Abstract
Three soilborne viruses transmitted by Polymyxa betae KESKIN in sugar beet have been described: Beet necrotic yellow vein virus (BNYVV), the agent of rhizomania, Beet soilborne virus (BSBV), and Beet virus Q (BVQ). A multiplex reverse transcription-PCR technique was developed to simultaneously detect BNYVV, BSBV, and BVQ, together with their vector, P. betae. The detection threshold of the test was up to 128 times greater than that of an enzyme-linked immunosorbent assay. Systematic association of BNYVV with one or two different pomoviruses was observed. BVQ was detected in samples from Belgium, Bulgaria, France, Germany, Hungary, Italy, Sweden, and The Netherlands but not in samples from Turkey.
Of the viral diseases of sugar beet, rhizomania is a major concern. Discovered 30 years ago in Italy (6), it now has a worldwide distribution (1, 37). The disease affects the sugar yield by diminishing the root weight and decreasing the sugar content of the sugar beet (1). The agent of rhizomania is the benyvirus Beet necrotic yellow vein virus (BNYVV) (32). It is transmitted to the plant by its vector, Polymyxa betae KESKIN. P. betae's resting spores, also called sporosori, can protect BNYVV in soil for more than 10 years, even without sugar beet being grown (1).
In the 1980s, another soilborne virus, Beet soilborne virus (BSBV), was found in sugar beet. First detected in the United Kingdom, BSBV was then reported in The Netherlands (14), Belgium, (38), the United States (J. E. Duffus, Abstr. 5th Int. Congr. Phytopathol., p. 452, 1988), Sweden, Germany (23), and France (24). Lesemann et al. (23) described two serotypes for BSBV, namely, the Ahlum and Wierthe serotypes. Recent data revealed that serotype Wierthe should be considered a distinct virus species named Beet virus Q (BVQ) (20). These two pomoviruses are also transmitted by P. betae (31). Although their contribution to rhizomania remains a matter of debate (12, 24, 31), it is not uncommon to find them associated with rhizomania-infested fields. Moreover, the occurrence of three different viruses, transmitted by a similar vector, within a single sugar beet raises questions regarding the epidemiology of rhizomania syndrome.
There is, therefore, a real need for a sensitive and specific technique for the detection of BNYVV, BSBV, BVQ, and their vector, P. betae. In recent years, reverse transcription (RT)-PCR, along with enzyme-linked immunosorbent assay (ELISA), has progressively become the prominent technique for the detection of BNYVV (15, 26). Multiplex RT (mRT)-PCR for the simultaneous detection of different viral targets has already been proposed for seed-borne legume viruses (2) and tree viruses (3, 34, 35). It has already been applied with success to the detection of poleroviruses infecting sugar beet (11) as well as soilborne viruses of potato (27) and cereals (10). This article describes an mRT-PCR for the simultaneous detection of BNYVV, BSBV, BVQ, and P. betae. Beet soilborne mosaic virus, another virus transmitted to sugar beet by P. betae, was not included in the analysis since this virus has been detected only in the United States to date (33).
Sampling, RNA extraction, primer design, and RT-PCR.
Sugar beets and soils were collected in different regions of Belgium, Bulgaria, Turkey, Hungary, The Netherlands, Italy, Germany, Sweden, and France based on rhizomania symptoms as described by Meunier et al. (25). Sugar beet seeds (Beta vulgaris cultivar cadyx) were planted in infected soil that was diluted 1/5 (vol/vol) with sterile quartz (diameter, 1 to 2 mm) and irrigated with Hoagland nutritive solution (36). After 6 weeks, 100 mg of rootlet tissue was used for total RNA or protein extraction. RNA extraction was performed with the RNeasy extraction kit (QIAGEN, Hilden, Germany) according to the kit protocol. In order to simplify the method and reduce the cost, direct use of ELISA extracts was checked as a template for mRT-PCR. One hundred milligrams of sugar beet rootlets was ground in 750 μl of ELISA extraction buffer (phosphate-buffered saline, 1% polyvinylpyrrolidone, 0.05% Tween, <1 g of NaN3/liter). The supernatant was used immediately for RT-PCR. A comparison of the extracts of different samples with RNeasy extracts showed that the direct use of ELISA extracts is feasible. Nevertheless, the storage of samples at −20°C, even for a short time, leads to nonreproducible results.
The primers used in RT-PCR and mRT-PCR were designed by using the PRIME program of the Genetics Computer Group (8). Primer-primer interactions were analyzed by using the GAP program (8), and specificity was checked with BLAST (29). Specifications for computer programs were default values. The primers selected were named according to the targeted organisms, followed by either “for” for forward primer or “rev” for reverse primer, and are listed as follows: BNYVV2(1)for, ACATTTCTATCCTCCTCCAC; BNYVV2(1)rev, ACCCCAACAAACTCTCTAAC; BSBV2for, CTTACGCTGTTCACTTTTATGCC; BSBV2rev, GTCCGCACTCTTTTCAACTGTTC; BVQ1(1)for, GCTGGAGTATATCACCGATGAC; BVQ1(1)rev, AAAATCTCGGATAGCATCCAAC; PB4for, CAAACGCCTGAAATCATCTAAC; and PB4rev GATGGCCCAATTCCTTACAC. The final concentration of each primer was 20 μM.
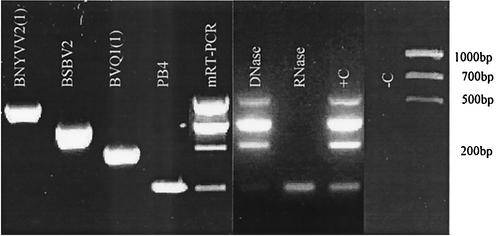
An RT reaction using Moloney murine leukemia virus (MMLV) reverse transcriptase (Promega, Madison, Wis.) and PCR using Taq polymerase (Promega) were performed as recommended by the manufacturer except as otherwise stated. Deoxynucleoside triphosphates (dNTPs) were used at final concentrations of 20 nmol/20 μl for the RT reaction and 15 nmol/50 μl for the PCR. Primers were used at final concentrations of 20 pmol/20 μl for the RT reaction and 20 pmol/50 μl for the PCR. The products were separated on 2.5% ethidium bromide agarose gels in 1× Tris-borate-EDTA buffer. The selected primer pairs gave the expected 545-bp (BNYVV), 399-bp (BSBV), 291-bp (BVQ), and 170-bp (P. betae) fragments for RT-PCR (Fig. 1). BNYVV primers gave the amplification profiles for strains of types A, B, and P (Table 1). No expected amplification bands were observed for the uninfected sugar beet, Chenopodium quinoa, or water controls.
FIG. 1.
Amplification of the different viruses and P. betae individually with the corresponding set of primers and combination of primers for mRT-PCR detection with or without DNase or RNase treatment. +C, positive control; −C, negative control.
TABLE 1.
Detection of BNYVV types A, B, and P and BSBV, BVQ, and P. betaea
| Soil originb | Presence of virus or vector determined byc:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| mRT-PCR
|
Single RT-PCR
|
|||||||
| BNYVV | BSBV | BVQ | P. betae | BNYVV | BSBV | BVQ | P. betae | |
| Unknown location (Bulgaria) | + | + | + | + | + | + | + | + |
| Hognoul (Belgium) | + | + | + | + | + | + | + | + |
| Jodoigne (Belgium) | + | + | + | + | + | + | + | + |
| Kabba (Hungary) | + | + | + | + | + | + | + | + |
| Magdeburg (Germany) | + | + | + | + | + | + | + | + |
| Mazy (Belgium) | + | + | + | + | + | + | + | + |
| Pecq (Belgium) | + | + | + | + | + | + | + | + |
| Pithiviers (France) | + | + | + | + | + | + | + | + |
| St. Germain (Belgium) | + | + | + | + | + | + | + | + |
| Steernkerke (Belgium) | + | + | + | + | + | + | + | + |
| Tompaladony (Hungary) | + | + | + | + | + | + | + | + |
| Bovesse (Belgium) | − | + | + | + | − | + | + | + |
| Gembloux (Belgium) | − | + | + | + | − | + | + | + |
| Kamperland (The Netherlands) | − | + | + | + | − | + | + | + |
| Oupeye (Belgium) | − | + | + | + | − | + | + | + |
| St. Jean de Losnes (France) | − | + | + | + | − | + | + | + |
| Bertinsart (France) | − | + | + | + | − | + | + | + |
| Gaytoy (Turkey) | + | + | − | + | + | + | − | + |
| Galkara (Turkey) | + | − | − | + | + | − | − | + |
| Gevreli (Turkey) | − | + | − | + | − | + | − | + |
| Kogaz (Turkey) | − | + | − | + | − | + | − | + |
| Lonzee 1 (Belgium) | − | + | − | + | − | + | − | + |
| Beclers (Belgium) | − | − | − | + | − | − | − | + |
| Lonzee 2 (Belgium) | − | − | − | + | − | − | − | + |
| Liguria (Italy) | − | − | − | − | − | − | − | − |
| Water | − | − | − | − | − | − | − | − |
| Chenopodium quinoa | − | − | − | − | − | − | − | − |
| Sugar beet (Beta vulgaris cv. cadyx) | − | − | − | − | − | − | − | − |
RNA was extracted from sugar beets 6 weeks after growth in soils from eight different countries (Bulgaria, Belgium, Turkey, Hungary, The Netherlands, Italy, Germany, and France). mRT-PCR and single RT-PCR with the corresponding primers were conducted on those samples.
Lonzee 1 and 2, two different samples from Lonzee, Belgium.
+, presence; −, absence.
mRT-PCR.
The four pairs of primers were combined in the mRT-PCR. For each RT reaction, 10 pmol of each of the four reverse primers was mixed with 1.2 μl of RNA and 7.3 μl of diethyl pyrocarbonate (DEPC)-treated water. The mixture was incubated at 65°C for 10 min and directly transferred into ice prior to the addition of 3.25 μl of DEPC-treated water, 2 μl of dNTPs (10 mM each), 4 μl of MMLV RT 5× buffer (Promega), and 0.25 μl of MMLV reverse transcriptase (200 U/μl) (Promega).
For the PCR, 18 pmol of each of the forward and reverse primers was added to 23.3 μl of DEPC-treated water, 10 μl of MgCl2 (25 mM) (Promega), 3.5 μl of Taq polymerase 10× buffer (Promega), 1.5 μl of dNTPs (10 mM each), 0.5 μl of Taq polymerase (5 U/μl) (Promega), and 4 μl of cDNA. Fresh dNTPs should be used for maximum efficiency (13). Amplification cycles were as follows: a first denaturation for 3 min at 94°C and then 35 cycles of denaturation for 30 s at 94°C, annealing for 30 s at 63°C, and elongation for 2 min at 72°C. A final elongation for 7 min at 72°C was added.
Interestingly, the detection of the P. betae repetitive EcoRI-like fragment (GenBank accession number X83745) directly in an RNA extract is feasible, thus rendering the detection of P. betae easier than that by the previously proposed PCR-based method (28) or the classical ELISA (7) and microscopical observation of sporosori. We suggest that this EcoRI-like fragment is amplified from mRNA and DNA, as observed from results of DNase or RNase treatments of RNA extract (Fig. 1). The different mRT-PCR products were extracted from the agarose gel and sequenced in both orientations. The fragments revealed between 99 and 100% similarity when the sequences obtained were compared with those present in GenBank.
mRT-PCR sensitivity.
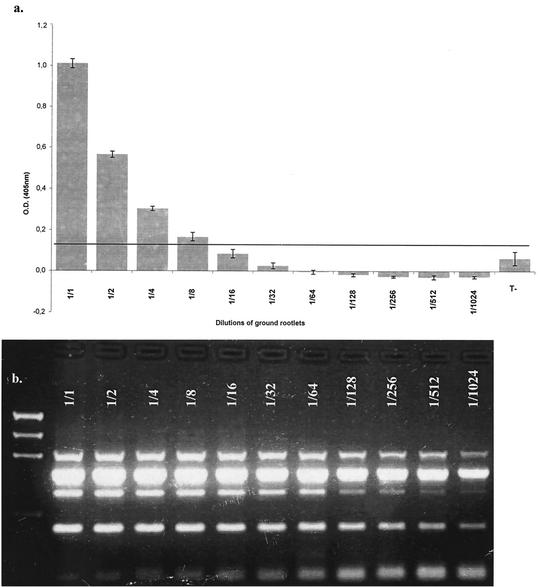
The sensitivity of mRT-PCR was compared to that of the double-antibody-sandwich ELISA (DAS-ELISA). A commercial kit (Sanofi Diagnostics Pasteur, Marnes-la-Coquette, France) was used for the detection of BNYVV by DAS-ELISA according to the manufacturer's protocol. Rootlets were ground in a mortar and resuspended in 900 μl of grinding buffer supplied with the kit. For the sensitivity test, the suspension was diluted to 1/2, 1/4, 1/8, 1/16, 1/32, 1/64, 1/128, 1/256, 1/512, and 1/1,024. The optical density at 405 nm (OD405) was measured with a Titertek MultiSkan MCC/340 spectrophotometer. OD405 values more than two times greater than that of healthy controls were considered positive. Figure 2 compares OD405 measurements for sample dilutions with the mRT-PCR results. mRT-PCR proved to be at least 128 times more sensitive than the DAS-ELISA.
FIG. 2.
(a) One hundred milligrams of BNYVV-infected rootlets was ground in ELISA grinding buffer (Sanofi Diagnostics Pasteur). The sample was diluted to 1/2, 1/4, 1/8, 1/16, 1/32, 1/64, 1/128, 1/256, 1/512, and 1/1,024 and tested according to the kit protocol. OD405 was plotted against the dilutions. The grey line represents two times the average OD of the negative control. (b) RNA from 100 mg of the same rootlets as described for panel a was extracted. This RNA was diluted as mentioned for panel a and tested with the mRT-PCR technique. mRT-PCR is at least 128 times more sensitive than DAS-ELISA in detecting BNYVV.
Validation of mRT-PCR.
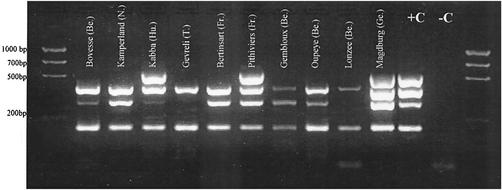
The results obtained by RT-PCR and mRT-PCR to determine the presence of BNYVV types A, B, and P are summarized in Table 1 for 25 soils from different origins. Samples received from Bulgaria, France, Hungary, Germany, Italy, Turkey, Sweden, and The Netherlands were treated in the same way as described above. Figure 3 illustrates the different patterns that were observed for 11 different samples. An interesting point is the systematic association in our samples of BNYVV with BSBV or of BSBV and BVQ. The most frequently detected virus was BSBV, followed by BVQ and then BNYVV. In no case was BVQ found alone. BVQ was found in Bulgaria, Belgium, France, Germany, Hungary, Italy, and The Netherlands, but it was not found in Turkey. From an epidemiological point of view, although the role of BNYVV in rhizomania syndrome is well established (21, 22), there is controversy regarding the role of P. betae in seedling growth (4) and in severe stunting of sugar beet (18) and still others reject the notion that P. betae has any effect on sugar beet growth (22). The controversy is emphasized by the study of two other viruses, which were possibly undetected in the P. betae used in previous studies (17). Our study confirms the ubiquitous presence of BSBV in sugar beet fields (24, 33). It was present in more than 80% of the analyzed samples, with a frequency much higher than that observed for BNYVV. The limited number of samples tested does not preclude further detection possibilities on different sugar beet cultivars or on different species. In all cases, BNYVV occurred with BSBV and often with BVQ as well. It would be very interesting to check a possible interaction of pomoviruses with BNYVV, as already pointed out by Lindsten (24).
FIG. 3.
Profiles obtained by mRT-PCR with 10 samples originating from different cities in Belgium (Be.), The Netherlands, (N.), Hungary (Hu.), Turkey (T.), France (Fr.), and Germany (Ge.). +C, positive control (Seneffe, Belgium); −C, negative control (RNA extracted from uninfected sugar beet cv. cadyx).
mRT-PCR tests have already been proposed to detect plasmodiophoromycete-transmitted viruses like Potato mop top virus (27), Soilborne wheat mosaic virus, and Wheat spindle streak mosaic virus (10). The proposed mRT-PCR technique allows the simultaneous detection of BNYVV, BSBV, BVQ, and their vector P. betae. The major advantage of the technique is this simultaneous detection, allowing insights into benyvirus and pomovirus epidemiologies and facilitating studies of recombination between viruses when the technique is combined with sequencing. The technique is a new tool for sugar beet breeders wanting to test their soils for the presence of different soilborne viruses. Though probably not as sensitive as a nested-PCR method for BNYVV detection (26), the new technique gives results that demonstrate that the sensitivity is much higher than that observed for the BNYVV DAS-ELISA and highly similar to that of RT-PCR. No RT-PCR technique has been described for the detection of BSBV and BVQ. Thus, the stated concentration limits of detectability of ELISA (16, 17, 24) emphasize the need for such a tool. Koenig and Lennefors (19) have emphasized that the diversity encountered among beet pomoviruses might be an explanation for the somewhat conflicting observations in their effects on yield. In the interaction of BNYVV with Beet soilborne mosaic virus, a synergy was shown (33). The recent advance in the field of viral suppressors of posttranscriptional gene silencing shed a new light on virus interactions among sugar beet and peanut viruses (5, 9, 30), underlining the interest in reexamining the rhizomania syndrome in light of the simultaneous presence of two or three different virus species.
Acknowledgments
A.M. is a fellow of the Belgian FRIA (Fonds pour la Recherche dans l'Industrie et l'Agriculture). J.-F.S. and C.B. thank the Ministry of Small Enterprises, Traders and Agriculture/DG6, Department Research and Development, Section Subsidized Research, for supporting this work.
We thank Sandor Kobza of the Plant Health and Soil Conservation Station of Budapest, Hungary, and Thomas Kuehne of the Federal Center for Breeding Research on Cultivated Plants (BAZ), Institute for Resistance Research and Pathogen Diagnostics, Aschersleben, Germany, for providing us with soil material.
REFERENCES
- 1.Asher, M. 1999. Sugar-beet rhizomania: the spread of a soilborne disease. Microbiol. Today 26:120-122.
- 2.Bariana, H. S., A. L. Shannon, P. W. G. Chu, and P. M. Waterhouse. 1994. Detection of five seed-borne legume viruses in one sensitive multiplex polymerase chain reaction. Phytopathology 84:1201-1205.
- 3.Bertolini, E., A. Olmos, M. C. Martinez, M. Gorris, and M. Cambra. 2001. Single-step multiplex RT-PCR for simultaneous and colourimetric detection of six RNA viruses in olive trees. J. Virol. Methods 96:33-41. [DOI] [PubMed] [Google Scholar]
- 4.Blunt, S. J., M. J. C. Asher, and C. A. Gilligan. 1991. Infection of sugar beet by Polymyxa betae in relation to soil temperature. Plant Pathol. 40:257-267. [Google Scholar]
- 5.Brault, V., S. Pfeffer, M. Erdinger, J. Mutterer, and V. Ziegler-Graff. 2002. Virus-induced gene silencing in transgenic plants expressing the minor capsid protein of beet western yellows virus. Mol. Plant-Microbe Interact. 15:799-807. [DOI] [PubMed]
- 6.Canova, A. 1959. Appunti di patologie della barbietola. Inf. Fitopatol. 9:390-396. [Google Scholar]
- 7.Delfosse, P., A. S. Reddy, A. Legreve, K. T. Devi, M. D. Abdurahman, H. Maraite, and D. V. R. Reddy. 2000. Serological methods for detection of Polymyxa graminis, an obligate root parasite and vector of plant viruses. Phytopathology 90:537-545. [DOI] [PubMed] [Google Scholar]
- 8.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunoyer, P., S. Pfeffer, C. Fritsch, O. Hemmer, O. Voinnet, and K. E. Richards. 2002. Identification, subcellular localization and some properties of a cysteine-rich suppressor of gene silencing encoded by peanut clump virus. Plant J. 29:555-567. [DOI] [PubMed] [Google Scholar]
- 10.Gitton, F., A. Diao, O. Ducrot, J. F. Antoniw, M. J. Adams, and H. Maraite. 1999. A two-step multiplex RT-PCR method for simultaneous detection of soil-borne wheat mosaic virus and wheat spindle streak mosaic virus from France. Plant Pathol. 48:635-641. [Google Scholar]
- 11.Hauser, S., C. Weber, G. Vetter, M. Stevens, M. Beuve, and O. Lemaire. 2000. Improved detection and differentiation of poleroviruses infecting beet or rape by multiplex RT-PCR. J. Virol. Methods 89:11-21. [DOI] [PubMed] [Google Scholar]
- 12.Heidel, G. B., and C. M. Rush. 1997. Characteristics of beet soil-borne mosaic virus, a furo-like virus infecting sugar beet. Plant Dis. 81:1070-1076. [DOI] [PubMed] [Google Scholar]
- 13.Henegariu, O., N. A. Heerema, S. R. Dlouhy, G. H. Vance, and P. H. Vogt. 1997. Multiplex PCR: critical parameters and step-by-step protocol. BioTechniques 23:504-511. [DOI] [PubMed] [Google Scholar]
- 14.Henry, C. M., I. Barker, R. A. C. Jones, and R. R. A. Coutts. 1986. Occurrence of a soil-borne virus of sugar beet in England. Plant Pathol. 35:585-591.
- 15.Henry, C. M., I. Barker, J. Morris, and S. A. Hugo. 1995. Detection of beet necrotic yellow vein virus using reverse transcription and polymerase chain reaction. J. Virol. Methods 54:15-28. [DOI] [PubMed] [Google Scholar]
- 16.Hutchinson, P. J., C. M. Henry, and H. A. Coutts. 1993. Mechanical inoculation of sugar beet roots with beet soil-borne virus in the absence of Polymyxa betae, p. 43-46. In J. L. Sherwood and C. M. Rush (ed.), Proceedings of the Second Symposium of the International Working Group on Plant Viruses with Fungal Vectors. American Society of Sugar Beet Technologists, Denver, Colo.
- 17.Kaufmann, A., R. Koenig, and H. Rohloff. 1993. Influence of beet soil-borne virus on mechanically inoculated sugar beet. Plant Pathol. 42:413-417. [Google Scholar]
- 18.Keskin, B. 1964. Polymyxa betae n. sp., ein parasit in den würzeln von Beta vulgaris Tournefort, besonders während der Jugendentwicklung der Zuckerrübe. Arch. Mikrobiol. 49:348-374. [DOI] [PubMed] [Google Scholar]
- 19.Koenig, R., and B. L. Lennefors. 2000. Molecular analyses of European A, B and P type sources of beet necrotic yellow vein virus and detection of the rare P type in Kazakhstan. Arch. Virol. 145:1561-1570. [DOI] [PubMed] [Google Scholar]
- 20.Koenig, R., C. W. Pleij, C. Beier, and U. Commandeur. 1998. Genome properties of Beet virus Q, a new furo-like virus from sugarbeet, determined from unpurified virus. J. Gen. Virol. 79:2027-2036. [DOI] [PubMed] [Google Scholar]
- 21.Koenig, R., W. Jarausch, Y. Li, U. Commandeur, W. Burgermeister, M. Gehrke, and P. Luddecke. 1991. Effect of recombinant beet necrotic yellow vein virus with different RNA compositions on mechanically inoculated sugar beets. J. Gen. Virol. 72:2243-2246. [DOI] [PubMed] [Google Scholar]
- 22.Koenig, R., and W. Burgermeister. 1989. Mechanical inoculation of sugar beet roots with isolates of Beet necrotic yellow vein virus having different RNA compositions. J. Phytopathol. 124:249-255. [Google Scholar]
- 23.Lesemann, D. E., R. Koenig, K. Lindsten, and C. Henry. 1989. Serotypes of beet soil-borne furovirus from FRG and Sweden. Bull. EPPO/OEPP 19:539-540. [Google Scholar]
- 24.Lindsten, K. 1993. Rhizomania—are both beet necrotic yellow vein virus (BNYVV) and beet soil-borne virus (BSBV) involved?, p. 67-70. In J. L. Sherwood and C. M. Rush (ed.), Proceedings of the Second Symposium of the International Working Group on Plant Viruses with Fungal Vectors. American Society of Sugar Beet Technologists, Denver, Colo.
- 25.Meunier, A., J. F. Schmit, A. Stas, A. Marlier, A. Wauters, S. Steyer, and C. Bragard. 2000. The status of rhizomania in Belgium. Parasitica 56:85-95. [Google Scholar]
- 26.Morris, J., G. R. Clover, V. A. Hraju, S. A. Hugo, and C. M. Henry. 2001. Development of a highly sensitive nested RT-PCR method for beet necrotic yellow vein virus detection. J. Virol. Methods 95:163-169. [DOI] [PubMed] [Google Scholar]
- 27.Mumford, R. A., K. Walsh, I. K. Barker, and N. Boonham. 2000. Detection of Potato mop top virus and Tobacco rattle virus using a multiplex real-time fluorescent reverse transcription polymerase chain reaction assay. Phytopathology 90:448-453. [DOI] [PubMed] [Google Scholar]
- 28.Mutasa, E. S., E. Ward, M. J. Adams, C. Collier, D. M. Chwarszczynska, and M. J. C. Asher. 1993. A sensitive DNA probe for the detection of Polymyxa betae in sugar beet roots. Physiol. Mol. Plant Pathol. 43:379-390. [Google Scholar]
- 29.Needleman, S., and C. Wunsch. 1970. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 48:443-453. [DOI] [PubMed] [Google Scholar]
- 30.Pfeffer, S., P. Dunoyer, F. Heim, K. E. Richards, G. Jonard, and V. Ziegler-Graff. 2002. P0 of beet western yellows virus is a suppressor of posttranscriptional gene silencing. J. Virol. 76:6815-6824. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Prillwitz, H., and E. Schlösser. 1992. Beet soil-borne virus: occurrence, symptoms and effect on development. Meded. Fac. Landbouwwet. Univ. Gent 57/2a:295-302.
- 32.Richards, K., and T. Tamada. 1992. Mapping functions on the multipartite genome of beet necrotic yellow vein virus. Annu. Rev. Phytopathol. 30:291-313. [Google Scholar]
- 33.Rush, C. M., and G. B. Heidel. 1995. Furovirus diseases of sugar beets in the United States. Plant Dis. 79:868-875. [Google Scholar]
- 34.Saade, M., F. Aparicio, J. Sanchez-Navarro, M. C. Herranz, A. Myrta, B. Di Terlizzi, and V. Pallas. 2000. Simultaneous detection of the three ilarviruses affecting stone fruit trees by nonisotopic molecular hybridization and multiplex reverse-transcription polymerase chain reaction. Phytopathology 90:1330-1336. [DOI] [PubMed] [Google Scholar]
- 35.Sharman, M., J. E. Thomas, and R. G. Dietzgen. 2000. Development of a multiplex immunocapture PCR with colourimetric detection for viruses of banana. J. Virol. Methods 89:75-88. [DOI] [PubMed] [Google Scholar]
- 36.Simon, L., T. J. Smalley, J. B. Jones, and F. T. Lasseigne. 1994. Aluminum toxicity in tomato. Part 2. Leaf gas exchange, chlorophyll content, and invertase activity. J. Plant Nutr. 17:307-311. [Google Scholar]
- 37.Tamada, T. 1999. Benyviruses, p. 154-160. In R. Webster and A. Granoff (ed.), Encyclopedia of virology, 2nd ed., vol. II. Academic Press, New York, N.Y.
- 38.Verhoyen, M., M. Van Den Bossche, and L. Van Steyvoort. 1987. Identification de nouveaux virus de la betterave en Belgique. Rev. Agric. (Brussels) 40:1463. [Google Scholar]