Abstract
Nine Listeria monocytogenes strains were treated individually with a continuous pulsed electric field (PEF) apparatus, and their sensitivities to the treatment were compared at 25 kV/cm. When cell suspensions of these strains in 0.1% NaCl (pH 7.0) were treated at 23°C for 144 μs, inactivation ranged from 0.7 to 3.7 log10 CFU/ml. Inactivation by 72-μs PEF treatments at 37°C ranged from 0.3 to 2.5 log10 CFU/ml. L. monocytogenes OSY-8578 was substantially more resistant than other strains when cells were PEF treated in 0.1% NaCl, whereas Scott A was one of the most sensitive strains. The superiority of OSY-8578's resistance to that of Scott A was confirmed in 50% diluted acid whey (pH 4.2). Changes in sensitivity to PEF during phases of growth were minimal in OSY-8578 and substantial in Scott A. Use of L. monocytogenes OSY-8578, therefore, is recommended in studies to optimize PEF processes that target L. monocytogenes. The nine L. monocytogenes strains were genotyped with pulsed-field gel electrophoresis (PFGE) and arbitrarily primed PCR (AP-PCR) techniques. These strains were better differentiated with PFGE than with AP-PCR. The target strain (OSY-8578) was characterized by both molecular typing techniques, but resistance to PEF, in general, was not associated with a particular genotype group.
The presence of Listeria monocytogenes in food constitutes a serious health hazard to consumers. This gram-positive pathogen has been detected in most raw foods, and its presence in processed products has caused numerous outbreaks of listeriosis, a disease with a high mortality rate (14, 35). Heat treatment is the most commonly used processing technique for inactivating L. monocytogenes in food. Alternative processing technologies such as pulsed electric field (PEF) are presently being developed to meet the increasing demand for safe food with fresh-like attributes (25). Gram-positive bacteria are more resistant to PEF processing than are gram-negative bacteria (e.g., reference 21). Reina et al. (34), for instance, reported a 4.2-log10 reduction in the count of L. monocytogenes Scott A when milk was treated for 600 μs at 30 kV/cm, whereas Grahl and Maerkl (18) found a similar degree of inactivation when Escherichia coli was treated in milk for only 200 μs at 22.4 kV/cm. Furthermore, the high PEF resistance of Listeria, compared to that of other gram-positive bacteria, was confirmed in various media (1, 12). Treatment with PEF for 360 μs at 20 kV/cm decreased L. monocytogenes and Staphylococcus aureus populations in phosphate buffer by 1 and 2 log10 CFU/ml, respectively (21). Therefore, destruction of L. monocytogenes is a critical measure of PEF efficacy in treatment of potentially contaminated food.
Resistant microbial strains are problematic to the food industry but are also useful as “targets” for designing safe processes. Limited information is available in published literature on variations in resistance to PEF among strains of the same microorganism. Published research on the efficacy of PEF against L. monocytogenes is generally based on single-strain studies. L. monocytogenes strains vary only slightly in resistance to heat (13), but the variability of these strains to inactivation by PEF processing is not known. Thermal inactivation comprises multiple simultaneous mechanisms, whereas PEF largely relies upon one. Heat concurrently alters membrane permeability, denatures cell proteins, causes single-strand breaks in DNA, and degrades RNA (3, 17). Lethality of PEF is mainly caused by membrane disruption (e.g., references 24, 25, and 40). Therefore, variation among strains in susceptibility to processing is believed likelier with PEF than with heat. The variability of L. monocytogenes strains' inactivation by PEF processing, to our knowledge, has not been studied before and should be assessed to identify potential target strains.
Genotyping of pathogens is widely used in epidemiological studies to facilitate tracking of virulent or processing-resistant strains in food-processing installations and to determine the contamination routes of these strains (9, 38). Genotyping is based on the detection of DNA polymorphism and is increasingly preferred to serotyping for its reproducibility and high discriminative power (2). Pulsed-field gel electrophoresis (PFGE) and ribotyping are generally the most sensitive genotyping methods (2, 20), but their cost, equipment unavailability, and poor adaptability to routine analysis are limiting the widespread use of these techniques (32). PFGE, however, is preferred to ribotyping in epidemiological studies of L. monocytogenes for its reproducibility and sensitivity (19, 20). Arbitrarily primed PCR (AP-PCR) is inexpensive to run, rapid, and easy to implement in most research laboratories (30). This study, therefore, compared AP-PCR and PFGE in identifying potential target strains for PEF processing.
To help design or optimize safe PEF processes, the first objective of this study was to identify an L. monocytogenes target strain. Since cell resistance to PEF may vary with initial treatment temperature (1, 11), this factor was considered during selection of the target strain. Resistance of the target strain to PEF was verified in simple (0.1% NaCl, pH 7.0) and food type (50% acid whey, pH 4.2) media. The second objective was to determine if the target strain remained PEF resistant at different ages of the culture. Maintaining this resistance is a desirable property of a target strain because cells of the exponential phase may lack resistance against environmental and processing stress (27, 33). Finally, all L. monocytogenes strains used in the screening were typed by AP-PCR and PFGE. The genotyping was carried out to select a method sufficiently discriminatory to recognize the target strain and to correlate molecular types with PEF sensitivity.
MATERIALS AND METHODS
Microorganisms.
L. monocytogenes Scott A (clinical isolate), California (milk isolate), and V7 (cheese isolate) were obtained from the culture collection of the Food Safety Laboratory at The Ohio State University. Strains OSY-8517, OSY-8576, OSY-8578, OSY-8579, OSY-8580, and OSY-8732 (isolated from meat) were provided by the Ohio Department of Agriculture (Reynoldsburg, Ohio). Stock cultures were stored at −80°C in tryptose broth (TB; Difco, Detroit, Mich.) containing 40% (vol/vol) glycerol. L. monocytogenes strains were propagated in TB by incubation at 37°C for 18 h. Cultures were transferred at least twice before use.
Cell suspension media.
Bacterial cells were suspended in 0.1% NaCl solution or in diluted acid whey for PEF treatment. The 0.1% (wt/vol) NaCl solution was at pH 7.0, with an electrical conductivity of 0.21 S/m. Acid whey was collected during cottage cheese production (Arps Dairy, Inc., Defiance, Ohio) and was stored at ∼4°C prior to filter sterilization. The whey was filter sterilized (0.4 μm pore size; Osmonics Inc., Westborough, Mass.) and was degassed with a vacuum pump prior to PEF treatment. The PEF equipment used for this experiment requires low-conductivity solutions for optimum operation. The filtered acid whey (pH 4.2), therefore, was diluted with an equal volume of sterile deionized water to lower the solution conductivity to 0.46 S/m; this product will be referred to as 50% acid whey.
PEF equipment.
The OSU 4-C PEF unit (Department of Food Science and Technology, Ohio State University, Columbus) was used (41). Samples with an initial temperature of 15, 23, or 37°C were passed through four co-field flow treatment chambers in series. The chamber diameter was 2.3 mm, and the electrode gap was 2.9 mm. Before and after passing through a pair of treatment chambers, the sample was adjusted to the predetermined initial temperature (15, 23, or 37°C) in coils submerged in a water bath. The flow rate was adjusted to 1 and 2 ml/s to achieve total treatment times of 144 and 72 μs, respectively. An electric field of 25 kV/cm, with bipolar pulses of 3 μs, was applied at a frequency of 1,000 Hz for cells suspended in NaCl and 667 Hz for cells suspended in the 50% acid whey. An oscilloscope (TDS 340A; Tektronix, Beaverton, Oreg.) monitored the square wave pulse, input voltage, and current.
PEF treatments.
Stock cultures of the nine L. monocytogenes strains were transferred (at 0.1% inoculation level) into TB and were incubated at 37°C. The cultures were sampled after 18 h of incubation to screen for a target strain at pretreatment temperatures of 23 and 37°C and to assess the PEF resistance of selected strains in 50% acid whey. Cultures of L. monocytogenes Scott A and OSY-8578 were also sampled after 4, 6, 8, 10, 12, 15, 18, and 24 h of incubation to determine age-related PEF sensitivity profiles at 15°C. For all treatments, cells were harvested by centrifugation at 5,000 × g for 10 min (Sorvall RC-5B; DuPont, Wilmington, Del.) at 4°C and were washed briefly by suspension in 0.1% NaCl, followed by centrifugation. Washed cells were resuspended in 0.1% NaCl at 15°C to determine the age sensitivity profile and at 23 or 37°C to screen for a target strain. The optical density of the suspensions at 600 nm was adjusted to an absorbance of ∼0.07, which corresponded to 107 to 108 CFU/ml. Cell suspensions were treated with PEF at 25 kV/cm for 144 μs when initial treatment temperatures (PEF inlet) were set at 15 and 23°C; the sample temperature increased momentarily to 52 to 56°C during the PEF treatment. Treated samples were rapidly cooled in a water-ice mixture. The processing time was reduced to 72 μs for samples with initial temperature of 37°C in order to maintain a similar sample temperature at the outlet of the PEF.
To establish survivor plots, washed cells of Scott A and OSY-8578 were suspended in 0.1% NaCl or 50% acid whey. Suspensions at 23°C were treated with PEF for 72 μs (0.1% NaCl) and 48 μs (50% acid whey). The sample temperature at the outlet of the PEF unit ranged between 37 and 40°C prior to cooling to 23°C in a water bath. A sample was taken for enumeration, and the rest of the processed suspension was treated again with PEF once in the case of NaCl and twice for the acid whey medium.
PEF inactivation data analysis.
Treated and untreated samples were serially diluted in 0.1% (wt/vol) peptone water and were plated onto tryptose agar (Difco) for enumeration. Two independent repeats of each experiment were executed, and inactivation during PEF treatment was calculated as follows: PEF inactivation = log10 CFU/milliliter before treatment − log10 CFU/milliliter after treatment.
All statistical analyses were made by using SAS statistical programs (Cary, N.C.). Averages of the PEF inactivation of the nine strains were compared by one-way analysis of variance (ANOVA) and Tukey's test. “Proc Mixed” procedure from SAS was used to compare the PEF inactivation of Scott A and OSY-8578 at different stages of growth. Analyses were done according to the following statistical model:
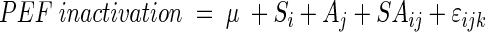 |
where Si is the fixed effect of strain (i = 1, 2); Aj is the fixed effect of culture age (j = 1,…, 8); SAij is the fixed effect of the ith strain by jth culture age; and ɛijk is the error term, assuming that εijk ∼ N(0, σij2). Error variance was not uniform at different stages of growth, and thus, a statistical procedure was followed to account for this variance inequality. Initially, individual error variances were fitted for each S × A subclass. A clear pattern of three magnitudes (groups) of variances emerged. Thus, a final model was fitted, where variance of each subclass belonged to one of the three variance groups. Comparisons of least-square means accounted for variance inequality and were done with Tukey's test.
The relationship between PEF inactivation and culture age (i.e., growth stage) was modeled by using orthogonal polynomials (36). To account for the uneven time intervals between tested ages, the interactive mixed-language procedure of SAS was used. This procedure generated appropriate orthogonal coefficients for linear, quadratic, and cubic contrasts. The highest significant polynomial term to be used with each strain was then identified. Parameters for the polynomial coefficients were estimated by using the SAS Proc Mixed procedure with culture age defined as a continuous variable (26).
AP-PCR analysis.
DNA phenol-chloroform extraction was adapted from Giraffa et al. (16). L. monocytogenes was inoculated, at 0.1% level, into 20 ml of TB and was grown at 37°C for 24 h. Cells were harvested by centrifugation as previously described, suspended in 800 μl of lysis buffer (10 mM Tris, pH 8.0; 12.5% [wt/vol] sucrose; and 5 mg of lysozyme/ml), and incubated at 37°C for 1 h. Cells suspensions were mixed with 100 μl of EDTA (0.5 M) and 100 μl of sodium dodecyl sulfate (10% [wt/vol]). After 1 h of incubation at ∼22°C, 5 μl of proteinase K (2 mg/ml) was added and the mixture was incubated at 60°C for 30 min and then centrifuged at 21,000 × g and 4°C for 15 min. DNA was extracted from the supernatant with a mixture of phenol, chloroform, and isoamyl alcohol (25:24:1 [vol/vol/vol]). The aqueous phase (∼500 μl) was collected, and DNA was precipitated with 1,000 μl of absolute ethanol and 50 μl of sodium acetate (3 M). The DNA was collected by centrifugation (21,000 × g and 4°C) for 15 min, washed with 70% (vol/vol) ethanol, and resuspended in 25 μl of Tris-EDTA buffer (10 mM Tris, pH 8.0; and 1 mM EDTA). Preparations were stored at −20°C until used for AP-PCR analysis. After preliminary trials using a variety of protocols, a modification of the procedure described by Louie et al. (28) was followed. Random oligonucleotide primers with an appropriate melting temperature and a low probability to anneal to themselves were used. The primers were PJ108 (5′-GCTTATTCTTGACATCCA-3′) and PJ118 (5′-TGTTCGTGCTGTTTCTG-3′). A mixture for amplification (50-μl final volume) was composed of 1.5 ng of purified genomic DNA into a PCR mix (10 mM Tris-HCl, pH 8.3; 50 mM KCl; 2.5 mM MgCl2; a 200 μM concentration each of dATP, dGTP, dCTP, and dTTP; a 3 μM concentration of a single primer; and 0.5 U of Taq polymerase). Sterile water (high-performance liquid chromatography grade) was used instead of the template DNA as a negative control. Genomic DNA was subjected to PCR in a PCR thermocycler (GeneAmp; Perkin-Elmer, Norwalk, Conn.). The thermocycler program included heating the amplification mixture for 3 min at 94°C, followed by 32 cycles at (i) 94°C for 1 min, (ii) 40°C for 2 min, and (iii) 72°C for 2 min. In the final cycle, samples were maintained at 72°C for 10 min. Amplified DNA products were resolved by electrophoresis on a 1.2% agarose-Tris-borate-EDTA (1× Tris-borate-EDTA) gel. A 1-kb DNA ladder (Promega, Madison, Wis.) was included in the gel to evaluate the molecular weight of the PCR products. Fragments were stained in ethidium bromide (0.5 μg/ml) and were visualized with a UV transilluminator (wavelength of incident light is 302 nm).
PFGE analysis.
The PFGE analysis procedure was adapted from Louie et al. (28). Briefly, L. monocytogenes was grown overnight in 5 ml of TB at 37°C, and cells were harvested as indicated earlier. Cells were suspended in 750 μl of buffer (10 mM Tris-HCl, pH 7.6, and 1 M NaCl). The cell suspension of each strain was mixed with an equal volume of 1.6% low-melting-point PFGE-grade agarose (SeaKem Gold; BMA, Rockland, Maine) to make agarose plugs. Plugs were incubated in lysis buffer (6 mM Tris-HCl, pH 7.6; 1 M NaCl; 100 mM EDTA, pH 8.0; 0.5% Brij 58; 0.2% deoxycholic acid; 0.5% Sarkosyl; 1 mg of lysozyme/ml; and 500 U of mutanolysin/ml) (Sigma, St. Louis, Mo.) at 35°C for 24 h. Lysed cells were then treated, at 35°C for 24 h, in 1 ml of pronase solution (1 mg of pronase/ml; 0.5% Sarkosyl [Sigma]; and 0.1 mM EDTA, pH 8.0). Plugs were washed five times with Tris-EDTA buffer (10 mM Tris-HCl, pH 7.6; and 0.1 mM EDTA, pH 8.0) and were then incubated for 24 h with an endonuclease (ApaI or SmaI; Invitrogen, Carlsbad, Calif.) at the temperatures recommended by the manufacturer. The DNA fragments entrapped in the plugs were subjected to electrophoresis in a 1% agarose (SeaKem Gold) gel in 0.5× Tris-borate-EDTA buffer at 6 V/cm and 12°C by using a contour-clamped homogeneous electric field PFGE system (DR-II; Bio-Rad, Hercules, Calif.). For ApaI restriction, the pulse times were linearly ramped from 2.5 to 35 s over 18 h. For SmaI digestion, the pulsed time linearly ramped from 1 to 25 s over 18 h. Banding patterns were compared with the Bionumerics software (Applied Maths, Kortrijk, Belgium). Clearly resolved bands only were counted. The Dice coefficient was used to analyze the similarities of the banding patterns. The unweighted pair group method with average linkages was used for cluster analysis.
RESULTS
Screening for PEF-resistant L. monocytogenes.
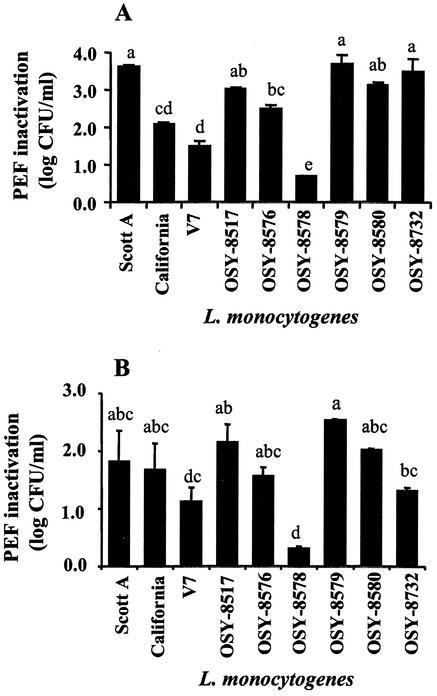
Sensitivity to 25-kV/cm PEF treatment in 0.1% NaCl (pH 7.0) at 23 and 37°C varied greatly (P < 0.01) among nine L. monocytogenes strains (Fig. 1). Treatment with PEF at 23°C for 144 μs inactivated 0.7 to 3.7 log10 CFU/ml and resulted in this order of increasing sensitivity (Fig. 1A): OSY-8578, V7, California, OSY-8576, OSY-8517, OSY-8580, OSY-8732, Scott A, and OSY-8579. Inactivation by PEF at 37°C for 72 μs ranged from 0.3 to 2.5 log10 CFU/ml, and sensitivity to the treatment was in this increasing order (Fig. 1B): OSY-8578, V7, OSY-8732, OSY-8576, California, Scott A, OSY-8580, OSY-8517, and OSY-8579. Strains OSY-8578 and V7 were the most resistant, whereas OSY-8579 and Scott A were among the most sensitive to the PEF treatment. The PEF inactivation value, at 23°C, for Scott A was approximately fivefold greater than that for OSY-8578. When the PEF inactivation values of the most sensitive strains were compared, no statistical difference (P > 0.05) was observed among Scott A, OSY-8517, OSY-8579, and OSY-8580; Scott A, however, is the best-known strain among this group. Scott A and OSY-8578, therefore, were selected for subsequent experiments to represent PEF-sensitive and PEF-resistant strains.
FIG. 1.
Decrease in log10 CFU per milliliter (PEF inactivation) of L. monocytogenes strains caused by PEF treatment at 25 kV/cm when cells were suspended in 0.1% NaCl. Cell suspensions were adjusted to ∼108 CFU/ml before treatment. (A) Pretreatment temperature, 23°C; and treatment time, 144 μs; (B) pretreatment temperature, 37°C; and treatment time, 72 μs. Different letter designations indicate strains with statistically different (P < 0.05) PEF inactivation levels, based on one-way ANOVA and Tukey mean comparison, within each treatment temperature data set.
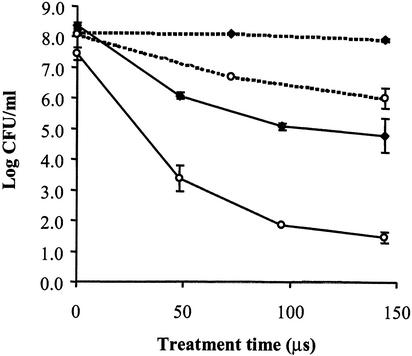
Cell suspensions of L. monocytogenes Scott A and OSY-8578, in 0.1% NaCl (pH 7.0) or 50% acid whey (pH 4.2), were processed with PEF at 25 kV/cm for 48 to 144 μs (pretreatment temperature 23°C), and survivor plots were constructed (Fig. 2). Temperature of the sample at the outlet of the PEF unit did not exceed 40°C. After PEF treatment for 144 μs, populations of the pathogen in 0.1% NaCl decreased by 2.1 log10 CFU/ml for Scott A and only 0.2 log10 CFU/ml for OSY-8578. Similar PEF treatments in 50% acid whey inactivated 6.0 log10 CFU of Scott A/ml and 3.6 log10 CFU of OSY-8578/ml. These data confirm the results of the previous experiment that sensitivity of L. monocytogenes to PEF is greater for Scott A than for OSY-8578. In addition, both strains were more sensitive to PEF when treated in 50% acid whey than when treated in 0.1% NaCl.
FIG. 2.
Survivor plot of L. monocytogenes Scott A and OSY-8578, treated with PEF at 25 kV/cm and 23°C, in 0.1% NaCl or 50% acid whey. Symbols: ○, Scott A; ♦, OSY-8578; dashed lines, 0.1% NaCl; and continuous line, 50% acid whey.
Stage of growth and sensitivity to PEF.
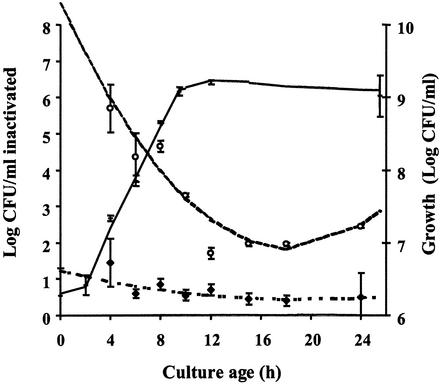
Strains Scott A and OSY-8578 showed similar growth patterns (Fig. 3). The lag phase lasted for 2 h, and the stationary phase started after 12 h of incubation. Samples from all stages of growth were PEF treated at 25 kV/cm for 144 μs with the initial temperature set at 15°C. Initial count and PEF treatment parameters were selected so that postprocessing viable counts were detectable by direct plating. Statistical analysis of the PEF inactivation data indicated heterogeneity of variances, and three groups of similar variances (0.875, 0.04, and 0.005) were identified. Larger variances, in general, were observed at the exponential phase of growth. Averages of PEF inactivation were analyzed by using SAS Proc Mixed to take into account the variance inequality. At each sampling time, Scott A was significantly (P < 0.01) more sensitive than was OSY-8578 to PEF treatment. Differences in PEF sensitivity between the two strains, however, were smaller at the stationary than at the exponential phase (Fig. 3). Scott A was markedly more sensitive during the exponential phase of growth than were cells of the same strain at the stationary phase (P < 0.01). The sensitivity to PEF, during growth of Scott A, reached its minimal value at the early stationary phase (12 to 18 h) and then increased slightly but significantly (P < 0.01) when the incubation period lengthened. Of interest, L. monocytogenes OSY-8578 remained highly resistant to PEF treatment and no statistically significant difference in PEF inactivation (P > 0.05) was found as the culture age varied from 4 to 24 h.
FIG. 3.
Growth (in log10 CFU/milliliter) and PEF inactivation (decrease in log10 CFU/milliliter) of L. monocytogenes Scott A and OSY-8578 after different incubation periods. Treatment with PEF was done with 0.1% NaCl suspension medium (15°C) with an electric field of 25 kV/cm and a total treatment time of 144 μs. For PEF inactivation data, symbols designate data points and lines represent these data after fitting with the second-order polynomial model. Symbols: +, total count during growth (data points for Scott A and OSY-8578 overlapped; therefore, both strains were represented by the same symbol); ○, PEF inactivation of Scott A; and ♦, PEF inactivation of OSY-8578.
The relation between PEF inactivation and time of incubation of Scott A and OSY-8578 was fitted by using a quadratic nonorthogonal regression model (as described earlier), and the estimated parameters for these strains were as follows:
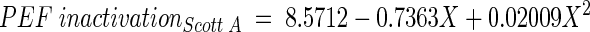 |
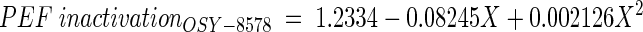 |
where X is culture age.
Derivation of these models indicated that PEF inactivation, particularly for Scott A, had the smallest value at the early stationary phase (∼18 h). At this age, PEF inactivation value was approximately fourfold larger for Scott A than for OSY-8578. Estimated model parameters for OSY-8578 were not significantly different from zero (P > 0.05), which confirmed that the relationship between age and PEF inactivation for this resistant strain could have been equally represented by a zero-order relationship.
Strain identification by genotyping.
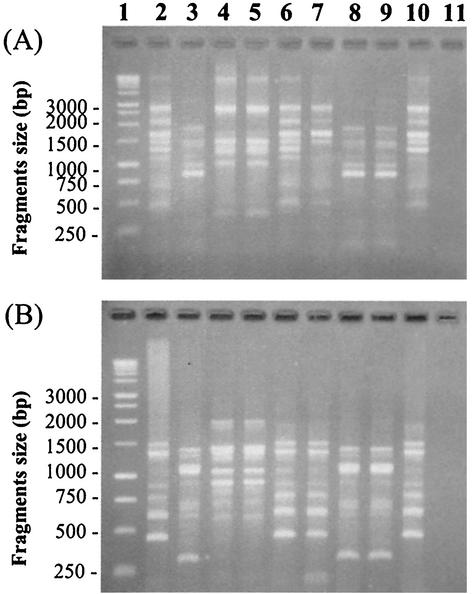
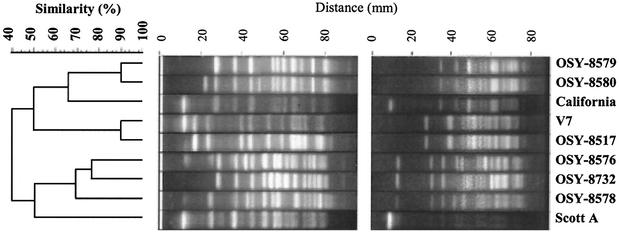
L. monocytogenes strains were first compared according to their AP-PCR banding patterns (Fig. 4). Strains were clustered into three groups of similar banding patterns: (i) Scott A, OSY-8576, OSY-8578, and OSY-8732; (ii) California, OSY-8579, and OSY-8580; and (iii) V7 and OSY-8517. Strain OSY-8578 had a missing band (∼1,300 bp) compared to the other strains from its group when DNA was amplified with the primer PJ108 (Fig. 4A). Other strains within each group were not clearly differentiated with this AP-PCR technique (Fig. 4). Repeats of this experiment with separate DNA extracts from the same strains showed that the banding patterns were reproducible (data not shown). A similar grouping was found when two additional primers (5′-TTATGTAAAACGACGGCCAGT-3′ and 5′-GGGCGTTGTCGGTGTTCATG-3′) that were proposed for Listeria typing by MacGowan et al. (29) were tested (data not shown).
FIG. 4.
Banding profiles of L. monocytogenes strains when genomic DNA was amplified by AP-PCR, with the primers PJ108 (A) and PJ118 (B). The PCR products were separated on agarose gel by electrophoresis. Tracks and the corresponding isolates are as follows: 1, 1-kb DNA ladder; 2, Scott A; 3, California; 4, V7; 5, OSY-8517; 6, OSY-8576; 7, OSY-8578; 8, OSY-8579; 9, OSY-8580; 10, OSY-8732; and 11, negative control.
The PFGE method was more discriminatory than AP-PCR, as all strains were differentiated (Fig. 5). Hierarchical cluster analysis identified three groups of closely related banding patterns that corresponded to the groups determined with AP-PCR analysis. No clear association was found between this grouping and strain sensitivity to PEF. For instance, strains V7 and OSY-8517 had similar banding patterns by AP-PCR (Fig. 4) and were the most closely related by PFGE (Fig. 5), with 90% similarity in banding patterns. These strains, however, differed significantly (P < 0.05) in sensitivity to PEF treatment (Fig. 1).
FIG. 5.
Dendrogram demonstrating the genetic relationship of the nine L. monocytogenes strains, based on DNA macrorestriction with ApaI (A) and SmaI (B).
DISCUSSION
Screening for a PEF-resistant strain.
Our objective was to select and identify a target L. monocytogenes strain that is most resistant to PEF for use in laboratory-scale evaluation or optimization of PEF processes. Considerable variations in PEF sensitivity were found at the subspecies level. Strain OSY-8578 was a suitable target Listeria strain for its high resistance to PEF at different phases of growth and under different pretreatment temperatures and medium compositions. Strain V7 also expressed PEF resistance and was noted in previous studies for its resistance to heat, which was slightly higher than those of Scott A and California (13). Identification of processing conditions sufficient to eradicate PEF-resistant strains was not the purpose of this study; instead, processing conditions that allow comparison of strains were chosen. High treatment intensity and combination with mild heating, as proposed by Iu et al. (22), may eliminate PEF-resistant strains, such as OSY-8578, from food samples.
Although PEF treatments are occasionally described as “nonthermal,” some heat is generated when food is processed by using this technology. Heat generation depends on equipment design and processing parameters such as total treatment time. In the screening experiments of this study, treatment times were varied when different initial temperatures were investigated. Thermally induced lethality of Listeria spp. was reportedly negligible in PEF treatments with an outlet temperature below 61 to 65°C (1, 11). Experimental conditions of this study were designed so that the outlet temperature was less than 56°C, regardless of samples' initial temperatures. Samples also were rapidly cooled on ice after the PEF treatments. Thermal contribution to lethality, if any, was therefore minimal. Population of L. monocytogenes V7 treated at 25 kV/cm for 144 μs, for example, decreased by 1.5 log10 CFU/ml, and the temperature increased from 23°C, initially, to 52 to 56°C during the PEF treatment (Fig. 1A). To achieve a similar reduction in the population of this strain by heating at 55°C, ∼6.7 min of thermal treatment would be needed (13).
Strain differences were more pronounced in the present PEF study than these reported earlier with heat (13); this is likely due to the more localized action of PEF, compared to that of heat, in bacterial cells (17, 24, 40). Rupture of the cell membrane and loss of cell membrane functionality are the primary causes of inactivation of Listeria with PEF (8, 24, 25, 40). The structure and composition of the cell membrane were likely involved in the high PEF resistance of the target strain, OSY-8578. Cell growth requires membrane fluidity, which is facilitated by the presence of fatty acids in a liquid-crystalline state (37). Excessive membrane fluidity may favor pore formation and hence PEF sensitivity (24).
Stage of growth.
The most PEF-resistant L. monocytogenes strain (OSY-8578) and one of the most sensitive strains (Scott A) were the subject of a detailed investigation. Sensitivity of L. monocytogenes Scott A to PEF was substantially greater at the stationary than at the exponential phase of growth (Fig. 3). These data are consistent with previous findings when L. monocytogenes was treated with different preservation factors (23, 27, 31) and when E. coli was treated with PEF (33). Variation in PEF inactivation among stationary-phase samples (12 to 24 h of incubation) was not appreciable, compared to the variability among samples from the exponential phase (Fig. 3). Rapid growth may have contributed to the larger variance among samples taken during the exponential phase, as dividing cells are likelier to be susceptible to membrane damage than are cells not undergoing division.
The increased PEF resistance of L. monocytogenes Scott A at the late stages of growth (Fig. 3) may have resulted from a shift in gene expression during cell transition from the exponential to the stationary phase. The physiology of transition to stationary phase bacteria has been thoroughly studied in Bacillus subtilis (6, 7). The alternative transcription factor, σB, is activated during the stationary phase and induces resistance of the culture to adverse conditions at this stage (5, 7). A similar transcription factor has been identified in L. monocytogenes and was found to contribute to acid resistance (39). It would, therefore, be of interest to investigate the contribution of σB induction to the resistance of L. monocytogenes to PEF. The high and constant PEF resistance of OSY-8578 during both exponential and stationary phases was unexpected. These findings suggest that an unidentified PEF resistance factor(s) is constitutively produced by the resistant strain. The higher resistance of OSY-8578 at all stages of growth emphasizes its suitability as a target strain for PEF process optimization. Such exceptional resistance to preservation factors may also explain the persistence of specific Listeria strains in the food-processing environment, despite efforts of food processors to eliminate the pathogen (10, 15).
Strain identification by genotyping.
Although both PFGE and AP-PCR characterized the target strain, OSY-8578, only PFGE was sufficiently discriminatory to produce specific banding patterns for all strains. This advantage of PFGE was also reported in earlier studies (e.g., references 20 and 32). The PFGE technique, however, is costly and technically challenging. Bender et al. (4) used PFGE to identify a genotype unique to multidrug-resistant isolates from Salmonella enterica serotype Typhimurium. In the present study, no obvious correlation was found between genotype groups and the sensitivity of L. monocytogenes strains to PEF; however, tracking the most PEF-resistant L. monocytogenes strain in the environment and processing facilities is possible through genotyping.
In conclusion, we demonstrated a procedure for identifying and characterizing a target L. monocytogenes strain for optimizing the efficacy of PEF processes. L. monocytogenes OSY-8578 was selected, among nine strains, for its superior resistance to the PEF treatments, regardless of the medium composition, pretreatment temperature, and stage of growth. The PFGE analysis clearly differentiated this target strain from the other strains, but the clustering pattern of this genotyping technique, as done in this study, could not be correlated with to the resistance to PEF processing.
Acknowledgments
This research was supported by the Ohio Agricultural Research and Development Center (OARDC), the Center for Advanced Food Processing and Packaging Studies (CAPPS), and the U.S. Army Natick Soldier Center, Natick, Mass.
We thank Normand St-Pierre, Charles Pretzmann, and Polly Courtney for their valuable technical assistance in conducting these experiments and analyzing the data. We also express our gratitude to Jenny Diehl and Steve Boomer, who provided the acid whey.
REFERENCES
- 1.Aronsson, K., M. Lindgren, B. R. Johansson, and U. Ronner. 2001. Inactivation of microorganisms using pulsed electric fields: the influence of process parameters on Escherichia coli, Listeria innocua, Leuconostoc mesenteroides and Saccharomyces cerevisiae. Innov. Food Sci. Emerg. Technol. 2:41-54. [Google Scholar]
- 2.Baloga, A. O., and S. K. Harlander. 1991. Comparison of methods for discrimination between strains of Listeria monocytogenes from epidemiological surveys. Appl. Environ. Microbiol. 57:2324-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bayles, D. O., M. H. Tunick, T. A. Foglia, and A. J. Miller. 2000. Cold shock and its effect on ribosomes and thermal tolerance in Listeria monocytogenes. Appl. Environ. Microbiol. 66:4351-4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bender, J. B., C. W. Hedberg, D. J. Boxrud, J. M. Besser, J. H. Wicklund, K. E. Smith, and M. T. Osterholm. 2001. Use of molecular subtyping in surveillance for Salmonella enterica serotype Typhimurium. N. Engl. J. Med. 344:189-195. [DOI] [PubMed] [Google Scholar]
- 5.Boylan, S. A., A. R. Redfield, M. S. Brody, and C. W. Price. 1993. Stress-induced activation of the σB transcription factor of Bacillus subtilis. J. Bacteriol. 175:7931-7937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boylan, S. A., A. R. Redfield, and C. W. Price. 1993. Transcription factor σB of Bacillus subtilis controls a large stationary-phase regulon. J. Bacteriol. 175:3957-3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boylan, S. A., A. Rutherford, S. M. Thomas, and C. W. Price. 1992. Activation of Bacillus subtilis transcription factor σB by a regulatory pathway responsive to stationary-phase signals. J. Bacteriol. 174:3695-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calderon-Miranda, M. L., G. V. Barbosa-Canovas, and B. G. Swanson. 1999. Transmission electron microscopy of Listeria innocua treated by pulsed electric fields and nisin in skimmed milk. Int. J. Food Microbiol. 51:31-38. [DOI] [PubMed] [Google Scholar]
- 9.Chasseignaux, E., M. T. Toquin, C. Ragimbeau, G. Salvat, P. Colin, and G. Ermel. 2001. Molecular epidemiology of Listeria monocytogenes isolates collected from the environment, raw meat and raw products in two poultry- and pork-processing plants. J. Appl. Microbiol. 91:888-899. [DOI] [PubMed] [Google Scholar]
- 10.Destro, M. T., M. F. F. Leitao, and J. M. Farber. 1996. Use of molecular typing methods to trace the dissemination of Listeria monocytogenes in a shrimp processing plant. Appl. Environ. Microbiol. 62:705-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunn, J. 2001. Pulsed electric field processing: an overview, p. 1-30. In G. V. Barbosa-Canovas and Q. H. Zhang (ed.), Pulsed electric fields in food processing: fundamental aspects and applications. Technomic Publishing Co., Lancaster, Pa.
- 12.Dutreux, N., S. Notermans, T. Wijtzes, M. M. Gongora-Nieto, G. V. Barbosa-Canovas, and B. G. Swanson. 2000. Pulsed electric fields inactivation of attached and free-living Escherichia coli and Listeria innocua under several conditions. Int. J. Food Microbiol. 54:91-98. [DOI] [PubMed] [Google Scholar]
- 13.El-Shenawy, M. A., A. E. Yousef, and E. H. Marth. 1989. Thermal inactivation and injury of Listeria monocytogenes in reconstituted nonfat dry milk. Milchwissenschaft 44:741-745. [Google Scholar]
- 14.Fenlon, D. R. 1999. Listeria monocytogenes in the natural environment, p. 21-38. In E. T. Ryser and E. H. Marth (ed.), Listeria, listeriosis and food safety. Marcel Dekker, Inc., New York, N.Y.
- 15.Giovannacci, I., C. Ragimbeau, S. Queguiner, G. Salvat, J. L. Vendeuvre, V. Carlier, and G. Ermel. 1999. Listeria monocytogenes in pork slaughtering and cutting plants: use of RAPD, PFGE and PCR-REA for tracing and molecular epidemiology. Int. J. Food Microbiol. 53:127-140. [DOI] [PubMed] [Google Scholar]
- 16.Giraffa, G., L. Rossetti, and E. Neviani. 2000. An evaluation of chelex-based DNA purification protocols for the typing of lactic acid bacteria. J. Microbiol. Methods 42:175-184. [DOI] [PubMed] [Google Scholar]
- 17.Gould, G. W. 1989. Heat-induced injury and inactivation, p. 11-42. In G. W. Gould (ed.), Mechanisms of action of food preservation procedures. Elsevier, London, United Kingdom.
- 18.Grahl, T., and H. Maerkl. 1996. Killing of microorganisms by pulsed electric fields. Appl. Microbiol. Biotechnol. 45:148-157. [DOI] [PubMed] [Google Scholar]
- 19.Graves, L. M. 1994. Comparison of ribotyping and multilocus enzyme electrophoresis for subtyping of Listeria monocytogenes isolates. J. Clin. Microbiol. 32:2936-2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graves, L. M., and B. Swaminathan. 2001. PulseNet standardized protocol for subtyping Listeria monocytogenes by macrorestriction and pulsed-field gel electrophoresis. Int. J. Food Microbiol. 65:55-62. [DOI] [PubMed] [Google Scholar]
- 21.Hulsheger, H., J. Potel, and E. G. Niemann. 1983. Electric field effects on bacteria and yeast cells. Radiat. Environ. Biophys. 22:149-162. [DOI] [PubMed] [Google Scholar]
- 22.Iu, J., G. S. Mittal, and M. W. Griffiths. 2001. Reduction in levels of Escherichia coli O157:H7 in apple cider by pulsed electric fields. J. Food Prot. 64:964-969. [DOI] [PubMed] [Google Scholar]
- 23.Jydegaard, A. M., A. Gravesen, and S. Knochel. 2000. Growth condition-related response of Listeria monocytogenes 412 to bacteriocin inactivation. Lett. Appl. Microbiol. 31:68-72. [DOI] [PubMed] [Google Scholar]
- 24.Kekez, M. M., P. Savic, and B. F. Johnson. 1996. Contribution to the biophysics of the lethal effects of electric field on microorganisms. Biochim. Biophys. Acta 1278:79-88. [DOI] [PubMed] [Google Scholar]
- 25.Lado, B. H., and A. E. Yousef. 2001. Alternative food preservation technologies: efficacy and mechanisms. Microbes Infect. 4:433-440. [DOI] [PubMed] [Google Scholar]
- 26.Littell, C. R., G. A. Milliken, W. W. Stroup, and R. D. Wolfinger. 1996. SAS system for mixed models. The SAS Institute, Cary, N.C.
- 27.Lou, Y., and A. E. Yousef. 1997. Adaptation to sublethal environmental stresses protects Listeria monocytogenes against lethal preservation factors. Appl. Environ. Microbiol. 63:1252-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Louie, M., P. Jayaratne, I. Luchsinger, J. Devenish, J. Yao, W. Schlech, and A. Simor. 1996. Comparison of ribotyping, arbitrarily primed PCR, and pulsed-field gel electrophoresis for molecular typing of Listeria monocytogenes. J. Clin. Microbiol. 34:15-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacGowan, A. P., K. O'Donaghue, S. Nicholls, J. McLauchlin, P. M. Bennett, and D. S. Reeves. 1993. Typing of Listeria spp. by random amplified polymorphic DNA (RAPD) analysis. J. Med. Microbiol. 38:322-327. [DOI] [PubMed] [Google Scholar]
- 30.Mazurier, S. I., and K. Wernars. 1992. Typing of Listeria strains by random amplification of polymorphic DNA. Res. Microbiol. 143:499-505. [DOI] [PubMed] [Google Scholar]
- 31.McClements, J. M., M. F. Patterson, and M. Linton. 2001. The effect of growth stage and growth temperature on high hydrostatic pressure inactivation of some psychrotrophic bacteria in milk. J. Food Prot. 64:514-522. [DOI] [PubMed] [Google Scholar]
- 32.O'Donoghue, K., K. Bowker, J. McLauchlin, D. S. Reeves, P. M. Bennett, and A. P. MacGowan. 1995. Typing of Listeria monocytogenes by random amplified polymorphic DNA (RAPD) analysis. Int. J. Food Microbiol. 27:245-252. [DOI] [PubMed] [Google Scholar]
- 33.Pothakamury, U. R., H. Vega, Q. H. Zhang, G. V. Barbosa-Canovas, and B. G. Swanson. 1996. Effect of growth stage and processing temperature on the inactivation of Escherichia coli by pulsed electric fields. J. Food Prot. 59:1167-1171. [DOI] [PubMed] [Google Scholar]
- 34.Reina, L. D., Z. T. Jin, Q. H. Zhang, and A. E. Yousef. 1998. Inactivation of Listeria monocytogenes in milk by pulsed electric field. J. Food Prot. 61:1203-1206. [DOI] [PubMed] [Google Scholar]
- 35.Slutsker, L., and A. Schuchat. 1999. Listeriosis in humans, p. 75-96. In E. T. Ryser and E. H. Marth (ed.), Listeria, listeriosis and food safety. Marcel Dekker, Inc., New York, N.Y.
- 36.Snedecor, G. W., and W. G. Cochran. 1980. Statistical methods, 7th ed. Iowa State University Press, Ames.
- 37.Suutari, M., and S. Laakso. 1994. Microbial fatty acids and thermal adaptation. Crit. Rev. Microbiol. 20:285-328. [DOI] [PubMed] [Google Scholar]
- 38.Vogel, B. F., H. H. Huss, B. Ojeniyi, P. Ahrens, and L. Gram. 2001. Elucidation of Listeria monocytogenes contamination routes in cold-smoked salmon processing plants detected by DNA-based typing methods. Appl. Environ. Microbiol. 67:2586-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wiedmann, M., T. J. Arvik, R. J. Hurley, and K. J. Boor. 1998. General stress transcription factor σB and its role in acid tolerance and virulence of Listeria monocytogenes. J. Bacteriol. 180:3650-3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wouters, P. C., A. P. Bos, and J. Ueckert. 2001. Membrane permeabilization in relation to inactivation kinetics of Lactobacillus species due to pulsed electric fields. Appl. Environ. Microbiol. 67:3092-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yin, Y., Q. H. Zhang, and S. K. Sastry. November1997. High voltage pulsed electric field chambers for the preservation of liquid food products. U.S. patent 5,690,978.