a source of biologically active secondary metabolites, p. 1-26. In L. G. Copping (ed.), Crop protection agents from nature: natural products and analogues. The Royal Society of Chemistry, London, United Kingdom.
- 32.Lanyi, B. 1987. Classical and rapid identification methods for medically important bacteria. Methods Microbiol. 19:1-67. [Google Scholar]
- 33.Leisinger, T., and R. Margraff. 1979. Secondary metabolites of the fluorescent pseudomonads. Microbiol. Rev. 43:422-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marquez, B., P. Verdier-Pinard, E. Hamel, and W. H. Gerwick. 1998. Curacin D, an antimitotic agent from the marine cyanobacterium Lyngbya majuscula. Phytochemistry 49:2387-2389. [DOI] [PubMed] [Google Scholar]
- 35.McFarland, J. W., H. L. Howes, L. H. Conover, J. E. Lynch, W. C. Austin, and D. H. Morgan. 1970. Novel anthelmintic agents. V. Thiazoline and dihydrothiazine analogues of pyrantel. J. Med. Chem. 13:113-119. [DOI] [PubMed] [Google Scholar]
- 36.Miller, C. M., R. V. Miller, D. G. Kenny, B. Redgrave, J. Sears, M. M. Condron, D. B. Teplow, and G. A. Strobel. 1998. Ecomycins, unique antimycotics from Pseudomonas viridiflava. J. Appl. Microbiol. 84:937-944. [DOI] [PubMed] [Google Scholar]
- 37.Nair, M. G., A. Chandra, and D. L. Thorogood. 1994. Gopalamicin, an antifungal macrolide produced by soil actinomycetes. J. Agric. Food Chem. 42:2308-2310. [Google Scholar]
- 38.Nair, M. G., S. K. Mishra, and A. R. Putnam. 1992. Antifungal anthracycline antibiotics, spartanamicins A and B from Micromonospora spp. J. Antibiot. 45:1738-1745. [DOI] [PubMed] [Google Scholar]
- 39.Nakajima, H., B. Sato, T. Fujita, S. Takase, H. Terano, and M. Okuhara. 1996. New antitumor substances, FR901463, FR901464 and FR90 1465. I. Taxonomy, fermentation, isolation, physico-chemical properties and biological activities. J. Antibiot. 49:1196-1203. [DOI] [PubMed] [Google Scholar]
- 40.Nannipieri, P. 1994. The potential use of soil enzymes as indicators of productivity, sustainability and pollution, p. 33-140. In C. E. Pankhurst, B. M. Doube, V. V. S. R. Gupta, and P. R. Grace (ed.), Soil biota-management in sustainable farming systems. CSIRO, Collingwood, Australia.
- 41.Nielsen, T. H., C. Thrane, C. Christophersen, U. Anthoni, and J. Sørensen. 2000. Structure, production characteristics and fungal antagonism of tensin—a new cyclic lipopeptide from Pseudomonas fluorescens strain 96.578. J. Appl. Microbiol. 89:992-1001. [DOI] [PubMed] [Google Scholar]
- 42.Nowak-Thompson, B., S. J. Gould, J. Kraus, and J. E. Loper. 1994. Production of 2,4-diacetylphloroglucinol by the biocontrol agent Pseudomonas fluorescens Pf-5. Can. J. Microbiol. 40:1064-1066. [Google Scholar]
- 43.Nyfeler, R., and P. Ackermann. 1992. Phenylpyrroles, a new class of agricultural fungicides related to the natural antibiotic pyrrolnitrin, p. 395-404. In D. R. Baker, J. G. Fenyes, and J. J. Steffens (ed.), Synthesis and chemistry of agrochemicals III. ACS symposium series 504. American Chemical Society, Washington, D.C.
- 44.O'Sullivan, D. J., and F. O'Gara. 1992. Traits of fluorescent Pseudomonas spp. involved in suppression of plant root pathogens. Microbiol. Rev. 56:662-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Palleroni, N. J. 1989. Gram-negative aerobic rods and cocci, p. 140-219. In N. R. Krieg and J.G. Holt (ed.), Bergey’s manual of systematic bacteriology. Williams & Wilkins, Baltimore, Md.
- 46.Pospiech, A., and B. Neumann. 1995. A versatile quick-prep of genomic DNA from gram-positive bacteria. Trends Genet. 11:217-218. [DOI] [PubMed] [Google Scholar]
- 47.Rosales, A. M., L. Thomashow, R. J. Cook, and T. W. Mew. 1995. Isolation and identification of antifungal metabolites produced by rice-associated antagonistic Pseudomonas sp. Phytopathology 85:1028-1032. [Google Scholar]
- 48.Sambrook, J., and D. W. Russel. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 49.Sasaki, T., Y. Igarashi, N. Saito, and T. Furumai. 2002. Watasemycins A and B, new antibiotics produced by Streptomyces sp. TP-A0597. J. Antibiot. 55:249-255. [DOI] [PubMed] [Google Scholar]
- 50.Shanahan, P., D. J. O'Sullivan, P. Simpson, J. D. Glennon, and F. O'Gara. 1992. Isolation of 2,4-diacetylphloroglucinol from a fluorescent pseudomonad and investigation of physiological parameters influencing its production. Appl. Environ. Microbiol. 58:353-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shoji, J., H. Hinoo, T. Kato, T. Hattori, K. Hirooka, K. Tawara, O. Shiratori, and T. Yoshihiro. 1990. Isolation of cepafungins I, II and III from Pseudomonas species. J. Antibiot. 43:783-787. [DOI] [PubMed] [Google Scholar]
- 52.Shoji, J., H. Hinoo, Y. Terui, J. Kikuchi, T. Hattori, K. Ishii, K. Matsumoto, and T. Yoshida. 1989. Isolation of azomycin from Pseudomonas fluorescens. J. Antibiot. 42:1513-1514. [DOI] [PubMed] [Google Scholar]
- 53.Slininger, P. J., K. D. Burkhead, D. A. Schisler, and R. J. Bothast. 2000. Isolation, identification, and accumulation of 2-acetamidophenol in liquid cultures of the wheat take-all biocontrol agent Pseudomonas fluorescens 2-79. Appl. Microbiol. Biotechnol. 54:376-381. [DOI] [PubMed] [Google Scholar]
- 54.Smibert, R. M., and N. R. Krieg. 1994. Phenotypic characterization, p. 607-654. In P. Gerhardt, R. G. E. Murray, W. A. Wood, and N. R. Krieg (ed.), Methods for general and molecular bacteriology. American Society for Microbiology, Washington, D.C.
- 55.Takeuchi, S., K. Hirayama, K. Ueda, H. Sasaki, and H. Yonehara. 1957. Blasticidin S, a new antibiotic. J. Antibiot. 11:1-5. [PubMed] [Google Scholar]
- 56.Thomashow, L. S., and D. M. Weller. 1988. Role of a phenizine antibiotic from Pseudomonas fluorescens in biological control of Gaeumannomyces graminis var. tritici. J. Bacteriol. 170:3499-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Umezawa, H., Y. Okami, T. Hashimoto, Y. Suhara, M. Hamada, and T. Takeuchi. 1965. A new antibiotic, kasugamycin. J. Antibiot. 18:101-103. [PubMed] [Google Scholar]
- 58.Voisard, C., C. Keel, D. Haas, and G. Defago. 1989. Cyanide production by Pseudomonas fluorescens helps suppress black root of tobacco under gnotobiotic conditions. EMBO J. 8:351-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weisburg, W. G., M. S. Barns, A. D. Pelletier, and J. D. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Woodruff, H. B. 1988. Natural products from microorganisms. Science 208:1225-1229. [DOI] [PubMed] [Google Scholar]
- 61.Wu, W., M. J. Welsh, P. B. Kaufman, and H. H. Zhang. 1997. Methods in gene biotechnology. CRC Press, New York, N.Y.
- 62.Yamaguchi, I. 1996. Pesticides of microbial origin and applications of molecular biology, p. 27-49. In L. G. Copping (ed.), Crop protection agents from nature: natural products and analogues. The Royal Society of Chemistry, London, United Kingdom.
- 63.Zamri, A., and M. A. Abdallah. 2000. An improved stereocontrolled synthesis of pyochelin, siderophore of Pseudomonas aeruginosa and Burkholderia cepacia. Tetrahedron 56:249-256. [Google Scholar]
- 64.Zunnundzhanov, A., I. A. Bessonova, N. D. Abdullaev, and D. K. Ogai. 1988. Structure of aerugine from Pseudomonas aeruginosa. Chem. Nat. Comp. 23:461-465. [Google Scholar]