Abstract
Expression of surface-associated and secreted protein MopE of the methanotrophic bacterium Methylococcus capsulatus (Bath) in response to the concentration of copper ions in the growth medium was investigated. The level of protein associated with the cells and secreted to the medium changed when the copper concentration in the medium varied and was highest in cells exposed to copper stress.
Copper ions are known to have an important role in the biology of the methanotrophic bacterium Methylococcus capsulatus (Bath). They regulate the switch between soluble methane monooxygenase (sMMO) and particulate methane monooxygenase (pMMO) (12, 13, 16). Copper ions are also required for the catalytic activity of pMMO (15, 19) and are essential for production of intracytoplasmic membranes (14). pMMO is associated with the intracellular membranes (2) and is produced when cells are grown at high copper-to-biomass ratios. At low copper-to-biomass ratios, cytoplasmic sMMO is the dominant MMO enzyme in methanotrophs that possess both forms of MMO (11).
The concentration of copper in biological habitats is often low (10), and accumulation of copper in M. capsulatus most likely requires active transport. Bacteria have evolved various mechanisms for scavenging essential metals (5, 8, 9, 17), but the exact mechanism of copper transport into bacterial cells is largely unknown. M. capsulatus produces small copper-binding peptides that are secreted into the spent medium. These peptides may be actively imported into the cells and bind to the catalytic site of pMMO (4, 19). The culture supernatant of M. capsulatus also contains high concentrations of the MopE protein. The secreted protein represents an N-terminally truncated form of a larger precursor protein that is associated with the surface of the cell (6, 7). Here we shall refer to the larger cell surface-associated protein as MopEC and to the secreted part of the protein as MopE*. MopE* displays sequence similarity to a copper-repressible protein of the methanotroph Methylomicrobium album (1), indicating that the protein may be involved in copper acquisition. In this study, we have investigated whether expression and secretion of MopE in M. capsulatus (Bath) are regulated by changes in the copper-to-biomass ratio in cultures.
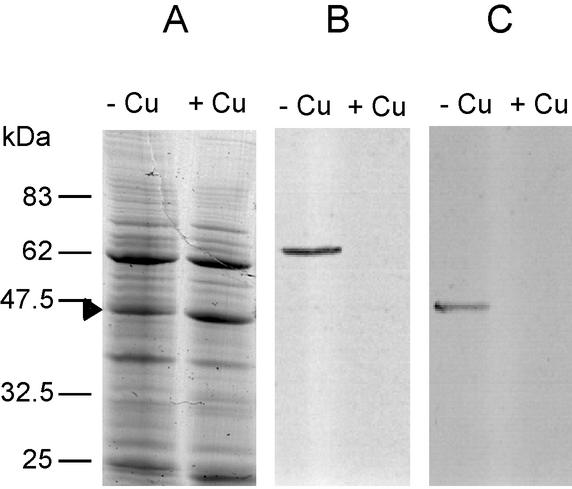
Initially, M. capsulatus (Bath) strain NCIMB 11132 was grown in batch culture with shaking (200 rpm) at 45°C with a headspace of methane, CO2, and air (40:2:48) in NMS medium as described by Whittenbury et al. (18). High-copper medium contained 1 mg of CuSO4 · 5H2O per liter, while no copper was added to low-copper media. Cells were harvested at an optical density at 540 nm (OD540) of 0.3 to 0.5 by centrifugation at 20,000 × g for 30 min at 4°C. The proteins in the resulting supernatant were precipitated in 80% (vol/vol) acetone and then analyzed, together with the whole-cell pellet, by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and protein immunoblotting (Fig. 1). In SDS-polyacrylamide gels, MopEC and MopE* migrate at approximately 66 and 46 kDa, respectively (7). Immunoreactive proteins corresponding to MopEC and MopE* were detected in the whole-cell lysate and also in spent medium isolated from low-copper-grown cells (Fig. 1B and C). The proteins were, however, not detected in cell lysate and supernatant isolated from high-copper-grown cells, indicating that MopE production and/or stability was affected by the higher copper concentration. Analysis of low-copper-grown cells on Coomassie-stained SDS-polyacrylamide gels did not reveal polypeptides corresponding to sMMO (Fig. 1A). Thus, although the PmoB polypeptide of pMMO was less abundant in low-copper-grown cells than in high-copper-grown cells, pMMO was apparently also the predominant MMO in low-copper-grown cells. sMMO activity was also not detected by the naphthalene assay (3), and thus, the copper-to-biomass ratio in the medium was apparently too high for induction of the sMMO genes under these growth conditions. Although no copper ions were added to this low-copper growth medium, there were probably sufficient trace amounts of copper ions to repress the expression of sMMO. This is a common observation in the field of methanotrophy.
FIG. 1.
Detection of MopE in M. capsulatus whole-cell lysates (A and B) and culture supernatants (C) isolated from cells grown in low (−Cu)- and high (+Cu)-copper media. The OD540 at harvest was 0.5 for both cultures. Proteins were resolved by SDS-PAGE and stained with Coomassie brilliant blue (A) or transferred to a nitrocellulose membrane for analysis with anti-MopE serum (B and C). The blots were developed as described previously (6). Each lane in panels A and B contains the same amount (approximately 7 μg) of protein. In panel C, an amount corresponding to 0.5 ml of culture supernatant was loaded onto the gel. Positions of molecular mass markers are shown to the left. The arrowhead indicates the position of the PmoB subunit of pMMO (19).
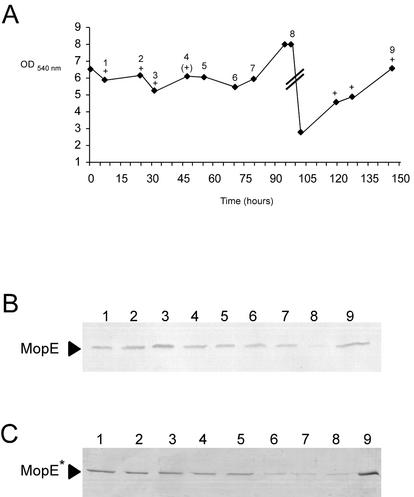
In order to examine MopE production as a response to variation in copper ion concentrations in the growth medium in more detail, M. capsulatus was grown at higher cell densities in a chemostat with various amounts of added copper. The cells were grown in NMS medium under an atmosphere of air-methane (5:1) in a 2-liter fermentor. The pH was continuously adjusted to 6.7 by automatic addition of 0.25 M HCl with constant agitation at 550 rpm. The dilution rate of the fermentor was 0.05 h −1, and oxygen was the limiting nutrient. During growth, cells and spent medium were collected at different time points (Fig. 2A) and analyzed by SDS-PAGE and protein immunoblotting (Fig. 2B and C and 3). All of the samples collected were screened for sMMO activity with the naphthalene assay (3).
FIG. 2.
(A) Growth of M. capsulatus in continuous culture (t = 0 to 79 h) and batch culture (t = 79 to 96 and 96 to 145 h) in medium supplemented with various concentrations of copper. Sample points for collection of cells for analyses by SDS-PAGE and protein immunoblotting are numbered 1 to 9. The symbols + and (+) indicate samples in which sMMO activity was detectable or barely detectable by the naphthalene test of Brusseau et al. (3). (B) Protein immunoblot of an SDS-polyacrylamide gel loaded with protein samples of cells collected at different time points (see panel A) during growth of M. capsulatus with NMS medium containing various concentrations of copper. The blot was treated with anti-MopE serum as described previously (6). (C) Proteins in the spent medium of cells collected at different time points (see panel A) during growth of M. capsulatus with NMS medium containing various concentrations of copper, resolved by SDS-PAGE, and stained with Coomassie brilliant blue. The supernatant proteins were concentrated by precipitation in 80% (vol/vol) acetone before being subjected to electrophoresis. The amounts of supernatant proteins loaded on the gel were adjusted to compensate for the different cell densities at the different stages of growth. The volumes were adjusted so that the ratio of spent medium (milliliters) to the OD540 of the culture was the same in each well.
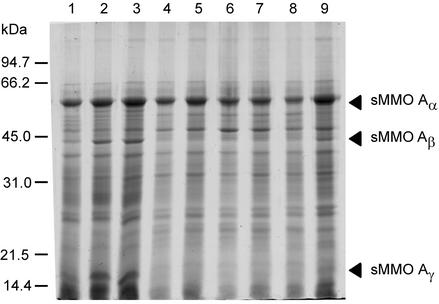
FIG. 3.
SDS-PAGE analysis of cells collected at different time points (see Fig. 2A) during growth of M. capsulatus with NMS medium containing various concentrations of copper. The positions of the three components (α, β, and γ) of the hydroxylase component of sMMO (11) are indicated by arrowheads to the right. The sMMO hydroxylase α subunit migrates nearly at the same position as the large subunit of methanol dehydrogenase, and thus, the increase in this component is seen as a broadening of the methanol dehydrogenase band. Each lane contains the same amount (approximately 7 μg) of protein. Positions of molecular mass markers are shown to the left. An identical gel was used in the immunoblotting experiments described in Fig. 2B.
Initially, the culture was supplied with NMS medium containing 0.25 mg of CuSO4 · 5H2O per liter. After 4 days, the batch culture reached an OD540 of 6.5 (t = 0; Fig. 2A). At this stage, MopEC was barely detectable on protein immunoblots (not shown) and no sMMO activity was detected (Fig. 2A). The batch culture was then switched to a continuous culture fed with NMS medium containing no added copper. Samples were then collected after 7 and 24 h of growth. MopEC and MopE* were detected after 7 h (t = 7) (Fig. 2B and C, lanes 1). sMMO activity was also detected at t = 7 h (Fig. 2A), indicating that the medium had been depleted of copper.
After 24 h of growth on low-copper medium (t = 24), the medium supply was switched to NMS medium containing 1 mg of CuSO4 · 5H2O per liter. Samples were collected after 31, 47, 55, and 79 h. At t = 31 h, the cells still produced high levels of both forms of MopE (MopEC and MopE*; Fig. 2) and sMMO (Fig. 3, lane 3), but over the next 48 h (t = 31 to 79 h), the amount of MopEC, and particularly that of MopE*, decreased significantly (Fig. 2B and C, lanes 3 to 7). During the same time interval, the cells switched from sMMO expression to pMMO expression, demonstrating that the copper-to-biomass ratio in the fermentor had increased (Fig. 2A and 3, lanes 3 to 7).
At t = 79 h, after approximately three volume changes, the growth medium supply was switched off, thus transforming the continuous culture to a batch culture in NMS medium containing 1 mg of CuSO4 · 5H2O (high-copper medium) per liter. When the OD540 reached 8.0 (t = 97 h), still only small amounts of MopEC and MopE* could be detected (Fig. 2B and C, lanes 8). The culture was then subjected to rapid copper depletion by removal of half of the volume of the fermentor and its replacement with low-copper NMS medium (containing no added copper). After a further 48 h (t = 145 h), the concentrations of MopEC and MopE* in the cells and medium supernatant, respectively, had increased dramatically (Fig. 2B and C, lanes 9). This was particularly notable for MopE*.
Taken together, the results of the growth experiments suggest that MopE synthesis and stability are regulated either at the level of transcription or translation by the availability of copper. This raises the possibility that MopE is involved in copper transport into the cell and/or in the physiological transition the cells make when they shift from copper sufficiency to copper limitation. The protein was detected at the cell surface and in the spent medium of copper-stressed cells expressing either pMMO or sMMO but seemed to be more abundant in the spent medium of sMMO-expressing cells. This is in contrast to the extracellular copper-binding peptides, which seem to be associated with expression of pMMO and are less abundant in the spent medium of sMMO-expressing cells (4), and it is tempting to speculate that M. capsulatus may have evolved multiple copper acquisition systems that operate at different copper concentrations. However, except for the CorA protein (1), MopE does not show sequence similarity to other proteins in the databases and the primary amino acid sequence of MopE does not appear to contain conserved copper-binding motifs (7). Thus, the ability of MopE to bind copper or copper-containing compounds remains to be investigated further by detailed biochemical studies.
Acknowledgments
This work was supported by the Norwegian Research Council (project 147714/432 to A.F.), GABI (SUP 140785/420), Norferm DA, and the Meltzer Foundation, University of Bergen. We thank BBSRC for a Ph.D. studentship for G.P.S., and we also thank the EC for funding through grant QLRT-1999-31528 to J.C.M.
REFERENCES
- 1.Berson, O., and M. E. Lidstrom. 1997. Cloning and characterization of corA, a gene encoding a copper-repressible polypeptide in the type I methanotroph, Methylomicrobium albus BG8. FEMS Microbiol. Lett. 148:169-174. [DOI] [PubMed]
- 2.Brantner, C. A., C. C. Remsen, H. A. Owen, L. A. Buchholz, and P. Collins. 2002. Intracellular localization of the particulate methane monooxygenase and methanol dehydrogenase in Methylomicrobium album BG8. Arch. Microbiol. 178:59-64. [DOI] [PubMed] [Google Scholar]
- 3.Brusseau, G. A., H. C. Tsien, R. S. Hanson, and L. P. Wackett. 1990. Optimization of trichloroethylene oxidation by methanotrophs and the use of a colorimetric assay to detect soluble methane monooxygenase activity. Biodegradation 1:19-29. [DOI] [PubMed] [Google Scholar]
- 4.DiSpirito, A. A., J. A. Zahn, D. W. Graham, H. J. Kim, C. K. Larive, T. S. Derrick, C. D. Cox, and A. Taylor. 1998. Copper-binding compounds from Methylosinus trichosporium OB3b. J. Bacteriol. 180:3606-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eitinger, T., and M. A. Mandrand-Berthelot. 2000. Nickel transport systems in microorganisms. Arch. Microbiol. 173:1-9. [DOI] [PubMed] [Google Scholar]
- 6.Fjellbirkeland, A., H. Kleivdal, C. Joergensen, H. Thestrup, and H. B. Jensen. 1997. Outer membrane proteins of Methylococcus capsulatus (Bath). Arch. Microbiol. 168:128-135. [DOI] [PubMed] [Google Scholar]
- 7.Fjellbirkeland, A., P. G. Kruger, V. Bemanian, B. T. Hogh, J. C. Murrell, and H. B. Jensen. 2001. The C-terminal part of the surface-associated protein MopE of the methanotroph Methylococcus capsulatus (Bath) is secreted into the growth medium. Arch. Microbiol. 176:197-203. [DOI] [PubMed] [Google Scholar]
- 8.Genco, C. A., and D. W. Dixon. 2001. Emerging strategies in microbial haem capture. Mol. Microbiol. 39:1-11. [DOI] [PubMed] [Google Scholar]
- 9.Hantke, K. 2001. Bacterial zinc transporters and regulators Biometals 14:239-249. [DOI] [PubMed] [Google Scholar]
- 10.Malmstrom, B. G., and J. Leckner. 1998. The chemical biology of copper. Curr. Opin. Chem. Biol. 2:286-292. [DOI] [PubMed] [Google Scholar]
- 11.Murrell, J. C., I. R. McDonald, and B. Gilbert. 2000. Regulation of expression of methane monooxygenases by copper ions. Trends Microbiol. 8:221-225. [DOI] [PubMed] [Google Scholar]
- 12.Nielsen, A. K., K. Gerdes, H. Degn, and J. C. Murrell. 1996. Regulation of bacterial methane oxidation: transcription of the soluble methane mono-oxygenase operon of Methylococcus capsulatus (Bath) is repressed by copper ions. Microbiology 142:1289-1296. [DOI] [PubMed] [Google Scholar]
- 13.Nielsen, A. K., K. Gerdes, and J. C. Murrell. 1997. Copper-dependent reciprocal transcriptional regulation of methane monooxygenase genes in Methylococcus capsulatus and Methylosinus trichosporium. Mol. Microbiol. 25:399-409. [DOI] [PubMed] [Google Scholar]
- 14.Prior, S., and H. Dalton. 1985. The effect of copper ions on the membrane content and methane monooxygenase activity in methanol-grown cells of Methylococcus capsulatus (Bath). J. Gen. Microbiol. 131:155-163. [Google Scholar]
- 15.Semrau, J. D., D. Zolandz, M. E. Lidstrom, and S. I. Chan. 1995. The role of copper in the pMMO of Methylococcus capsulatus bath: a structural vs. catalytic function. J. Inorg. Biochem. 58:235-244. [DOI] [PubMed] [Google Scholar]
- 16.Stanley, S. H., S. D. Prior, D. J. Leak, and H. Dalton. 1983. Copper stress underlies the fundamental change in intracellular location of methane mono-oxygenase in methane-oxidizing organisms: studies in both batch and continuous cultures. Biotechnol. Lett. 5:487-492. [Google Scholar]
- 17.Wandersman, C., and I. Stojiljkovic. 2000. Bacterial heme sources: the role of heme, hemoprotein receptors and hemophores. Curr. Opin. Microbiol. 3:215-220. [DOI] [PubMed] [Google Scholar]
- 18.Whittenbury, R., K. C. Phillips, and J. F. Wilkinson. 1970. Enrichment, isolation and some properties of methane-utilizing bacteria. J. Gen. Microbiol. 61:205-218. [DOI] [PubMed] [Google Scholar]
- 19.Zahn, J. A., and A. A. DiSpirito. 1996. Membrane-associated methane monooxygenase from Methylococcus capsulatus (Bath). J. Bacteriol. 178:1018-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]