Abstract
For 30 years it has been assumed that a single species of cyanobacteria, Phormidium corallyticum, is the volumetrically dominant component of all cases of black band disease (BBD) in coral. Cyanobacterium-specific 16S rRNA gene primers and terminal restriction fragment length polymorphism analyses were used to determine the phylogenetic diversity of these BBD cyanobacteria on coral reefs in the Caribbean and Indo-Pacific Seas. These analyses indicate that the cyanobacteria that inhabit BBD bacterial mats collected from the Caribbean and Indo-Pacific Seas belong to at least three different taxa, despite the fact that the corals in each case exhibit similar signs and patterns of BBD mat development.
Black band disease (BBD) is a globally distributed coral disease that affects a variety of different coral species (3, 16). It is manifested as a bacterial mat, ranging from 0.1 mm to a few centimeters wide, that migrates across infected corals at a rate as high as 1 cm/day from top to bottom, killing healthy tissue and leaving behind dead skeletal surfaces stripped clean of tissue (3, 13, 15). Because BBD preferentially infects massive framework-building corals (3, 6), the aftermath of the disease has a profound influence on the ecological and geological structures of coral reef ecosystems (6, 9). Since BBD was first reported in the 1970s, it has been known that the BBD microbial mat is dominated by a filamentous cyanobacterium (2, 3) that was originally optically identified as Oscillatoria submembranaceae (3) and later reclassified as Phormidium corallyticum (16). P. corallyticum has been the focus of many BBD studies (9, 13, 14, 15) and has also been proposed to be the disease pathogen (16). However, it has not yet been proven by a systematic fulfillment of Koch's postulates that P. corallyticum is a BBD pathogen. P. corallyticum cyanobacteria create a ropy network that structurally supports the BBD mat and houses a complex, phylogenetically diverse community of at least 40 species of bacteria (8). Moreover, the microbial consortium of the BBD mat is 92% distinct from the microbial communities that inhabit overlying seawater, healthy coral tissue, and dead coral surfaces (8). Cyanobacteria produce low-molecular-weight toxins that can kill fish and other animals during bacterial blooms (4). However, as a group, cyanobacteria have not generally been considered to be associated with the infection of eukaryotic tissue. Prior to the present study, culture-independent molecular techniques have not specifically targeted the filamentous cyanobacteria associated with BBD. Therefore, it is unknown whether BBD mats growing on infected coral surfaces in different ocean basins actually contain the same species of cyanobacteria. Difficulties in growing pure cultures of BBD cyanobacteria have proven to be a significant obstacle to completing the 16S rRNA gene sequence for identification purposes. The goal of the present study was to determine the phylogenetic identity of the cyanobacteria that volumetrically dominate BBD mats and to determine whether a single species occurs on the coral reefs within an ocean basin and on those in different ocean basins.
Phylogenetic diversity of cyanobacteria present in BBD.
Comparative analyses were performed on BBD mat samples collected from infected corals at three different locations along the southwestern coast of Curaçao, Netherlands Antilles, in the Caribbean Sea, and at two different locations on the northern coast of New Britain, Papua New Guinea, in the Indo-Pacific Sea. On Curaçao, samples were collected from three back reef sites at water depths of 4 to 5 m along the southern coast of the island in August 2000 and in January and August 2001. These sites included the sea aquarium, the water plant, and Playa Kalki (Fig. 1). On New Britain, BBD mat samples were collected from an equivalent back reef environment off the northern coast at Father's Reef (Fig. 1) at a water depth of 5 m in August 2000. A natural segregation of coral species exists between the Caribbean and Indo-Pacific Seas; the framework-building corals of the Caribbean belong to the family Faviidae, and those in the Indo-Pacific belong to the family Poritidae (17). Samples were taken from actively migrating BBD mats on 12 different coral colonies representing four different coral species. These included three coral species from Curaçao (six colonies of Diploria strigosa, two colonies of Montastrea cavernosa, and one colony of Montastrea annularis) and one coral species from New Britain (two colonies of Porites lutea). Only two cases of BBD were observed on New Britain corals along almost 1,200 km of coastline, while over 50 cases of BBD were observed on Curaçao corals along only 50 km of coastline (7). In addition, a nonaxenic culture of P. corallyticum provided by L. Richardson (Florida International University, Miami) and collected in the Florida Keys was analyzed. Finally, data from samples taken from two other locations each in the Caribbean, Barbados, and the U.S. Virgin Islands (5) were compared with the results obtained in the present work.
FIG. 1.
Map of sampling sites on the islands of Curaçao, Netherlands Antilles, and New Britain, Papua New Guinea.
Visual analyses of our samples by fluorescence and scanning electron microscopies confirmed the dominance of a filamentous cyanobacterium in each BBD mat sample (16). Although the bacterium thought responsible for BBD had been optically identified previously as P. corallyticum (16), recent 16S rRNA gene analyses of BBD mats did not detect the genus Phormidium in either healthy or BBD-infected coral tissues (5, 8). The molecular sampling, extraction, cloning, and sequencing techniques used in this study have been presented previously (8). In the present study, two sets of primers specific for the cyanobacterial 16S rRNA gene (11) were used on environmental BBD mat samples that contained complex microbial communities. In addition, terminal restriction fragment length polymorphism (T-RFLP) analyses of PCR products obtained by using one set (CYA106) of cyanobacterium-specific primers used previously (10) provided relative estimates of the most abundant cyanobacterium present in selected samples of the BBD mat.
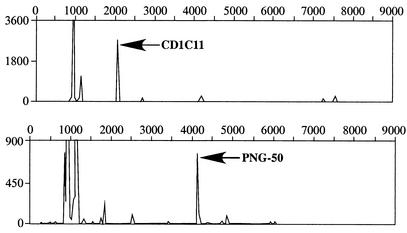
A total of 31 clones from the six colonies of D. strigosa, 8 clones from the two colonies of M. cavernosa, and 8 clones from the one colony of M. annularis yielded identical 16S rRNA gene sequences, regardless of the coral species sampled and its location. Sequences ranging from 300 to 600 bp were used for the analysis. A partial gene sequence for this cyanobacterium had previously been reported and designated CD1C11 (8) (Fig. 2). A new sequence has been identified in the present study from both of the cases of BBD observed in New Britain, and this clone is herein designated PNG-50. The relative abundance of cyanobacterium CD1C11 and PNG-50 in each BBD mat was determined by T-RFLP analyses as described previously (10). T-RFLP analyses were applied in this study to assess the relative abundance of microorganisms. The results indicate that the sequences obtained for the cyanobacterial clones CD1C11 and PNG-50 are the most abundant in the BBD mats. The use of cyanobacterium-specific primers proves that this result is not the product of random amplification of some other cyanobacterial 16S rRNA genes. The forward primer was labeled at the 5′ end with 6-carboxyfluorescein. Aliquots of amplified 16S rRNA genes were digested with HhaI, RsaI, and MspI. The actual sizes of the peaks obtained by T-RFLP were compared with the expected sizes generated by a theoretical digest of the previously obtained sequences from CD1C11 and PNG-50. These analyses document that with respect to the cyanobacterial portion of the community, CD1C11 and PNG-50 were the most abundant (10) cyanobacteria found in the BBD mats collected from the Caribbean and Indo-Pacific reefs, respectively (Fig. 3). Furthermore, samples from the Caribbean showed the presence of at least four additional types of cyanobacteria at much lower concentrations than that of CD1C11. Samples from the Indo-Pacific showed the presence of at least six other types of cyanobacteria, and two of these were present in high concentrations.
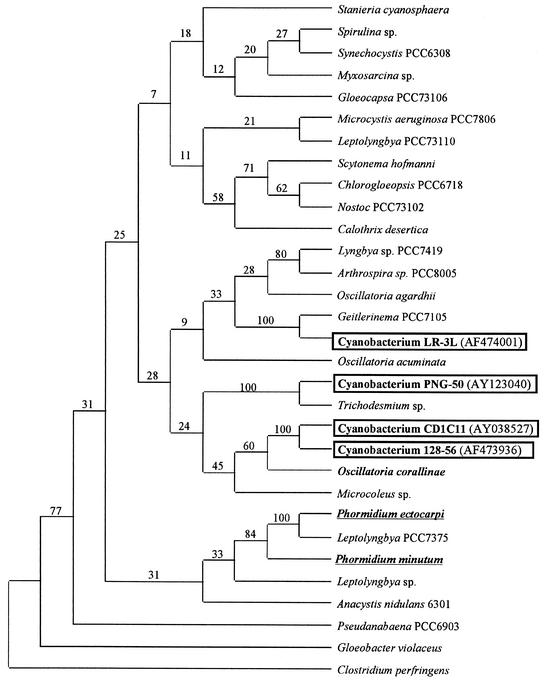
FIG. 2.
Phylogenetic consensus tree based on parsimony analysis of 16S rRNA gene sequences of BBD cyanobacteria and other cyanobacteria. The tree is based on sequences of 300 to 600 bp in length. These represent a total of 31 clones from six colonies of D. strigosa, 8 clones from two colonies of M. cavernosa, and 8 clones from one colony of M. annularis. The numbers at the nodes are the bootstrap values based on a total of 10,000 replicate resamplings. GenBank accession numbers are included. The underlined strains represent marine Phormidium spp.
FIG. 3.
Results of T-RLFP analysis of BBD samples from Curaçao (top) and New Britain (bottom) by using specific primers for the cyanobacterial 16S rRNA gene. PCR analyses of products from the BBD mats were obtained by using the primers indicated in the text for amplifying cyanobacterial 16S rRNA genes.
Phylogenetic analysis of the samples was performed by using the program CLUSTALX to align the sequences. Phylogenetic analysis and trees were generated by using the program PAUP. Trees were constructed by parsimony analysis by bootstrapping 10,000 trees from resampled data. Results of the phylogenetic analyses showed that the 16S rRNA gene sequence from clone CD1C11 was 98% similar to cyanobacterial sequence 128-56. This uncultured cyanobacterium has been reported to be present in BBD mats on D. strigosa and M. annularis in other Caribbean sites, including Barbados and the U.S. Virgin Islands (5). This relationship is supported by a 100% bootstrap correlation of the phylogenetic trees. In addition, cyanobacterium CD1C11 and cyanobacterium 128-56 form a cluster with Oscillatoria corallinae, a marine benthic strain isolated from the surface of a worm tube whose sequence was supported in only 60% of the bootstrap trees. Importantly, sequences from two marine Phormidium species appear to be more distantly related to this Oscillatoria strain than to each other (Fig. 2), which suggests the need for future reclassification of P. corallyticum as a member of the genus Oscillatoria.
In addition, the 14 clones obtained from the nonaxenic culture of P. corallyticum, herein called cyanobacterium LR-3L (5), yielded unique sequences that were not affiliated with cyanobacteria detected at other sites in the Caribbean and Indo-Pacific Seas. However, the bootstrap values supporting the separation of these branch points were only 9 and 24% (Fig. 2). The closest related genus to cyanobacterium LR-3L was Geitlerinema, which also belongs to the order Oscillatoriales, in which P. corallyticum has been grouped. Identical phylogenetic relationships for cyanobacterium LR-3L have previously been reported (5). Eleven clones generated from BBD mats collected in the Indo-Pacific Sea yielded gene sequences, herein designated PNG-50, that are affiliated with a different genus of filamentous cyanobacterium which is closely related to Trichodesmium spp. These results indicate that at least three taxonomically distinct filamentous cyanobacteria—two taxa from the Caribbean Sea and one taxon from the Indo-Pacific Sea—are associated with the BBD mats that were analyzed.
Cyanobacterium CD1C11 was the most common cyanobacterium detected in Caribbean corals in this study, occurring in three of the four sample sites analyzed. The presence or absence of this cyanobacterium does not appear to be coral species specific. The absence of CD1C11 sequences in the BBD mat samples from New Britain, as well as its presence in all but one sample from the Caribbean, suggests that the cyanobacteria associated with BBD may have distinct geographic distributions. If that is the case, this biogeographic distribution is significantly different from that observed for Trichodesmium, one of the most abundant genera of filamentous cyanobacteria in the world's oceans. Trichodesmium spp. from the Indian Ocean, the Caribbean Sea, and the Indo-Pacific Sea exhibit essentially identical 16S rRNA gene sequences (12). The phylogenetic diversity of cyanobacteria associated with BBD may therefore at least partially explain the variable environmental conditions in which BBD has been observed (1). Little is known about the environmental factors that lead to the development of BBD. The genetic diversity among the cyanobacteria that are part of the BBD mat is a new discovery that may help explain why BBD has been observed under many different environmental conditions (1, 7, 14). BBD may be a consequence of combined coral and microbial responses to environmental stress. Each of the different cyanobacteria that we have detected may have a different environmental tolerance with regard to temperature, heavy metal contamination, organic pollution, and other factors.
Occurrence of cyanobacterium CD1D11 on healthy coral tissues, BBD mats, dead coral surfaces, and overlying seawater.
Questions about how BBD is transmitted and what the natural habitat for cyanobacterium CD1D11 is remain unanswered. One previous study that used microscopy to detect P. corallyticum showed that this cyanobacterium is present in healthy corals, although at a low frequency (14). The sequence for CD1C11 was used in the present study as a template for designing primers to make PCR a more sensitive method to detect the presence of this cyanobacterium in different marine subenvironments (including overlying seawater, healthy coral tissues, BBD mats, and dead coral surfaces) and therefore better elucidate its potential source (8). We chose this sequence because it was detected in all of our BBD samples from the Caribbean Sea and because its presence has now been reported in two other Caribbean locations (5). The specific primers used for detecting P. corallyticum were PhcF (5′-CTGTAGGTGGCCAGCT-3′) and PhcR (5′-TTCCCTTCGCAGGTTCGCTGC-3′). The specificity of these primers was tested by comparing them to a clone library of several representative cyanobacterial 16S rRNA gene sequences, including clones CD1C11, PNG-50, and LR-3L (Fig. 2). Results indicated that the primers amplified DNA exclusively from clone CD1C11. The conditions for PCR have been described previously (8). A total of 33 different samples were analyzed. All BBD mat samples except for the New Britain samples were positive for the presence of clone CD1C11 (Table 1). In the case of the dead zone left behind by the BBD mat, half of the samples analyzed were positive for the presence of clone CD1C11. However, none of the healthy coral species or the seawater analyzed gave a positive result. This low frequency of occurrence of CD1C11 agreed with previous results (14). Whatever the natural habitat for CD1C11 might be, its presence in the different marine environments sampled appeared to be at low concentrations except when growing in the BBD mat.
TABLE 1.
Occurrence of cyanobacterium CD1C11 in different coral samples and seawater
| Coral and condition or seawater | No. of samples | Positivity for CD1C11
|
|
|---|---|---|---|
| No. of samples | Frequency (%) | ||
| Montastrea cavernosa, BBD infected | 2 | 2 | 100 |
| Montastrea annularis, BBD infected | 1 | 1 | 100 |
| Diploria strigosa, BBD infected | 6 | 6 | 100 |
| Porites lutea, BBD infected | 1 | 0 | 0 |
| Montastrea cavernosa, BBD infected, healthy | 1 | 0 | 0 |
| Montastrea annularis, healthy | 3 | 0 | 0 |
| Diploria strigosa, healthy | 3 | 0 | 0 |
| Porites lutea, healthy | 3 | 0 | 0 |
| Porites furcata, healthy | 2 | 0 | 0 |
| Porites astreoides, healthy | 1 | 0 | 0 |
| Montastrea annularis, dead | 1 | 1 | 100 |
| Montastrea cavernosa, dead | 1 | 1 | 100 |
| Diploria strigosa, dead | 3 | 1 | 33 |
| Porites astreoides, dead | 1 | 0 | 0 |
| Seawater | 4 | 0 | 0 |
Conclusion.
It had previously been hypothesized that a single cyanobacterial species was the primary microbial constituent of BBD mats. The present work shows that more than one species of cyanobacteria constitute BBD mats and that these species differ not only between the Caribbean and Indo-Pacific Seas but also within the Caribbean Sea. In the Caribbean, each of the cyanobacteria is present on multiple coral species of the family Faviidae that are infected with BBD. These results provide the microbial baseline needed for future studies that will attempt to identify environmental controls of BBD transmission through seawater and the resulting initiation and development of infection on healthy corals.
Acknowledgments
This work was supported by a research grant from the Office of Naval Research.
The invitation of P. Cruz and his research team to join their Natural Products expedition to Papua New Guinea was greatly appreciated. We thank J. Klaus for his review of the manuscript.
The conclusions in this report are those of the authors and do not necessarily reflect those of the funding agencies.
REFERENCES
- 1.Al-Moghrabi, S. M. 2001. Unusual black band disease (BBD) outbreak in the northern tip of the Gulf of Aqaba (Jordan). Coral Reefs 19:330-331. [Google Scholar]
- 2.Antonius, A. 1981. Coral reef pathology—a review, p. 3-6. In E. D. Gomez et al. (ed.), The reef and man. Proceedings of the Fourth International Coral Reef Symposium, vol. 2. Marine Sciences Center, University of the Philippines, Quezon City.
- 3.Antonius, A. 1981. The ‘band’ diseases in coral reefs, p. 7-14. In E. D. Gomez et al. (ed.), The reef and man. Proceedings of the Fourth International Coral Reef Symposium, vol. 2. Marine Sciences Center, University of the Philippines, Quezon City.
- 4.Codd, G. A., C. J. Ward, and S. G. Bell. 1997. Cyanobacterial toxins: occurrence, modes of action, health effects and exposure route. Arch. Toxicol. Suppl. 19:399-410. [DOI] [PubMed] [Google Scholar]
- 5.Cooney, R. P., O. Pantos, M. D. A. Le Tissier, M. R. Barer, A. G. O'Donell, and J. C. Bythell. 2002. Characterization of the bacterial consortium associated with black band disease in coral using molecular microbiological techniques. Environ. Microbiol. 4:401-413. [DOI] [PubMed] [Google Scholar]
- 6.Edmunds, P. J. 1991. Extent and effect of black band disease on a Caribbean reef. Coral Reefs 10:161-165. [Google Scholar]
- 7.Fortwengler, M. R. 2002. Distribution and frequency of black band disease and partial mortality of Diploria strigosa on Curaçao, Netherland Antilles. M.S. thesis. University of Illinois Urbana-Champaign, Urbana.
- 8.Frias-Lopez, J., A. L. Zerkle, G. T. Bonheyo, and B. W. Fouke. 2002. Partitioning of bacterial communities between seawater and healthy, black band diseased, and dead coral surfaces. Appl. Environ. Microbiol. 68:2214-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuta, K., and L. L. Richardson. 1996. Abundance and distribution of black band disease of corals in the northern Florida Keys. Coral Reefs 15:219-223. [Google Scholar]
- 10.Liu, W.-T., T. L. Marsh, H. Cheng, and L. J. Forney. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nübel, U., F. Garcia-Pichel, and G. Muyzer. 1997. PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl. Environ. Microbiol. 63:3327-3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orcutt, K. M., U. Rasmussen, E. A. Webb, J. B. Waterbury, K. Gundersen, and B. Bergman. 2002. Characterization of Trichodesmium spp. by genetic techniques. Appl. Environ. Microbiol. 68:2236-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richardson, L. L. 1996. Motility patterns of Phormidium corallyticum and Beggiatoa spp. associated with black band disease of corals. Microb. Ecol. 32:323-335. [DOI] [PubMed] [Google Scholar]
- 14.Richardson, L. L. 1997. Occurrence of the black-band disease cyanobacterium on healthy corals of the Florida Keys. Bull. Mar. Sci. 61:485-490. [Google Scholar]
- 15.Richardson, L. L. 1998. Coral diseases: what is really known? Trends Ecol. Evol. 13:438-443. [DOI] [PubMed] [Google Scholar]
- 16.Rutzler, K., and D. Santavy. 1983. The black band disease of Atlantic reef corals. I. Description of a cyanophyte pathogen. Mar. Ecol. 4:301-319. [Google Scholar]
- 17.Veron, J. E. N. 2000. Corals of the world. Odyssey Publishing, El Cajon, Calif.