Abstract
We studied the effects of cocultivation with either Euglena gracilis (Euglenophyta), Microcystis aeruginosa (Cyanophyta), Chlamydomonas neglecta (Chlorophyta), or Carteria inversa (Chlorophyta) on the production of extracellular plasmid DNA by Escherichia coli LE392(pKZ105). Dot blot hybridization analysis showed a significant release of plasmid DNA by cocultivation with all the algae tested. Further analysis by electrotransformation confirmed the release of transformable plasmid DNA by cocultivation with either E. gracilis, M. aeruginosa, or C. inversa. These results suggest algal involvement in bacterial horizontal gene transfer by stimulating the release of transformable DNA into aquatic environments.
Dissolved DNA (dDNA), DNA that passes through a 0.2-μm-pore-size filter, is a form of extracellular DNA commonly observed in natural aquatic environments and has been found at concentrations from 0.2 to 88 μg liter−1 in both freshwater and seawater (7, 16, 27, 28, 32). dDNA was initially thought to be an attractive source of nutrients for microorganisms (15, 31), but finding naturally competent bacterial strains suggested that the occurrence of bacterial dDNA might also act as a source for the uptake of genetic material (9, 14). We recently reported on the stability of the transformability of plasmid DNA in natural lake water and suggested that dDNA may have a genetic function as a gene pool for further natural transformation (22).
DNA from bacteria has been shown to form a major component of the dDNA pool (27, 33), while algal DNA may also be a significant constituent (25). The production of dDNA is controlled by both physicochemical (26) and biological factors (27). Grazing by bacteriovores (11, 17, 33) and cell lysis by viral infection (1, 29, 35) seem to be the main forces releasing bacterial cellular DNA into the surrounding water. However, the presence of bacterial chromosomal DNA in culture liquids (3, 10) and the release of transformable plasmid and chromosomal DNA from cultured bacteria (20) have indicated that active DNA excretion may occur without disruption of the cells.
Photosynthetic algae are major biological constituents of aquatic environments and are also important biological factors in stimulating bacterial growth via their secretion of dissolved organic carbon (5). Although the influence of algae on bacterial growth has been studied, there has been no study of their impact on the production of dDNA by bacteria. Algal blooms are frequently observed in eutrophic waters where antibiotic-resistant coliform bacteria, including Escherichia coli, are delivered via wastewater (13) and are probably an important reservoir of antibiotic resistance genes. Thus, one effect of algae on the production of dDNA by bacteria may be involved in the dissemination of bacterial antibiotic resistance genes via further natural transformation. In this study, we examine the involvement of algae in the production of extracellular plasmid DNA by E. coli and the ability of the DNA released to transform in the bacteria.
A 974-bp PvuII-EcoRI fragment of pEGFP (Clontech, Palo Alto, Calif.) containing the lac promoter and egfp, a green fluorescent protein gene, was inserted into the SmaI-EcoRI site of pHY300PLK (12), a shuttle vector between Bacillus subtilis and E. coli. The resulting plasmid, pKZ105, which is expressed as green fluorescence in E. coli but not in B. subtilis, was transferred into E. coli LE392 (30) and used to trace the excretion of cellular plasmid DNA. A slightly modified version of C medium (8) was used as a culture medium with the following composition (in milligrams per liter of distilled water): Ca(NO3)2 · H2O, 150; KNO3, 100; β-Na2 glycerophosphate, 50; MgSO4 · 7H2O, 40; vitamin B12, 0.0001; biotin, 0.0001; thiamine HCl, 0.01; and yeast extract, 200. C medium also contained 3 ml of a solution of trace metals (pH adjusted to 7.5). The trace metal solution consisted of the following (in milligrams per liter of distilled water): FeCl3 · 6H2O, 196; MnCl2 · 4H2O, 36; ZnSO4 · 7H2O, 22; CoCl2 · 6H2O, 4; Na2MoO4 · 2H2O, 2.5; and Na2EDTA · 2H2O, 1,000. Yeast extract was sterilized separately from the other components, and they were mixed together before use. The medium was decanted into a series of autoclaved test tubes (10 ml each) before inoculation of the organisms. E. coli LE392(pKZ105) was precultured overnight at 37°C in the modified C medium and directly inoculated into each test tube at 105 CFU ml−1. E. coli LE392(pKZ105) was also cocultured with either euglenophyte Euglena gracilis Z, cyanophyte Microcystis aeruginosa (NIES 298), or chlorophyte Chlamydomonas neglecta (NIES 439) or chlorophyte Carteria inversa (NIES 422) to evaluate the effects of algae on DNA excretion by this bacterium. E. gracilis was kindly provided by N. Nakano, Department of Agricultural Chemistry, University of Osaka Prefecture, Sakai, Japan, and the other algal strains were obtained from the National Institute for Environmental Studies (NIES), Environmental Agency, Tsukuba, Japan. All algal strains were axenic and had been precultured for 10 days in the modified C medium. Precultured algae were concentrated by centrifugation (3,000 × g, 10 min) and then directly inoculated into each test tube at 4.5 × 104 organisms ml−1 (E. gracilis), 1.8 × 106 organisms ml−1 (M. aeruginosa), 4.2 × 105 organisms ml−1 (C. neglecta), and 7.7 × 103 organisms ml−1 (C. inversa). All the test tubes were incubated for 17 days at 25°C with a light regimen of 12 h of light and 12 h of dark under cool-white fluorescent lamps.
The population of each organism was assessed by counting E. coli CFU on Luria-Bertani (LB) plates (30) and by directly counting algae under a microscope. At each observation time, extracellular DNA in the culture fluids was separated from the microorganisms by brief centrifugation (3,000 × g, 5 min), followed by filtration through a 0.22-μm-pore-size Milex-GV25 filter unit (Millipore, Bedford, Mass.). The filtrate was concentrated using 30,000-molecular-weight-cutoff centrifugal filter devices (CENTRIPLUS; Millipore) and then purified by utilizing QIAquick PCR purification kit (QIAGEN K. K., Tokyo, Japan). The concentration of total extracellular DNA was determined by a modified version of the fluorescence method of DeFlaun et al. (7), using λ-HindIII digest (Takara Shuzo, Tokyo, Japan) as standard DNA. To increase the sensitivity of the measurement, we used SYBR green I (Molecular Probes, Eugene, Oreg.) instead of Hoechst 33258 dye. pKZ105 was quantified by dot blot hybridization, using part of the egfp fragment as a probe, by the method of Matsui et al. (22). Significant differences in the release of plasmid DNA were examined statistically by one-way analysis of variance with post hoc Fisher's protected least significant difference test, using Stat View 5.0 for Macintosh (SAS Institute Inc. Cary, N.C.).
An aliquot of purified DNA, equivalent to the DNA in 0.25 ml of culture, was also used to electrotransform E. coli JM109 by the method of Sambrook and Russell (30). A series of transformations with different amounts of pure, isolated pKZ105 from E. coli LE392(pKZ105) was also conducted to obtain a standard curve of transformation. Isolation of pKZ105 was conducted by alkaline lysis (30) using the CONCERT rapid plasmid purification system (Life Technologies, Inc., Rockville, Md.). Fish sperm DNA (molecular biology grade; Roche Diagnostics, Tokyo, Japan) was mixed with pKZ105 as a mimic of total extracellular DNA to test the interference effects on transformation. Transformants were selected for resistance to 20 μg of tetracycline per liter in LB agar medium. Expression of the green fluorescence gene was confirmed by color change of colonies and expression of green fluorescence.
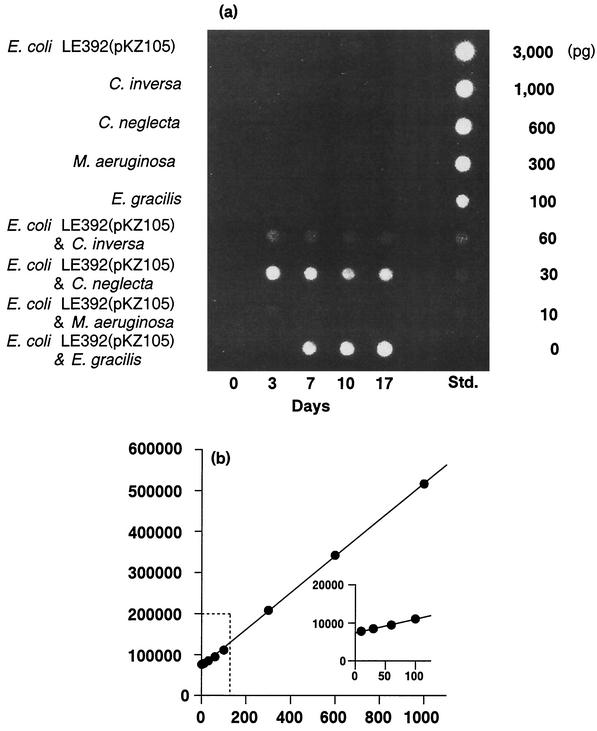
A dot blot and the standard curve of dot blot hybridization are presented in Fig. 1. The DNA detection limits of the methods employed were 10 pg ml−1 (dot blot hybridization analysis; plasmid pKZ105) and 200 pg ml−1 (fluorometric analysis; total extracellular DNA). The time courses of population density and the release of extracellular DNA were examined in monocultures and cocultures of E. coli LE392(pKZ105) (Fig. 2). Significant extracellular DNA production was observed from cocultures of E. coli LE392(pKZ105) with algae, while the amount of DNA detected and the timing of DNA production varied with the algal species.
FIG. 1.
(a) Dot blots of plasmid from experimental cultures. Each dot contains DNA (equivalent to the amount of DNA in 1 ml of culture). No signal was detected from the samples of monospecies cultures. Pure isolated plasmid DNA was used as a standard (Std.), and the amount of DNA applied on each dot is shown to the right of the gel. (b) Regressions of the intensity of hybridization signals (y axis) against the amount of pKZ105 DNA in each dot (x axis). The correlation coefficient for the linear regression was 0.996 over the range of 10 to 1,000 pg of standard pKZ105 DNA.
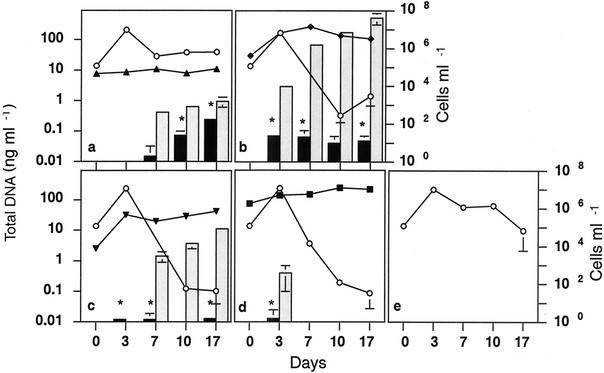
FIG. 2.
Changes in the concentration of total extracellular DNA (shaded bars) and pKZ105 aliquot (black bars) and changes in the number of each organism (lines) in cultures of E. coli LE392(pKZ105) (○) grown with E. gracilis (▴) (a), C. neglecta (⧫) (b), C. inversa (▾) (c), or M. aeruginosa (▪) (d) and in E. coli LE392(pKZ105) monoculture (e). The means ± standard deviations (error bars) from three experiments are shown. Statistically significant release of pKZ105 (at 5% level) is denoted by an asterisk.
When E. coli was cocultured with E. gracilis, release of total extracellular DNA was observed after 7 days of cultivation (Fig. 2a), and all the total DNA detected must have originated from E. coli LE392(pKZ105), since no DNA was detected in the monoculture of E. gracilis (Fig. 3a). The data from dot blot hybridization demonstrated that the pKZ105 plasmid was also present in the extracellular DNA fraction and confirmed the release of DNA from E. coli LE392(pKZ105) when it was cocultivated with E. gracilis. Extracellular DNA production was not observed in the monoculture of E. coli LE392(pKZ105), and the population changes of E. coli LE392(pKZ105) were no different when E. gracilis was included in the culture (Fig. 2a and e). Thus, the production of extracellular DNA would not have been caused by cell lysis of E. coli, and indirect effects, such as the involvement of metabolite-mediated effects of E. gracilis, were indicated.
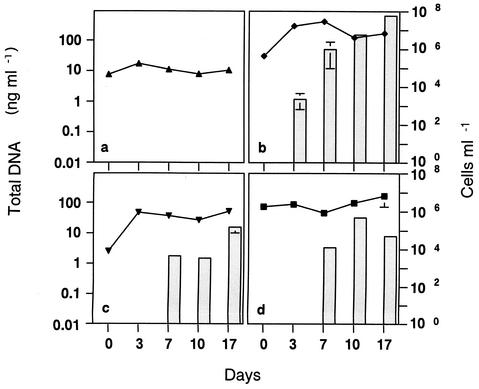
FIG. 3.
Changes in the concentration of total extracellular DNA (shaded bars) and in the number of organisms (lines) in monocultures of E. gracilis (▴) (a), C. neglecta (⧫) (b), C. inversa (▾) (c), and M. aeruginosa (▪) (d). Note that there was no E. coli(pKZ105) in these cultures, so no pKZ105 was recorded. The means ± standard deviations (error bars) from three experiments are shown.
Coculturing E. coli LE392(pKZ105) with C. neglecta or C. inversa also resulted in a gradual increase of total extracellular DNA during cultivation (Fig. 2b and c), but the detection of similar total amounts of extracellular DNA in the monocultures of C. neglecta and C. inversa suggested that in this case the majority of the extracellular DNA in the cocultures was released from the algae (Fig. 3b and c). However, detection of pKZ105 DNA by dot blot hybridization indicated that the plasmid DNA was also released from E. coli LE392(pKZ105) when it was cultivated with C. neglecta or C. inversa. The sudden decline in the cell density of E. coli after 7 days of culture suggested the release of E. coli DNA by cell lysis (Fig. 2b and c). It is possible that C. neglecta or C. inversa produced antibacterial substances, since some algal species are known to produce substances which inhibit the growth and respiration of bacteria (4). However, the detection of E. coli plasmid DNA after 3 days of culture, when the E. coli population density was at its highest, suggested that there was an alternative effect of these cocultured algae on the release of extracellular DNA. In either case, extracellular DNA was released from E. coli LE392(pKZ105) by cocultivation with C. neglecta or C. inversa.
When E. coli LE392(pKZ105) was cocultured with M. aeruginosa, extracellular DNA was detected only after the first 3 days of culture (Fig. 2d). The increase in the population density of E. coli during the first 3 days and the fact that no extracellular DNA was detected after 7 days of cultivation (when the E. coli cell density had declined) suggested that cocultivation with M. aeruginosa had a stimulative effect on the release of E. coli DNA. In the M. aeruginosa monoculture, extracellular DNA was detected at 7, 10, and 17 days of culture but not at 3 days (Fig. 3d). Although the cellular condition of the M. aeruginosa cocultured with E. coli LE392(pKZ105) seemed different from the M. aeruginosa in monoculture, the detection of pKZ105 DNA in the coculture did indicate that there had been some release of extracellular DNA from E. coli LE392(pKZ105) when it was cocultured with M. aeruginosa.
No extracellular DNA was detected in monocultures of E. coli LE392(pKZ105), and the culture conditions employed here were clearly not suitable for E. coli LE392(pKZ105) to release extracellular DNA, at least for the 17 days of cultivation (Fig. 2e). Thus, all of the four algal species tested were involved in the production of extracellular DNA from E. coli under our experimental conditions.
Recent advances in studies of molecular microbial ecology and genome analysis indicate that horizontal gene transfer is involved in bacterial evolution and adaptation to changing environments (6, 24). As a means of gene transfer, transformation may be the most important mechanism by which bacteria acquire DNA from distantly related organisms, including eukaryotes (23). We think that there may be natural transformation-mediated horizontal gene transfer in an aquatic ecosystem from a study of a naturally transformable bacterium, B. subtilis (K. Matsui, N. Ishii, and Z. Kawabata, submitted for publication). The availability of a transformable gene is a necessity for successful horizontal gene transfer by transformation and for completion of the transformation event (21).
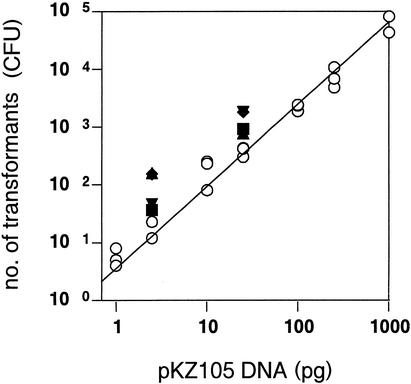
To determine whether the detected plasmid (pKZ105) maintained its transformability, we conducted further transformation experiments (Table 1). A standard curve was obtained by a series of transformations with pKZ105 which were isolated from E. coli LE392(pKZ105) (Fig. 4). By applying the amount of pKZ105 detected to the standard curve, the number of transformants on each electrotransformation was also estimated. No transformants were obtained with the DNA from cocultivation with C. neglecta, while significant amounts of extracellular pKZ105 DNA were detected. Since the total extracellular DNA concentration was high in coculture with C. neglecta, that level of total extracellular DNA might suppress the transformation event. This hypothesis was tested by conducting the transformation experiments using pure pKZ105 DNA mixed with fish sperm DNA (Fig. 4). The addition of fish sperm DNA slightly enhanced, and did not reduce, the transformation efficiency. Thus, the total extracellular DNA would not have interfered with the transformation, and failure to transform with the DNA from cocultivation with C. neglecta would result in the release of no transformable DNA by cocultivation with this strain. However, in contrast to the result from cocultivation with C. neglecta, the successful electrotransformation did indicate the release of transformable DNA from E. coli LE392(pKZ105) when in cocultivation with E. gracilis, C. inversa, or M. aeruginosa.
TABLE 1.
Electrotransformation of E. coli JM109 with concentrated extracellular DNA from 0.25 ml of cocultured E. coli LE392(pKZ105)
| Culture condi- tion of E. coli LE392(pKZ105) | Cultivation period (no. of days) | No. of transformants (CFU) (mean ± SD)
|
|
|---|---|---|---|
| Expecteda | Detected | ||
| Cocultured with E. gracilis | 3 | 0 | 0 |
| 7 | (5.7 ± 2.7) × 101 | (1.0 ± 0.5) × 100 | |
| 10 | (2.7 ± 0.5) × 102 | (4.0 ± 2.0) × 100 | |
| 17 | (1.2 ± 0.2) × 103 | 0 | |
| Cocultured with C. neglecta | 3 | (2.1 ± 0.2) × 102 | 0 |
| 7 | (1.9 ± 0.9) × 102 | 0 | |
| 10 | (1.0 ± 0.5) × 102 | 0 | |
| 17 | (1.2 ± 0.3) × 102 | 0 | |
| Cocultured with C. inversa | 3 | (2.6 ± 0.2) × 101 | (2.8 ± 1.3) × 101 |
| 7 | (2.7 ± 0.8) × 101 | 0 | |
| 10 | (1.6 ± 0.1) × 101 | 0 | |
| 17 | (1.8 ± 0.1) × 101 | 0 | |
| Cocultured with M. aeruginosa | 3 | (2.7 ± 1.7) × 101 | (1.0 ± 0.5) × 100 |
| 7 | 0 | 0 | |
| 10 | 0 | 0 | |
| 17 | 0 | 0 | |
Expected numbers of transformants were calculated from the equation of linear regression of the standard curve, using the amount of pKZ105 detected from each coculture.
FIG. 4.
Relationship between the number of transformants and the amount of applied pKZ105 DNA mixed with fish sperm DNA. pKZ105 DNA was mixed with the following amounts of fish sperm DNA: 0 (○), 0.1 (▴), 1 (▾), 10 (⧫), and 100 (▪) ng. The solid line indicates the regression; the correlation coefficient was 0.997. The means ± standard deviations (error bars) from three experiments are shown.
Previous studies have also suggested active excretion of DNA from several bacteria, but the mechanism by which those bacteria excrete cellular DNA is not yet clear (3, 10, 20, 26). Lorenz et al. (20) reported the active release of transformable plasmids and chromosomal DNA from competent B. subtilis during growth in minimal media. Their observation suggested that DNA excretion was influenced by the physiological state of the bacterial cells. In aquatic environments, both biotic and abiotic factors are involved in determining the physiological state of bacteria. A recent study has demonstrated that extracellular products from a green alga, Chlamydomonas reinhardtii, suppress heterocyst formation in the cyanobacterium Anabaena flos-aquae (18) and suggested that there are chemical interactions which can alter the physiological state of other organisms and even organisms in other kingdoms. Paul and David (26) showed that the production of extracellular DNA from E. coli HB101(R388) was altered by physiochemical factors such as salinity changes. These studies suggested that the existence of chemical interactions which alter the physiological state of E. coli LE392(pKZ105) and that the subsequent release of transformable DNA could have occurred by cocultivation with algae.
Compounds, such as acylated homoserine lactones and oligopeptides, are known to be involved in cell-to-cell communication and the alternation of both intra- and interspecific behaviors in bacteria (2, 34). As one example of interspecies interaction, compounds produced from the macroalga Delisea pulchra alter the behavior of bacteria by interfering with the bacterial signaling system mediated by acylated homoserine lactones (19, 34). Thus, there may be a signal compound which stimulates DNA excretion from E. coli. Characterization of this compound(s) would clarify this hypothesized-chemical-mediated interaction between algae and bacteria.
The reason why algae stimulate the release of bacterial DNA is another question to which there is no clear answer. One possibility is that stimulation of the release of bacterial DNA, which is rich in phosphorus, may be an alternative way for algae to ameliorate the competition with bacteria for nutrients, since uptake of phosphate is commonly dominated by bacteria (>50 to nearly 100%) in many freshwater environments (36).
The exact mechanisms by which, and the reasons why, the algae tested could induce the excretion of E. coli plasmid DNA still remain to be clarified, but this study has clearly demonstrated algal involvement in the production of transformable plasmid DNA by E. coli bacteria, which may suggest a new ecological interaction facilitating horizontal gene transfer in aquatic environments.
Acknowledgments
This work was supported in part by a Japanese Ministry of Education, Culture, Sports, Science and Technology Grant-in-Aid for Creative Basic Research (09NP1501) and a Grant-in-Aid for Basic Research (A) (13309009). K.M. was also supported by a Research Fellowship of the Japan Society for the Promotion of Science for Young Scientists.
We thank Masaya Ueki for cultivation of algae, Atsushi Maruyama and Yasushi Miyamoto for statistical analysis, and Mary Morris for improving the English.
REFERENCES
- 1.Alonso, M. C., V. Rodriguez, J. Rodriguez, and J. J. Borrego. 2000. Role of ciliates, flagellates and bacteriophages on the mortality of marine bacteria and on dissolved-DNA concentration in laboratory experimental systems. J. Exp. Mar. Biol. Ecol. 244:239-252. [Google Scholar]
- 2.Bassler, B. L. 2002. Small talk: cell-to-cell communication in bacteria. Cell 109:421-424. [DOI] [PubMed] [Google Scholar]
- 3.Catlin, B. W. 1956. Extracellular deoxyribonucleic acid of bacteria and a deoxyribonuclease inhibitor. Science 124:441-442. [DOI] [PubMed] [Google Scholar]
- 4.Chróst, R. J. 1975. Inhibitors produced by algae as an ecological factor affecting bacteria in water. II. Antibacterial activity of algae during blooms. Acta Microbiol. Pol. B 7:167-176. [PubMed] [Google Scholar]
- 5.Chróst, R. J. 1983. Plankton photosynthesis, extracellular release and bacterial utilization of released dissolved organic carbon (RDOC) in lakes of different trophy. Acta Microbiol. Pol. 32:275-287. [PubMed] [Google Scholar]
- 6.Davison, J. 1999. Genetic exchange between bacteria in the environment. Plasmid 42:73-91. [DOI] [PubMed] [Google Scholar]
- 7.DeFlaun, M. F., J. H. Paul, and D. Davis. 1986. Simplified method for dissolved DNA determination in aquatic environments. Appl. Environ. Microbiol. 52:654-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erata, M. 1997. Global Environmental Forum (GEF) list of strains, 5th ed., p. 128. National Institute for Environmental Studies, Environmental Agency, Tsukuba, Japan.
- 9.Frischer, M. E., G. J. Stewart, and J. H. Paul. 1994. Plasmid transfer to indigenous marine bacterial populations by natural transformation. FEMS Microbiol. Ecol. 15:127-136. [Google Scholar]
- 10.Hara, T., and S. Ueda. 1981. A study on the mechanisms of DNA excretion from P. aeruginosa KYU-1: effect of mitomycin C on extracellular DNA production. Agric. Biol. Chem. 45:2457-2461. [Google Scholar]
- 11.Ishii, N., Z. Kawabata, S. Nakano, M. G. Min, and R. Takata. 1998. Microbial interactions responsible for dissolved DNA production in a hypereutrophic pond. Hydrobiologia 380:67-76. [Google Scholar]
- 12.Ishiwa, H., and H. Shibahara. 1985. New shuttle vectors for Escherichia coli and Bacillus subtilis. II. Plasmid pHY300PLK, a multipurpose cloning vector with a polylinker, derived from pHY460. Jpn. J. Genet. 60:235-243. [Google Scholar]
- 13.Iwane, T., T. Urase, and K. Yamamoto. 2001. Possible impact of treated wastewater discharge on incidence of antibiotic resistant bacteria in river water. Water. Sci. Technol. 43:91-99. [PubMed] [Google Scholar]
- 14.Jeffrey, W. H., J. H. Paul, and G. J. Stewart. 1990. Natural transformation of a marine Vibrio species by plasmid DNA. Microb. Ecol. 19:259-269. [DOI] [PubMed] [Google Scholar]
- 15.Jørgensen, N. O. G., and C. S. Jacobsen. 1996. Bacterial uptake and utilization of dissolved DNA. Aquat. Microb. Ecol. 11:263-270. [Google Scholar]
- 16.Karl, D. M., and M. D. Bailiff. 1989. The measurement and distribution of dissolved nucleic acids in aquatic environments. Limnol. Oceanogr. 34:543-558. [Google Scholar]
- 17.Kawabata, Z., N. Ishii, M. Nasu, and M. G. Min. 1998. Dissolved DNA produced through a prey-predator relationship in a species-defined aquatic microcosm. Hydrobiologia 385:71-76. [Google Scholar]
- 18.Kearns, K. D., and M. D. Hunter. 2002. Algal extracellular products suppress Anabaena flos-aquae heterocyst spacing. Microb. Ecol. 43:174-180. [DOI] [PubMed] [Google Scholar]
- 19.Kjelleberg, S., P. Steinberg, M. Givskov, L. Gram, M. Manefield, and R. de Nys. 1997. Do marine natural products interfere with prokaryotic AHL regulatory systems? Aquat. Microb. Ecol. 13:85-93. [Google Scholar]
- 20.Lorenz, M. G., D. Gerjets, and W. Wackernagel. 1991. Release of transforming plasmid and chromosomal DNA from two cultured soil bacteria. Arch. Microbiol. 156:319-326. [DOI] [PubMed] [Google Scholar]
- 21.Lorenz, M. G., and W. Wackernagel. 1994. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol. Rev. 58:563-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsui, K., M. Honjo, and Z. Kawabata. 2001. Estimation of the fate of dissolved DNA in thermally stratified lake water from the stability of exogenous plasmid DNA. Aquat. Microb. Ecol. 26:95-102. [Google Scholar]
- 23.Mazodier, P., and J. Davies. 1991. Gene transfer between distantly related bacteria. Annu. Rev. Genet. 25:147-171. [DOI] [PubMed] [Google Scholar]
- 24.Ochman, H., J. G. Lawrence, and E. A. Groisman. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299-304. [DOI] [PubMed] [Google Scholar]
- 25.Paul, J. H., L. Cazares, and J. Thurmond. 1990. Amplification of rbcL gene from dissolved and particulate DNA from aquatic environments. Appl. Environ. Microbiol. 56:1963-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paul, J. H., and A. W. David. 1989. Production of extracellular nucleic acids by genetically altered bacteria in aquatic-environment microcosms. Appl. Environ. Microbiol. 55:1865-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paul, J. H., W. H. Jeffrey, and M. F. DeFlaun. 1987. Dynamics of extracellular DNA in the marine environment. Appl. Environ. Microbiol. 53:170-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paul, J. H., S. C. Jiang, and J. B. Rose. 1991. Concentration of viruses and dissolved DNA from aquatic environments by vortex flow filtration. Appl. Environ. Microbiol. 57:2197-2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paul, J. H., J. B. Rose, S. C. Jiang, C. A. Kellogg, and L. Dickson. 1993. Distribution of viral abundance in the reef environment of Key Largo, Florida. Appl. Environ. Microbiol. 59:718-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook, J., and D. W. Russell. 2000. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 31.Siuda, W., R. J. Chróst, and H. Güde. 1998. Distribution and origin of dissolved DNA in lakes of different trophic states. Aquat. Microb. Ecol. 15:89-96. [Google Scholar]
- 32.Siuda, W., and H. Güde. 1996. Determination of dissolved deoxyribonucleic acid concentration in lake water. Aquat. Microb. Ecol. 11:193-202. [Google Scholar]
- 33.Turk, V., A. S. Rehnstam, E. Lundberg, and Å. Hagström. 1992. Release of bacterial DNA by marine nanoflagellates, an intermediate step in phosphorus regeneration. Appl. Environ. Microbiol. 58:3744-3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whitehead, N. A., A. M. L. Barnard, H. Slater, N. J. L. Simpson, and G. P. C. Salmond. 2001. Quorum-sensing in gram-negative bacteria. FEMS. Microbiol. Rev. 25:365-404. [DOI] [PubMed] [Google Scholar]
- 35.Weinbauer, M. G., D. Fuks, and P. Peuzzi. 1993. Distribution of viruses and dissolved DNA along a coastal trophic gradient in the northern Adriatic Sea. Appl. Environ. Microbiol. 59:4074-4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wetzel, R. G. 2001. Limnology. Academic Press, San Diego, Calif.