Abstract
The study of free-living nitrogen-fixing organisms in bulk soil is hampered by the great diversity of soil microbial communities and the difficulty of relating nitrogen fixation activities to individual members of the diazotroph populations. We developed a molecular method that allows analysis of nifH mRNA expression in soil in parallel with determinations of nitrogen-fixing activity and bacterial growth. In this study, Azotobacter vinelandii growing in sterile soil and liquid culture served as a model system for nifH expression, in which sucrose served as the carbon source and provided nitrogen-limited conditions, while amendments of NH4NO3 were used to suppress nitrogen fixation. Soil RNA extraction was performed with a new optimized direct extraction protocol that yielded nondegraded total RNA. The RNA extracts were of high purity, free of DNA contamination, and allowed highly sensitive and specific detection of nifH mRNA by a reverse transcription-PCR. The level of nifH gene expression was estimated by PCR amplification of reverse-transcribed nifH mRNA fragments with A. vinelandii-specific nifH primers. This new approach revealed that nifH gene expression was positively correlated with bulk nitrogen fixation activity in soil (r2 = 0.72) and in liquid culture (r2 = 0.84) and therefore is a powerful tool for studying specific regulation of gene expression directly in the soil environment.
Fixation of atmospheric nitrogen (N) by free-living soil microorganisms is considered a minor source of bioavailable nitrogen compared to systems such as the Rhizobium-legume and Frankia-alder symbioses (22, 40). However, a capacity for asymbiotic N fixation is present in most soils (25, 42, 47, 51, 56), and potential soil diazotrophs are found in diverse bacterial groups, such as the Proteobacteria, the cyanobacteria, and the Firmicutes (42, 56, 57). Anthropogenic activities have led to increased nitrogen fertilization and atmospheric deposition of bioavailable nitrogen (NH4+, NO3−), thus reducing the extent of N limitation in many ecosystems. On the other hand, N fixation activities of free-living microorganisms can be induced or stimulated locally by increasing the C/N ratios or availability of suitable easily degradable C sources (21, 24, 43), such as decaying plant material or root exudates (40). Nitrogen-limited conditions may also be induced after application of organic amendments in agricultural systems (e.g., after straw incorporation into soil) (33, 46). The temporal and spatial niches and nutritional requirements for optimal activity of free-living diazotrophs remain largely unknown, and the role of these organisms in soil nutrient cycling is still poorly understood.
Most methods in classical microbiology require cultivation of the organism being studied and provide only limited information on in situ activities. In contrast, DNA- and RNA-targeting techniques allow in situ analysis of microbial community structures and activities in the soil environment (3, 17). DNA sequences with high phylogenetic information contents, like rRNA genes, have been used for community description and have greatly increased our knowledge of microbial diversity in the environment (37). More recently, functional genes (e.g., the nifH gene as a marker for N fixation) have also been used for this purpose (41, 56, 59). While DNA-based studies mainly provide information on community structure, studies of RNA and specifically analyses of mRNA expression provide additional information on activities of specific populations (17). Many studies have focused on describing gene expression and regulatory mechanisms in laboratory cultures. However, culture conditions differ greatly from the conditions in the natural soil environment; therefore, there is an increasing need to investigate gene expression directly in soil.
A more detailed understanding of the dynamics of N-fixing populations, the activities of specific groups, and the conditions required for induction of their N-fixing activities in soil may aid in devising and optimizing nitrogen management strategies for sustainable low-input farming and forestry. Although strategies based on deliberate release (4, 22) or stimulation of the natural diazotroph population (20, 21) have yielded variable results in the past, such strategies have considerable potential (21, 22). In order to advance research in this area, development of tools that allow in situ studies of gene expression and activities in the soil matrix is required.
In order to study gene expression in soil, a robust protocol for extraction of total RNA (more specifically, nondegraded mRNA) is essential. However, only a small number of studies have reported successful analyses of mRNA isolated from soil or sediment (9, 15, 19, 27, 29, 35, 53), and there have been no reports that directly link expression of genes involved in N fixation to an assay of nitrogenase activity in soil. Moreover, in most previous investigations high-density inoculation or very active communities were required in order to reliably detect mRNA. Reliable extraction of mRNA from soil is still considered a challenge in soil microbiological research (17). Recent progress in extraction technology, however, has shown that the approach is feasible (19).
Here we describe an effective total RNA extraction protocol based on a previously described direct extraction procedure for total nucleic acids (8). Azotobacter vinelandii, an aerobic free-living soil diazotroph, was cultivated in a previously sterilized soil and in liquid culture. This system was used to establish and verify a method for nifH mRNA extraction and detection by reverse transcription (RT) and PCR. N fixation was either induced by providing excess organic carbon (sucrose) or repressed by providing excess bioavailable N (NH4NO3). Population growth, bulk N-fixing activities, and nifH mRNA expression were monitored and compared in order to link nifH gene expression to N-fixing activity in a soil environment.
MATERIALS AND METHODS
Preparation of A. vinelandii inoculum.
Inoculum for liquid medium and sterile soil microcosms was prepared by growing A. vinelandii strain DSM 85 (German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany) to the late log phase (approximately 5 × 107 cells ml−1) in ATCC 14 medium (American Type Culture Collection, Manassas, Va.). Cells were centrifuged (10 min at 550 × g and 10°C) and resuspended in sterile saline (0.1% NaCl). The cell concentration of the inoculum was determined by direct cell counting with a Zeiss Axioplan microscope (Zeiss, Oberkochen, Germany) and a bright line counting chamber (Hausser Scientific, Horsham, Pa.). The cell concentration of the inoculum was adjusted to 105 cells ml−1 with sterile saline.
Preparation of liquid medium microcosms.
Liquid medium microcosms were prepared in 250-ml baffled Erlenmeyer flasks closed with cotton stoppers. The culture medium (ATCC 14 medium) contained 2% sucrose as a carbon source. One milliliter of inoculum was added to 99 ml of liquid medium, resulting in a starting concentration of 1 × 103 cells ml−1. Liquid culture treatments that received 1 ml of filter-sterilized 2 M NH4NO3 are referred to below as LC+N. The LC−N treatments received 1 ml of sterile water instead of NH4NO3. A sterile control (LC control) was identical to the LC+N treatment but received 1 ml of sterile saline instead of inoculum (Table 1).
TABLE 1.
Starting conditions for the experimental treatments and controls
| Treatment | Matrixa | A. vinelandii concn (cells ml−1 or cells g−1)b | Sucrose concn (%)c | NH4NO3 concn (μmol ml−1 or μmol g−1)d | No. of replicates |
|---|---|---|---|---|---|
| LC+N | Liquid medium | 1 × 103 | 2 | 20 | 2 |
| LC−N | Liquid medium | 1 × 103 | 2 | 0 | 2 |
| SC+N | Sterile soil | 1 × 103 | 2 | 20 | 2 |
| SC−N | Sterile soil | 1 × 103 | 2 | 0 | 2 |
| LC control | Liquid medium | 0 | 2 | 20 | 1 |
| SC control | Sterile soil | 0 | 2 | 20 | 1 |
| Reference soil | Nonsterile soil | NAe | 0 | 0 | 1 |
The Liquid medium was ATTC 14 medium, and the soil was Pappelacker (see text).
Strain DSM 85.
The concentration in liquid medium was 2% (wt/vol), and the concentration in soil was 2% (wt/wt).
Concentration of NH4NO3 added. The soil contained additional indigenous nitrogen.
NA, not applicable.
Preparation of soil microcosms.
Soil (Pappelacker) was obtained from Les Barges, Valais, Switzerland. The physical and chemical properties of this soil are as follows: soil unit, calcaric fluvisol (Food and Agriculture Organization); texture, sandy loam (U.S. Department of Agriculture); clay content, 4.2% (54); silt content, 20.3% (54); sand content, 75.5% (54); organic C content, 0.8% (54); organic N content, 0.09% (54); pH 7.5 (1 M KCl) (this study); water-holding capacity, 44.4% (54); and cell content (4′,6-diamidino-2-phenylindole [DAPI]-stained cells), 9 × 108 ± 1 × 108 cells g−1 (this study). Soil microcosms were prepared in 500-ml infusion bottles. One hundred grams of soil was placed in each bottle, and the bottles were closed with cotton stoppers and sterilized by autoclaving them three times for 30 min at 121°C with 2 days of incubation between autoclaving treatments. A nonsterilized sample was stored at 10°C and used as a reference soil for RNA extraction.
Sucrose crystals (molecular biology grade; Fluka, Buchs, Switzerland) were sterilized by exposure to UV light on a transilluminator (TS-36; UVP Inc., San Gabriel, Calif.) for 20 min. The treatments that received 2 g of sucrose crystals and 1 ml of sterile NH4NO3 (2 M) are referred to below as SC+N (Table 1). One milliliter of inoculum was sprinkled over the soil, resulting in a final concentration of 1 × 103 cells g of soil−1. The SC−N microcosms received 1 ml of sterile water instead of the NH4NO3 solution. A sterile control (SC control) was prepared as described above for the SC+N treatment, but it received 1 ml of sterile saline instead of inoculum (Table 1). The contents of all microcosms were thoroughly mixed. The final soil water content after inoculation was approximately 40% of the water-holding capacity and remained constant throughout the experiment.
Microcosm incubation conditions and sampling.
All microcosms (Table 1) were incubated in a climate chamber at 25°C for 1 week in the dark. The liquid medium was agitated on a rotary shaker at 150 rpm.
Samples for direct cell counting, determination of N fixation activity, and determination of ammonium concentration were taken immediately after inoculation and then every 24 h for 3 days. An endpoint measurement was obtained after 7 days. Nucleic acid extractions were performed 2 and 3 days after inoculation and also with the endpoint samples. On days 3 and 7 the liquid cultures were very dense and had accumulated high concentrations of exopolymeric substances. This made both centrifugation and filtration difficult or impossible. Therefore, determination of ammonium concentrations and extraction of nucleic acids could not be performed for some of the samples.
Cell counting.
To determine cell counts, a sample of soil or liquid culture was harvested from each microcosm. Direct cell counts in soil were determined by staining with the DNA-specific dye DAPI by using the method of Zarda et al. (58). Cell numbers and standard deviations were expressed as cells per gram of soil or cells per milliliter of medium.
Nitrogen fixation.
Nitrogen fixation was estimated by the acetylene (C2H2) reduction assay (2). Samples of soil (10 g) or culture (10 ml) were transferred into 60-ml serum bottles sealed with butyl stoppers. Approximately 10% of the remaining air volume in each bottle (50 ml) was replaced with 5 ml of acetylene. Incubation was performed under the conditions described above. The ethylene (C2H4) concentrations in 0.5-ml headspace samples were determined with an 8000 GC gas chromatograph equipped with a flame ionization detector (Carlo Erba Instruments, Milan, Italy) and a Hayesep N column (100/120 mesh; 2 m by 1.0 mm; BGB Analytik, Anwil, Switzerland) running at 75°C. Three measurements were obtained; the first measurement was obtained 5 min after addition of C2H4, and then two measurements were obtained at 1-h intervals. The rate of C2H4 production was determined from the slope of the linear regression of time versus C2H4 concentration and was expressed as nanomoles of C2H4 per hour per gram of soil or milliliter of medium. The significance of each rate determined was tested by using Excel's regression analysis tool.
Ammonium concentrations.
To determine the soluble ammonium concentration in soil, 1 g of soil was extracted with 1 ml of distilled H2O (45). In brief, the slurry was vortexed for 10 s, incubated for 30 min on ice, and vortexed again for 10 s. The supernatant was collected after centrifugation for 10 min at 9,500 × g and 4°C. For liquid medium microcosm samples, cells were removed by centrifugation, and the supernatants were filtered (pore size, 0.2 μm). The ammonium ion concentrations in 15-μl extract samples were determined as described by Nickel et al. (32) by ion chromatography with a Dionex DX-100 ion chromatograph equipped with an Ionpac SC12 column (Dionex, Sunnyvale, Calif.) and were quantified with Chrom-Card for Windows (Fision Instruments, Rodano, Italy). The results were expressed as nanomoles per gram (dry weight) of soil or nanomoles per milliliter of medium.
RNA extraction.
Three subsamples from each soil sample were extracted by a bead beating-based total nucleic acid extraction method (8), with minor changes. Briefly, 0.5 g of soil, 1.25 ml of extraction buffer (0.2% hexadecyltrimethylammonium bromide [CTAB], 1 mM 1,4-dithio-dl-threitol [DTT], 0.2 M sodium phosphate buffer [pH 8], 0.1 M NaCl, 50 mM EDTA), and 0.75 g of silica beads (diameter, 0.1 mm; B. Braun Biotech International GmbH, Melsungen, Germany) were processed in a bead beater (Bio 101/Savant, Farmingdale, N.Y.) for 45 s at 6 m s−1. After centrifugation, 850 μl of the supernatant was extracted with 1.5 ml of a 1:1 mixture of phenol (pH 8) and chloroform-isoamyl alcohol (24:1) (CIA) and then with 750 μl of CIA. Each extracted soil pellet was extracted again with 850 μl of fresh buffer and processed as described above. The extracts obtained from the subsamples of each microcosm and by using both extraction procedures were pooled in a 15-ml sterile Falcon tube. Aliquots (800 μl) were stored at −80°C until the end of the 7-day incubation period. Separate aliquots were processed for total nucleic acid and RNA extraction. Nucleic acid extracts were precipitated from thawed aliquots for 1 h at 37°C with 850 μl of RNase-free precipitating solution (20% polyethylene glycol, 2.5 M NaCl). After centrifugation at 16,000 × g for 30 min, the pellets were washed once with cold ethanol (70%). For direct use in PCR, pellets containing total nucleic acid extracts were dissolved in 100 μl of Tris-EDTA buffer. For preparation of pure RNA extracts, pellets were dissolved in 30 μl of 1× reaction buffer containing 3 U of RNase-free DNase I (Promega, Madison, Wis.) and incubated for 30 min at 25°C. DNA digestion was stopped by adding 125 μl of RNA extraction reagent (0.6% CTAB, 1 mM DTT, 50 mM sodium acetate [pH 4.5]; treated with 0.1% diethyl pyrocarbonate and autoclaved [10]). RNA extraction reagent should be at room temperature to avoid CTAB precipitation. Each solution was extracted once with 250 μl of a 1:1 mixture of phenol (pH 4.5) and CIA and once with 200 μl of CIA. The supernatant (150 μl) was transferred to a fresh tube, and then 1 μl of glycogen (10 mg ml−1; Amersham Biosciences Europe GmbH, Dübendorf, Switzerland) was added before the RNA was precipitated with 200 μl of isopropanol by incubation for 3 min at room temperature and for 3 min on ice. The tubes were centrifuged at 0°C for 10 min at 16,000 × g. The pellets were washed once with 500 μl of cold ethanol (70%), air dried, dissolved in 30 μl of cold RNase-free water, and stored at −80°C.
Cells were harvested from liquid medium microcosms by pipetting 10 ml of liquid culture into sterile 15-ml tubes on ice. To stop cell growth and conserve the RNA content, 1 ml of an ethanol-phenol mixture (5% phenol in 96% ethanol) was added. Samples were centrifuged (550 × g, 15 min, 4°C), and the pellets were stored at −80°C until the end of the experiment. For nucleic acid extraction, samples were thawed on ice, and the cells were resuspended in 1 ml (final volume) (day 2) or 5 ml (final volume) (days 3 and 7) of sterile H2O. Aliquots of each suspension (200 μl) were lysed with 100 μl of proteinase K (6 mg ml−1) and 3 μl of sodium dodecyl sulfate (10%) and incubated for 5 min at 37°C. The lysate was extracted once with 300 μl of a 1:1 mixture of phenol (pH 8) and CIA and once with 300 μl of CIA. Nucleic acids were precipitated with 750 μl of ethanol and 30 μl of sodium acetate (3 M, pH 5.2) at −80°C overnight. Centrifugation, RNA cleanup, and storage were then performed as described above for soil.
Quantification and quality of RNA.
RNA was quantified with a RiboGreen kit (Molecular Probes, Eugene, Oreg.) by using a method analogous to the method developed for quantification of DNA in soil extracts using Picogreen (8, 48); 1 μl of RNA extract or RNA standard and 400 μl of RiboGreen (1:4,000 in Tris-ETDA buffer) were combined. Fluorescence was determined with a luminescence spectrophotometer (LS 50 B; Perkin-Elmer, Rotkreuz, Switzerland) by using an excitation wavelength of 480 nm and an emission wavelength of 520 nm. Standards were prepared from the RNA standard solution supplied with the RiboGreen kit.
RNA quality was checked by electrophoresis of 6-μl RNA extracts (equivalent to RNA extracted from 0.1 g of soil or 0.4 ml of liquid culture) in 2% agarose gels together with a molecular size marker (1-kb DNA ladder; Life Technologies AG, Basel, Switzerland).
Primer design.
A. vinelandii-specific nifH primers were designed at the positions defined by Widmer et al. (56) for nested universal nifH PCR. The primer sequences were designed by using previously published nifH sequences for A. vinelandii (GenBank accession numbers M11579 and M20568). The sequence of primer nifH-g1-forB (forward) was GGTTGTGACCCGAAAGCTGA, and the sequence of primer nifH-g1-rev (reverse) was GCGTACATGGCCATCATCTC. Comparison of these primer sequences with sequences in the GenBank database verified the specificity of the primer set for previously published A. vinelandii nifH sequences.
RT and PCR.
Most RNA samples were diluted to a concentration of 3 μg ml−1 prior to RT; the only exceptions were reference soil samples (reference soil DNA concentration, 66 ng μl−1; reference soil RNA concentration, 32 ng μl−1). To remove secondary structure, a 5-μl aliquot of each of the RNA extracts was heated for 3 min at 65°C and immediately placed on ice. For cDNA synthesis, 20 μl of the RT reaction mixture (13) was added. The RT reaction mixture contained 50 mM KCl, 10 mM Tris (pH 8), 1.5 mM MgCl2, each deoxynucleoside triphosphate at a concentration of 0.1 mM, 15 U of avian myeloblastosis virus (AMV) reverse transcriptase (Amersham Biosciences), and 0.1 μM primer. The primer used for cDNA synthesis from nifH mRNA was nifH-γ1-rev, and the primer used for cDNA synthesis from small-subunit (SSU) rRNA was uni-b-rev (3, 7) (GACGGGCGGTGTGTRCAA; Microsynth, Balgach, Switzerland). The reaction mixtures were incubated for 5 min at 25°C and then at 42°C for 1 h. AMV was inactivated by heating the reaction mixtures at 95°C for 5 min before samples were stored at −20°C. To test for DNA contamination of the RNA extracts, control reaction mixtures were prepared as described above, but AMV reverse transcriptase was not added.
PCR conditions were adjusted to allow for sensitive detection of A. vinelandii nifH (data not shown). To allow for relative quantification of nifH mRNA, preliminary experiments were carried out with stepwise reduction of the number of PCR cycles to determine the maximum cycle number at which none of the samples reached the amplification plateau. For amplification of nifH cDNA, 1 μl of the RT reaction product was used in a subsequent PCR. The reaction mixture (final volume, 50 μl) contained 0.8 mM MgCl2, each deoxynucleoside triphosphate at a concentration of 0.2 mM, 0.2 μM primer nifH-g1-forB, 0.2 μM primer nifH-g1-rev, 5 mg of bovine serum albumin per ml, and 1× PCR buffer (Amersham Biosciences). Taq DNA polymerase (1 U) was added to the PCR mixture under hot-start conditions (56). The reaction conditions were 43 cycles of denaturation at 95°C for 11 s and at 92°C for 15s, annealing at 58°C for 8 s and at 60°C for 30 s, and extension at 74°C for 8 s and at 72°C for 10 s, followed by a final extension at 72°C for 5 min.
cDNA of SSU rRNA was amplified with bacterial SSU rRNA gene-specific primers (EUB338-for [3] and uni-b-rev) by using a previously described protocol (8), except that 35 cycles were used. Reaction products (nifH, 0.37 kb; bacterial SSU rRNA, 1.07 kb) were visualized with ethidium bromide DNA staining following agarose gel electrophoresis. Fragment sizes were confirmed by comparison with a 1-kb DNA ladder molecular weight marker (Life Technologies AG).
The intensities of RT-PCR products on agarose gels were quantified with the GelDoc 2000 system and the volume tool of the QuantityOne software (Bio-Rad, Hercules, Calif.). A background value was subtracted by using an average background reading from an empty lane at the position of the PCR product band. Data were expressed in arbitrary units of intensity per square millimeter.
Restriction fragment length polymorphism analyses (56) of RT-PCR-amplified bacterial SSU rRNA extracted after 7 days (soil) or after 3 days (liquid culture) indicated that all treatments still contained pure cultures of A. vinelandii (data not shown).
RESULTS AND DISCUSSION
RNA extraction.
In a number of studies, mRNA extracted from soil or sediment has been analyzed by using both specific detection of functional genes (5, 15, 19, 27, 35, 53) and differential display based on arbitrary primers (15). However, in a recent review Sayler et al. (49) noted the small number of studies that have been initiated to relate mRNA levels to specific microbial activities in soil (14, 15, 36). It is clear that the use of mRNA analysis as a standard tool in soil microbial ecology is still hindered by methodological difficulties. The objective of the present study was to develop and validate a set of methods which allow a combination of molecular analysis of nifH mRNA and measurement of cell numbers and enzymatic activities.
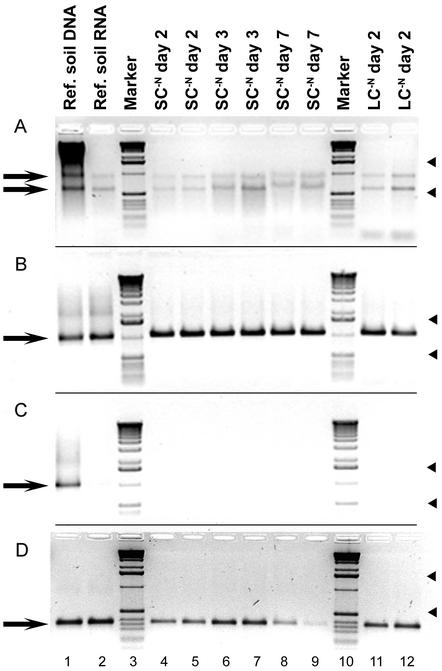
The protocol developed for soil RNA extraction yielded high-quality total RNA, as indicated by sharp bands of SSU and large-subunit rRNA and a lack of visible DNA contamination in the extracts (Fig. 1A). The suitability of extracted RNA for RT was demonstrated by RT-PCR of bacterial SSU rRNA. All samples yielded strong amplicons of the expected size (1,070 bp) (Fig. 1B). Control reactions in which the mixtures did not contain AMV reverse transcriptase in the RT step were performed to test for DNA contamination and were always negative (Fig. 1C). Only reference soil total nucleic acid extract yielded the expected positive PCR amplification. The presence of nifH mRNA was demonstrated by A. vinelandii-specific nifH RT-PCR, which yielded amplicons of the expected size (approximately 371 bp) with all samples (Fig. 1D).
FIG. 1.
Gel images illustrating the quality of RNA extracts and their suitability for RT-PCR. In each panel the arrows on the left indicate the position(s) of the expected band(s) on the gel. The arrowheads on the right indicate the positions of the 1,636- and 517- or 506-bp fragments of the molecular weight marker. Lane 1, total nucleic acid extract from reference soil; lane 2, total RNA extract from reference (Ref.) soil; lanes 4 to 9, duplicate SC−N RNA extracts (lanes 4 and 5, day 2; lanes 6 and 7, day 3; lanes 8 and 9, day 7); lanes 11 and 12, LC−N liquid culture RNA extracts from day 2; lanes 3 and 10, 1-kb DNA marker. (A) Six microliters (equivalent to RNA from 0.1 g of soil or 0.4 ml of liquid culture) of nucleic acid extract sample (2% agarose). (B) Six-microliter portions of 50-μl bacterial SSU rRNA RT-PCR products (1,070 bp; 1% agarose). Reference soil extracts were used undiluted, and all other samples were diluted to a concentration of 3 ng μl−1 prior to RT. (C) Six-microliter portions of 50-μl bacterial SSU rRNA PCR products used to test for DNA contamination (1% agarose). The reaction mixtures were identical to those shown in panel B except that they did not contain AMV reverse transcriptase in the RT step. (D) Six-microliter portions of 50-μl A. vinelandii nifH RT-PCR products (370 bp; 2% agarose).
Complete removal of DNA is crucial for highly sensitive RT-PCR assays, especially considering that high cycle numbers are necessary for amplification of nifH transcripts from soil. Therefore, several alternative methods for DNA removal were tested. To obtain pure RNA from the initial total nucleic acid extracts, DNA was selectively removed by combined use of DNase digestion and acid phenol extraction (6, 13), which proved to be more efficient than either method used individually. Skipping the DNase digestion step resulted in RNA extracts that appeared to be DNA free upon visual inspection after gel electrophoreses. However, positive amplification was observed in control RT-PCR mixtures not containing AMV reverse transcriptase, indicating that there was residual contamination with DNA (data not shown). Replacing the acid phenol extraction step with heat inactivation of the added DNase resulted in extracts with which the RT-PCR procedure was frequently unsuccessful. Similarly, the use of commercially available spin columns for DNA removal from the initial extracts also resulted in unsatisfactory RNA cleanup (data not shown).
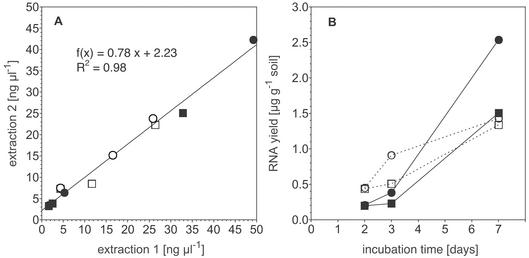
RNA quantification with RiboGreen was very reliable and sensitive. Addition of the RNA standard to RNA extracts indicated that there was a small quenching effect (<15%) that increased with cell density and was similar for soil and liquid culture extracts (data not shown). The reproducibility of the cleanup procedure and the potential for RNA degradation during storage were evaluated by repeating the protocol with raw extracts that had been stored for 4 months. Although the RNA concentrations in the second extraction were about 78% of those in the initial extraction, the concentration differences between samples were highly consistent (Fig. 2A). The RNA yield from soil increased with time for all treatments (Fig. 2B). The SC−N treatment with induced N-fixing populations yielded higher RNA concentrations at days 2 and 3. However, the highest RNA yields, 1.5 and 2.5 μg g−1, were obtained on day 7 with the NH4NO3-amended SC+N treatment (Fig. 2B). The RNA contents in soil were strongly determined by cell density (r2 = 0.83, P = 0.01). However, the extractable RNA content per cell was highest for both soil treatments on day 2 (SC−N, 41 and 36 ng of RNA per 106 cells; SC+N, 14 and 13 ng of RNA per 106 cells) and subsequently decreased to about 5 ng of RNA per 106 cells at days 3 and 7 for all treatments. The protocol described here emphasizes purity and quality over a high yield of extracted RNA. Nevertheless, when our RNA extraction method was tested with the nonsterile reference soil, the RNA yield (4.0 μg g of soil−1) was comparable to the yields obtained by previously described methods, which were between 1 and 3 μg g of soil−1 (6, 30, 53). Higher yields were reported by Hurt et al. (19), who obtained yields ranging from 7.3 to 56.1 μg g−1 for several soil samples. Sessitsch et al. (50) compared three different methods and various pretreatments and reported yields that were between 0.64 and 9.94 μg g−1. However, data for cell numbers or biomass in these soils are not available, and different methods of RNA quantification were used, making a direct comparison difficult. A more detailed comparison of RNA yields obtained by different methods and with different soils may help define optimal extraction protocols for specific systems and research questions.
FIG. 2.
Quantification of RNA extracted from soil after 2, 3, and 7 days of incubation. (A) Correlation of RNA concentrations determined with RiboGreen in extracts purified from raw total nucleic acid extracts 1 week (extraction 1) and 4 months (extraction 2) after the end of the experiment, showing the reproducibility of the RNA cleanup protocol. (B) Total RNA yield from extraction of A. vinelandii-inoculated soil over time (extraction 1). Symbols: ▪, SC+N replicate 1; •, SC+N replicate 2; □, SC−N replicate 1; ○, SC−N replicate 2.
Our method for soil RNA extraction has a number of advantages over previously described protocols. It is based on optimized nucleic acid extraction with a highly efficient and fast bead beating method (8), and only small amounts (0.5 g) of soil are used. The extraction buffer allows both soil DNA and total soil RNA isolation due to the nuclease (protein)-denaturing capacities of CTAB and DTT and the rapid application of the phenol-chloroform extraction procedure (10, 13). Since the RNA cleanup procedure is a separate step of the protocol, the method is well suited for simultaneous analyses of DNA and RNA from the same total nucleic acid extract. In addition, successful nifH mRNA amplification after 4 months of storage of raw extracts (data not shown) showed that mRNA is protected in phenol-chloroform-treated raw extracts kept at −80°C. Temporally separating the initial extraction procedure from the cleanup protocol should facilitate future experiments with higher numbers of samples or field studies.
Physiological response: growth and ammonium uptake by A. vinelandii.
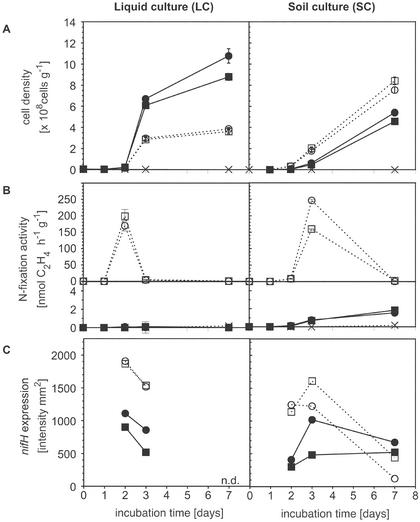
Data from all sampling dates revealed that NH4NO3 addition induced different growth responses in soil and liquid culture. Between day 2 and day 3 the cell densities of A. vinelandii in soil were still lower than those in liquid culture (Fig. 3A). In soil, however, considerable growth was observed between day 3 and day 7, and for both soil treatments the cell densities were higher than the cell densities for the LC−N treatment at day 7 (Fig. 3A). In soil, the cell densities were higher for the SC−N treatment than for the SC+N treatment throughout the experiment, but the reverse was true for the liquid medium microcosms (Fig. 3A). No cell growth was observed in the sterile controls included for soil and for liquid cultures.
FIG. 3.
Cell density, nitrogen fixation activity, and intensity of nifH mRNA expression plotted against time. (A) Cell counts for DAPI-stained bacteria in liquid culture and soil. (B) Nitrogen fixation activity determined by the acetylene reduction method with liquid cultures and soil. Upper plot, SC−N and LC−N; lower plot (reduced scale), SC+N and LC+N. (C) Intensities of nifH mRNA RT-PCR products determined by image analyses of gel images. Left panel, liquid culture treatments; right panel, soil culture treatments. The error bars in panels A and B indicate the standard errors of the measurements. Symbols: ▪, LC+N or SC+N replicate 1; •, LC+N or SC+N replicate 2; □, LC−N or SC−N replicate 1; ○, LC−N or SC−N replicate 2; ×, LC control or SC control. n.d., not determined.
Direct cell counting with DAPI staining provided a simple means for accurately and comparably determining cell densities in both liquid and soil cultures. A. vinelandii grew well in both systems. While there has been considerable work on growth and nitrogen fixation by A. vinelandii in liquid cultures in defined and undefined media (1, 31, 34, 38), the physiology of this organism in soil has not been described in detail. Experiments with Azotobacter chroococcum revealed growth characteristics similar to those observed for A. vinelandii in this study (44).
In treatments to which NH4NO3 was not added (LC−N, SC−N), no ammonium was detected during the first 2 days of incubation. After 3 days, the ammonium concentration increased slightly in the duplicate SC−N treatments, to 1.3 and 1.4 μmol g−1. In one of the duplicates that received the SC+N treatment the concentration of ammonium decreased continuously from 15.3 to 12.3 μmol g−1 over the course of the experiment, while in the other duplicate there was not a clear trend and overall the concentration decreased from 17.6 to 15.6 μmol g−1. The ammonium concentration in the SC control was 18.5 μmol g−1 by the end of the experiment. In the preparations that received the LC+N treatment the concentration of ammonium decreased more rapidly, from approximately 20.1 and 19.2 μmol ml−1 to 9.1 and 8.1 μmol ml−1 on day 2. The LC control contained 17.4 μmol of ammonium ml−1 on day 7.
The ammonium data indicated that throughout the experiment nitrogen-limiting conditions were maintained in SC−N and LC−N treatments and that an excess of nitrogen was present in the LC+N and SC+N treatments. The ammonium concentrations decreased much more rapidly in liquid cultures than in soil, and the differences in the availability of ammonium in soil and in liquid cultures may have been responsible in part for the different growth rates of A. vinelandii described above. Assuming a cell dry weight of 0.2 ng per 106 cells and a nitrogen content of 10% (39), we calculated the expected reduction in the ammonium concentration based on the measured cell densities. Based on this calculation, the N content of the biomass after 7 days represented about 50% of the added ammonium for both the LC+N and SC+N treatments, indicating that the added ammonium was sufficient to account for the observed biomass production. The relatively high ammonium concentration in soil even after 7 days indicated that there was resupply of the extractable ammonium pool from soil-bound ammonium.
These data suggested that a model system was established which provided suitable growth conditions for A. vinelandii while maintaining either nitrogen-limited or nitrogen-excess conditions in the different treatments. The different growth responses to NH4NO3 addition depending on the matrix (soil or liquid culture) demonstrated the importance of in situ studies of physiology and gene regulation in soil habitats.
N fixation and detection of nifH gene expression.
In all preparations to which NH4NO3 was added (LC+N and SC+N), the N fixation activity remained very low (<2 nmol of C2H4 h−1 g−1) throughout the experiment (Fig. 3B). For the SC+N treatment, low N fixation activity was observed, and the activity increased slightly over time; for the LC+N treatment and sterile controls the rates of ethylene formation remained below 0.1 nmol of C2H4 h−1 g−1 (Fig. 3B) and were not significant (P = 0.05). The highest activities were observed during the early growth phase for the LC−N and SC−N treatments, on day 2 for the LC−N treatments (199 and 169 nmol h−1 ml−1) and on day 3 for the SC−N treatments (160 and 247 nmol h−1 g−1). After these maxima, the N fixation rates decreased to 0.3 and 0.2 nmol h−1 g−1 for the LC−N treatments and to 2.5 and 0.9 nmol h−1 g−1 for the SC−N treatments at day 7. Our observations are in agreement with previous studies showing that additions of ammonium and nitrate result in strong regulatory effects on the N fixation process (12, 23, 31).
Direct quantification of PCR products from band intensities on gels or blots is frequently used as a simple method for quantification of DNA template concentrations in soil (5, 16, 55). A number of precautions were taken to ensure that RT-PCR could be used as a semiquantitative method to determine expression of nifH in soil. The RT conditions were constant for all samples. By adjusting the number of PCR cycles in preliminary experiments we ensured that PCR amplification did not reach a plateau in any of the samples. In addition, a separate dilution experiment showed that the measured product intensity of A. vinelandii nifH PCR products was linearly related to the logarithm of the template concentration (r2 = 0.97) (data not shown). In a pure-culture system such as the one used in this study, the intensity of the A. vinelandii nifH-specific RT-PCR product can be used to directly obtain a semiquantitative measure of the amount of nifH mRNA, while amplification from community DNA may require the use of specific probes to ensure specific quantification (5). Due to the great range of cell densities and the presumably different RNA extraction efficiencies for soil and liquid cultures, it is not feasible to use absolute quantification of mRNA templates. Consequently, mRNA samples from all sampling dates and treatments were adjusted to a total RNA concentration of 3 μg ml−1 prior to RT-PCR detection of A. vinelandii nifH. Quantification of RT-PCR amplicons from the diluted extracts was therefore relative to the total RNA content and thus a measure of the relative expression of the functional gene.
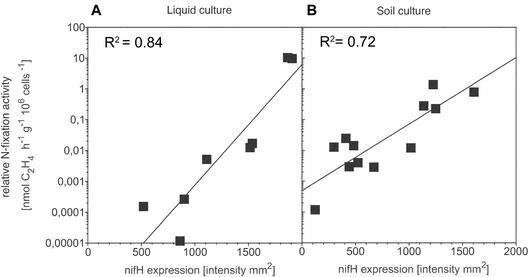
Transcripts of nifH were detected in diluted RNA samples analyzed from day 2 onward (Fig. 3C). Transcripts were detected in treatments with and without NH4NO3 amendment, but the intensities of the signals for the nitrogen-fixing LC−N and SC−N treatments were approximately twice the intensities of the signals for the nonamended treatments on days 2 and 3 (Fig. 3C). According to the regression analysis for the template dilution experiment, a doubling of the signal strength corresponded to a template increase of approximately 2.5 orders of magnitude. The intensity of the RT-PCR product correlated significantly (P = 0.01) with the logarithm of the observed specific N fixation activity (Fig. 4), and the correlation coefficients for the exponential fitting functions were comparable in liquid cultures (r2 = 0.84) and soil (r2 = 0.72).
FIG. 4.
Relationship of A. vinelandii nifH RT-PCR product intensity quantified from ethidium bromide-stained agarose gels to specific N fixation activity determined by acetylene reduction. (A) Liquid culture treatments. (B) Soil culture treatments. The correlation coefficients of the exponential fitting functions were highly significant (P = 0.01) for both treatment types.
It is well known that expression of all nitrogenase systems is repressed by high cellular concentrations of fixed nitrogen (28). We therefore expected strongly reduced N fixation activities and reductions in nifH mRNA levels in the LC+N and SC+N treatments. Although the N fixation activities remained below the detection limit for the LC+N treatments and never reached more than 1% of the maximum activity for the SC+N treatments (Fig. 3B), considerable levels of nifH mRNA were detected (Fig. 3C). This observation was consistent with reports indicating that expression of nifH can take place in the presence of relatively high levels of fixed nitrogen, as previously demonstrated for Rhodobacter capsulatus, in which nifH expression could be detected at ammonium concentrations of up to 12.5 mM (18). A possible explanation for this observation is the presence of posttranslational regulation mechanisms that have been described for many known diazotrophs (26). Whether transcription of nifH is more stringently regulated in natural soil systems, which are more oligotrophic in nature than our sucrose-amended system, remains to be studied.
Despite the relatively small number of samples and low resolution on the time scale, we were able to clearly show effects of the presence of ammonium in a soil system on the expression of nifH mRNA in A. vinelandii. This represents an important step toward the study of gene regulation and a more detailed understanding of microbial processes in soil. The combination of specific mRNA and bulk activity detection in natural soils provide a powerful tool to examine activities and processes in complex soil microbial communities. The combination of specific real-time PCR and the methods that have been developed should provide an even more powerful method of quantification for RT-PCR, but the procedure requires considerable optimization (11, 52).
Conclusions.
Our results obtained with the newly developed methods suggested that detection of active members of natural soil microbial communities is feasible. However, detection of background levels of nifH mRNA from A. vinelandii populations that did not exhibit detectable N fixation also suggests that results obtained from environmental samples must be interpreted with caution. A combined approach in which both molecular tools and measurement of enzymatic activity are used is necessary when complex microbial communities are studied. The A. vinelandii pure culture in combination with the methods developed for RNA extraction and nifH mRNA detection provided a suitable model system for relating cell number, nifH mRNA expression, and enzymatic activity in soil by using N fixation as a model function.
Acknowledgments
We thank Manuel Pesaro for critical and stimulating discussions and technical support throughout this study. We are grateful to Annatina Zarda for assistance with DAPI cell counting. Two anonymous reviewers are acknowledged for comments that helped improve the manuscript.
This research was supported in part by Swiss Federal Office for Education and Science grant C99.0036 in the framework of COST Action 831 (Biotechnology of Soil: Monitoring, Conservation and Remediation) and the Indo-Swiss Collaboration in Biotechnology (project SA 5).
REFERENCES
- 1.Abdalla, M. H. 1994. Utilization of some phenolic compounds by Azotobacter chroococcum and their effect on growth and nitrogenase activity. Folia Microbiol. 39:57-60. [PubMed] [Google Scholar]
- 2.Alef, K. 1995. Nitrogen mineralization in soils, p. 234-270. In K. Alef and P. Nannipieri (ed.), Methods in applied soil microbiology and biochemistry. Academic Press, London, United Kingdom.
- 3.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becking, J. H. 1992. The family Azotobacteriaceae, p. 3145-3170. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K. Schleifer (ed.), The prokaryotes, 2nd ed., vol. IV. Springer, New York, N.Y.
- 5.Bogan, B. W., B. Schoenike, R. T. Lamar, and D. Cullen. 1996. Manganese peroxidase mRNA and enzyme activity levels during bioremediation of polycyclic aromatic hydrocarbon-contaminated soil with Phanerochaete chrysosporium. Appl. Environ. Microbiol. 62:2381-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borneman, J., and E. W. Triplett. 1997. Rapid and direct method for extraction of RNA from soil. Soil Biol. Biochem. 29:1621-1624. [Google Scholar]
- 7.Bundt, M., F. Widmer, M. Pesaro, J. Zeyer, and P. Blaser. 2001. Preferential flow paths: biological ‘hot spots' in soils. Soil Biol. Biochem. 33:729-738. [Google Scholar]
- 8.Bürgmann, H., M. Pesaro, F. Widmer, and J. Zeyer. 2001. A strategy for optimizing quality and quantity of DNA extracted from soil. J. Microbiol. Methods 45:7-20. [DOI] [PubMed] [Google Scholar]
- 9.Burke, D. J., E. P. Hamerlynck, and D. Hahn. 2002. Interactions among plant species and microorganisms in salt marsh sediments. Appl. Environ. Microbiol. 68:1157-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheung, A. L., K. J. Eberhardt, and V. A. Fischetti. 1994. A method to isolate RNA from gram-positive bacteria and mycobacteria. Anal. Biochem. 222:511-514. [DOI] [PubMed] [Google Scholar]
- 11.Deprez, R. H. L., A. C. Fijnvandraat, J. M. Ruijter, and A. F. M. Moorman. 2002. Sensitivity and accuracy of quantitative real-time polymerase chain reaction using SYBR green I depends on cDNA synthesis conditions. Anal. Biochem. 307:63-69. [DOI] [PubMed] [Google Scholar]
- 12.Drozd, J. W., R. S. Tubb, and J. R. Postgate. 1972. Chemostat study of effect of fixed nitrogen sources on nitrogen fixation, membranes and free amino-acids in Azotobacter chroococcum. J. Gen. Microbiol. 73:221-232. [DOI] [PubMed] [Google Scholar]
- 13.Farrell, R. E. 1998. RNA methodologies, 2nd ed. Academic Press, San Diego, Calif.
- 14.Fleming, J. T., J. Sanseverino, and G. S. Sayler. 1993. Quantitative relationship between naphthalene catabolic gene frequency and expression in predicting PAH degradation in soils at town gas manufacturing sites. Environ. Sci. Technol. 27:1068-1074.
- 15.Fleming, J. T., W. H. Yao, and G. S. Sayler. 1998. Optimization of differential display of prokaryotic mRNA: application to pure culture and soil microcosms. Appl. Environ. Microbiol. 64:3698-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia-Pichel, F., A. Lopez-Cortes, and U. Nubel. 2001. Phylogenetic and morphological diversity of cyanobacteria in soil desert crusts from the Colorado Plateau. Appl. Environ. Microbiol. 67:1902-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greer, C. W., L. G. Whyte, J. R. Lawrence, L. Masson, and R. Brousseau. 2001. Genomics technologies for environmental science. Environ. Sci. Technol. 35:364A-370A. [DOI] [PubMed]
- 18.Hubner, P., B. Masepohl, W. Klipp, and T. A. Bickle. 1993. nif gene expression studies in Rhodobacter capsulatus: ntrC-independent repression by high ammonium concentrations. Mol. Microbiol. 10:123-132. [DOI] [PubMed] [Google Scholar]
- 19.Hurt, R. A., X. Y. Qiu, L. Y. Wu, Y. Roh, A. V. Palumbo, J. M. Tiedje, and J. H. Zhou. 2001. Simultaneous recovery of RNA and DNA from soils and sediments. Appl. Environ. Microbiol. 67:4495-4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanungo, P. K., B. Ramakrishnan, and V. R. Rao. 1997. Placement effects of organic sources on nitrogenase activity and nitrogen-fixing bacteria in flooded rice soils. Biol. Fertil. Soils 25:103-108. [Google Scholar]
- 21.Keeling, A. A., J. A. Cook, and A. Wilcox. 1998. Effects of carbohydrate application on diazotroph populations and nitrogen availability in grass swards established in garden waste compost. Biores. Technol. 66:89-97. [Google Scholar]
- 22.Kennedy, I. R., and N. Islam. 2001. The current and potential contribution of asymbiotic nitrogen fixation to nitrogen requirements on farms: a review. Aust. J. Exp. Agric. 41:447-457. [Google Scholar]
- 23.Kleiner, D. 1975. Ammonium uptake by nitrogen-fixing bacteria. 1. Azotobacter vinelandii. Arch. Microbiol. 104:163-169. [DOI] [PubMed] [Google Scholar]
- 24.Limmer, C., and H. L. Drake. 1996. Non-symbiotic N2-fixation in acidic and pH-neutral forest soils: aerobic and anaerobic differentials. Soil Biol. Biochem. 28:177-183. [Google Scholar]
- 25.Lovell, C. R., Y. M. Piceno, J. M. Quattro, and C. E. Bagwell. 2000. Molecular analysis of diazotroph diversity in the rhizosphere of the smooth cordgrass, Spartina alterniflora. Appl. Environ. Microbiol. 66:3814-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ludden, P. W., and G. P. Roberts. 1989. Regulation of nitrogenase activity by reversible ADP ribosylation. Curr. Top. Cell. Regul. 30:23-56. [DOI] [PubMed] [Google Scholar]
- 27.Mendum, T. A., R. E. Sockett, and P. Hirsch, R. 1998. The detection of gram-negative bacterial mRNA from soil by RT-PCR. FEMS Microbiol. Lett. 164:369-373. [Google Scholar]
- 28.Merrick, M. J. 1992. Regulation of nitrogen fixation genes in free-living and symbiotic bacteria, p. 835-876. In G. Stacey, R. H. Burris, and H. J. Evans (ed.), Biological nitrogen fixation. Chapman & Hall, New York, N.Y.
- 29.Miskin, I. P., P. Farrimond, and I. M. Head. 1999. Identification of novel bacterial lineages as active members of microbial populations in a freshwater sediment using a rapid RNA extraction procedure and RT-PCR. Microbiology 145:1977-1987. [DOI] [PubMed] [Google Scholar]
- 30.Moran, M. A., V. L. Torsvik, T. Torsvik, and R. E. Hodson. 1993. Direct extraction and purification of ribosomal RNA for ecological studies. Appl. Environ. Microbiol. 59:915-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moreno, J., and C. Vargasgarcia. 1995. Growth and nitrogenase activity of Azotobacter vinelandii in chemically defined media containing glucose and p-hydroxybenzoic acid. Chemosphere 31:2605-2610. [Google Scholar]
- 32.Nickel, A., D. Hahn, K. Zepp, and J. Zeyer. 1999. In situ analysis of introduced Frankia populations in root nodules of Alnus glutinosa grown under different water availability. Can. J. Bot. 77:1231-1238. [Google Scholar]
- 33.Ocio, J. A., P. C. Brookes, and D. S. Jenkinson. 1991. Field incorporation of straw and its effects on soil microbial biomass and soil inorganic N. Soil Biol. Biochem. 23:171-176. [Google Scholar]
- 34.Oelze, J. 1991. Diazotrophic mixed culture of Azotobacter vinelandii and Rhodobacter capsulatus. Plant Soil 137:135-138. [Google Scholar]
- 35.Ogram, A., W. Sun, F. J. Brockman, and J. K. Fredrickson. 1995. Isolation and characterization of RNA from low-biomass deep-subsurface sediments. Appl. Environ. Microbiol. 61:763-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogunseitan, O. A., and B. H. Olson. 1993. Effect of 2-hydroxybenzoate on the rate of naphthalene mineralization in soil. Appl. Microbiol. Biotechnol. 38:799-807. [Google Scholar]
- 37.Pace, N. R. 1996. New perspective on the natural microbial world: molecular microbial ecology. ASM News 62:463-470. [Google Scholar]
- 38.Papadelli, M., A. Roussis, K. P. A. Venieraki, I. Chatzipavlidis, P. Katinakis, and K. Ballis. 1996. Biochemical and molecular characterization of an Azotobacter vinelandii strain with respect to its ability to grow and fix nitrogen in olive mill wastewater. Int. Biodeterior. Biodegrad. 38:179-181. [Google Scholar]
- 39.Paul, E. A., and F. E. Clark. 1996. Soil microbiology and biochemistry, 2nd ed. Academic Press, San Diego, Calif.
- 40.Peoples, M. B., and E. T. Craswell. 1992. Biological nitrogen fixation: investments, expectations and actual contributions to agriculture. Plant Soil 141:13-39. [Google Scholar]
- 41.Poly, F., L. J. Monrozier, and R. Bally. 2001. Improvement in the RFLP procedure for studying the diversity of nifH genes in communities of nitrogen fixers in soil. Res. Microbiol. 152:95-103. [DOI] [PubMed] [Google Scholar]
- 42.Poly, F., L. Ranjard, S. Nazaret, F. Gourbiere, and L. J. Monrozier. 2001. Comparison of nifH gene pools in soils and soil microenvironments with contrasting properties. Appl. Environ. Microbiol. 67:2255-2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Postgate, J. R. 1982. The fundamentals of nitrogen fixation. Cambridge University Press, Cambridge, England.
- 44.Pozo, C., M. V. Martinez-Toledo, V. Salmeron, B. Rodelas, and J. Gonzalez-Lopez. 2000. Effects of benzidine and benzidine analogues on the growth and nitrogenase activity of Azotobacter. Appl. Soil Ecol. 14:183-190. [Google Scholar]
- 45.Rhoades, J. D. 1982. Soluble salts, p. 167-179. In A. L. Page, R. H. Miller, and D. R. Keeney (ed.), Methods of soil analyses, part 2, 2nd ed. ASA-SSSA, Madison, Wis.
- 46.Roper, M. M., and J. K. Ladha. 1995. Biological N2 fixation by heterotrophic and phototrophic bacteria in association with straw. Plant Soil 174:211-224. [Google Scholar]
- 47.Roper, M. M., J. E. Turpin, and J. P. Thompson. 1994. Nitrogenase activity (C2H2 reduction) by free-living bacteria in soil in a long-term tillage and stubble management experiment on a vertisol. Soil Biol. Biochem. 26:1087-1091. [Google Scholar]
- 48.Sandaa, R. A., O. Enger, and V. Torsvik. 1998. Rapid method for fluorometric quantification of DNA in soil. Soil Biol. Biochem. 30:265-268. [Google Scholar]
- 49.Sayler, G. S., J. T. Fleming, and D. E. Nivens. 2001. Gene expression monitoring in soils by mRNA analysis and gene lux fusions. Curr. Opin. Biotechnol. 12:455-460. [DOI] [PubMed] [Google Scholar]
- 50.Sessitsch, A., S. Gyamfi, N. Stralis-Pavese, A. Weilharter, and U. Pfeifer. 2002. RNA isolation from soil for bacterial community and functional analysis: evaluation of different extraction and soil conservation protocols. J. Microbiol. Methods 51:171-179. [DOI] [PubMed] [Google Scholar]
- 51.Shaffer, B. T., F. Widmer, L. A. Porteous, and R. J. Seidler. 2000. Temporal and spatial distribution of the nifH gene of N2 fixing bacteria in forests and clearcuts in western Oregon. Microb. Ecol. 39:12-21. [DOI] [PubMed] [Google Scholar]
- 52.Stults, J. R., O. Snoeyenbos-West, B. Methe, D. R. Lovley, and D. P. Chandler. 2001. Application of the 5′ fluorogenic exonuclease assay (TaqMan) for quantitative ribosomal DNA and rRNA analysis in sediments. Appl. Environ. Microbiol. 67:2781-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsai, Y. L., M. J. Park, and B. H. Olson. 1991. Rapid method for direct extraction of messenger RNA from seeded soils. Appl. Environ. Microbiol. 57:765-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Widmer, F., A. Fliessbach, E. Laczko, J. Schulze-Aurich, and J. Zeyer. 2001. Assessing soil biological characteristics: a comparison of bulk soil community DNA-, PLFA-, and Biolog-analyses. Soil Biol. Biochem. 33:1029-1036. [Google Scholar]
- 55.Widmer, F., R. J. Seidler, and L. S. Watrud. 1996. Sensitive detection of transgenic plant marker gene persistence in soil microcosms. Mol. Ecol. 5:603-613. [Google Scholar]
- 56.Widmer, F., B. T. Shaffer, L. A. Porteous, and R. J. Seidler. 1999. Analysis of nifH gene pool complexity in soil and litter at a Douglas fir forest site in the Oregon Cascade mountain range. Appl. Environ. Microbiol. 65:374-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Young, J. P. W. 1992. Phylogenetic classification of nitrogen-fixing organisms, p. 43-86. In G. Stacey, R. H. Burris, and H. J. Evans (ed.), Biological nitrogen fixation. Chapman & Hall, New York, N.Y.
- 58.Zarda, B., D. Hahn, A. Chatzinotas, W. Schoenhuber, A. Neef, R. Amann, and J. Zeyer. 1997. Analysis of bacterial community structure in bulk soil by in situ hybridization. Arch. Microbiol. 168:185-192. [Google Scholar]
- 59.Zehr, J. P., and L. A. McReynolds. 1989. Use of degenerate oligonucleotides for amplification of the nifH gene from the marine cyanobacterium Trichodesmium thiebautii. Appl. Environ. Microbiol. 55:2522-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]