Abstract
Clostridium difficile is the principal etiologic agent of pseudomembranous colitis and is a major cause of nosocomial antibiotic-associated diarrhea. A limited degree of success in controlling C. difficile infection has been achieved by using probiotics; however, prebiotics can also be used to change bacterial community structure and metabolism in the large gut, although the effects of these carbohydrates on suppression of clostridial pathogens have not been well characterized. The aims of this study were to investigate the bifidogenicity of three nondigestible oligosaccharide (NDO) preparations in normal and antibiotic-treated fecal microbiotas in vitro and their abilities to increase barrier resistance against colonization by C. difficile by using cultural and molecular techniques. Fecal cultures from three healthy volunteers were challenged with a toxigenic strain of C. difficile, and molecular probes were used to monitor growth of the pathogen, together with growth of bifidobacterial and bacteroides populations, over a time course. Evidence of colonization resistance was assessed by determining viable bacterial counts, short-chain fatty acid formation, and cytotoxic activity. Chemostat studies were then performed to determine whether there was a direct correlation between bifidobacteria and C. difficile suppression. NDO were shown to stimulate bifidobacterial growth, and there were concomitant reductions in C. difficile populations. However, in the presence of clindamycin, activity against bifidobacteria was augmented in the presence of NDO, resulting in a further loss of colonization resistance. In the absence of clindamycin, NDO enhanced colonization resistance against C. difficile, although this could not be attributed to bifidobacterium-induced inhibitory phenomena.
The term colonization resistance is commonly used to describe the ability of the colonic microbiota to resist invasion by exogenous microorganisms (43). The mechanisms involved in colonization resistance include host factors such as bile acids, as well as gastrointestinal proteases, motility, and bacterial factors such as competition for adhesion sites and nutrients, together with production of toxic metabolites and antagonistic compounds (42, 44). Antibiotic therapy breaks down some of these normal defense systems, leaving individuals more susceptible to infection with opportunistic pathogens, such as Clostridium difficile.
In several studies workers have attempted to identify bacteria in the healthy intestinal microbiota that are responsible for suppression of C. difficile. One of the simplest approaches to antagonize growth of this organism is to allow a nontoxigenic strain to colonize an animal before challenge with a toxigenic isolate. Under these conditions, mortality can be dramatically reduced for several weeks before the toxigenic strain is able to become established (47). The use of synthetic microbiotas in attempts to identify the organisms responsible for colonization resistance has shown that various species are able to suppress the growth of C. difficile through competition for nutrients, lowered pH, and production of inhibitory metabolites (44).
In studies in which the abilities of over 400 fecal isolates to prevent multiplication of C. difficile in vitro were investigated (33), the majority of the bacteria were found to have no suppressive effect on the pathogen, although a few species belonging to six different genera displayed some inhibitory activity. Lactobacilli and group D enterococci displayed the most antagonistic activity; however, some bifidobacteria and bacteroides were also found to be effective. Populations of lactobacilli have been reported to be significantly smaller in C. difficile-positive hospitalized patients, while patients in whom the populations of lactobacilli remained relatively unchanged after antimicrobial treatment were less likely to be colonized by the pathogen (29).
While antibiotics are used to alleviate the symptoms of C. difficile diarrhea, they do not prevent relapse, the rate of which can be as high as 20%, and they have no role in the treatment of the majority of antibiotic-associated diarrheas in which C. difficile cannot be identified as the cause. Probiotic agents, such as saccharomyces, bifidobacteria, and lactobacilli, have therefore been viewed as being attractive treatments for antibiotic-associated diarrhea and C. difficile infection, and a number of workers have investigated their potential in this respect (31, 41).
The use of nondigestible oligosaccharides (NDO) as prebiotic agents may provide a useful approach for restoring or improving colonization resistance in compromised patients, since members of a host's indigenous microbiota would be stimulated instead of having foreign organisms introduced, as would be the case with probiotics. The ability of NDO to stimulate growth of bifidobacteria in the human colon has been well documented (14, 19, 21), and several authors have reported beneficial effects of these carbohydrates with respect to prevention or alleviation of diarrheal symptoms. In a recent study Oli et al. evaluated the use of oral electrolyte solutions in combination with fructooligosaccharides (FOS) for the treatment of acute secretory diarrhea induced by cholera toxin in pigs (30). While FOS supplementation did not result in a reduction in the duration of diarrhea, pigs treated with an oral electrolyte solution alone were found to have significantly higher lumen and mucosal populations of enterobacteria, indicating that FOS accelerated recovery of beneficial microorganisms to the detriment of potentially pathogenic species.
FOS have also been investigated to determine their ability to increase colonization resistance against C. difficile infection in animal models, in which they increased the mean survival time of animals with antibiotic-induced colitis; this was attributed to a reduction in toxin A titers (13, 48). An inhibitory effect of lactulose on growth of C. difficile in human fecal cultures has also been reported (20). Lactobacilli predominated in these cultures, while bacteroides, bifidobacteria, and enterobacteria were suppressed. However, bifidobacteria have tended to be the organisms that exhibited the greatest degree of stimulation in human studies with a variety of NDO (14, 19, 22), and it is for this reason that these bacteria were the focus of the present study.
Molecular analyses have been used to assess the effects of NDO on colonic bacterial populations (36, 38). These methods allow specific groups of bacteria to be targeted with a high degree of confidence without a need to isolate the organisms from complex communities, and they provide an indication of the relative metabolic potentials of different bacterial groups. The objectives of this study were to investigate the effects of NDO on colonization resistance against C. difficile in normal and antibiotic-treated fecal cultures by using 16S rRNA and viable count methods and to determine whether bifidobacteria play a role in this process.
MATERIALS AND METHODS
Organisms.
Fresh feces provided by three healthy male volunteers (ages, 25 to 31 years) were homogenized separately in the sterile, anaerobic, semidefined growth medium described below (pH 5.5) and were sieved through a 500-μm wire mesh to produce 20% (wt/vol) slurries. C. difficile NCTC 11223 was obtained from the National Collection of Type Cultures, Public Health Laboratory Service, London, England. The bacterium was stored at −80°C and maintained on the same semidefined culture medium supplemented with 1% (wt/vol) bacteriological agar.
Batch culture growth conditions.
Fermentation vessels with working volumes ranging from 260 to 282 ml were used to investigate growth and toxin formation by C. difficile in control and antibiotic-treated fecal cultures. The growth medium contained (per liter of distilled water) 4.0 g of starch (soluble), 1.0 g of xylan (oat spelt), 2.0 g of pectin (citrus), 1.0 g of guar gum, 4.0 g of mucin (type II; Sigma), 1.0 g of arabinogalactan (larchwood), 5.0 g of peptone water, 4.0 g of tryptone, 4.0 g of yeast extract, 2.0 g of casein, 4.5 g of NaCl, 0.4 g of K2HPO4, 0.15 g of MgCl2 · 6H2O, 0.4 g of NH4Cl, 0.25 g of KCl, 0.5 g of cysteine, 0.005 g of vitamin B12, and 0.01 g of hemin. One milliliter of Tween 80 and 1 ml of a trace mineral solution (3) were added to each 1 liter of culture medium. The culture medium was sterilized by autoclaving. The fermentors were automatically pH controlled at pH 5.5, flushed with oxygen-free nitrogen gas, and maintained at 37°C by using a circulating bath connected to external water jackets.
One milliliter of a log-phase C. difficile culture (106 viable cells) was added to each fermentation vessel containing 200 ml of the semidefined culture medium described above at zero time together with 50 ml of fecal slurry. Eight fermentors were operated simultaneously to test the oligosaccharides Raftiline LS (inulin; Orafti, Tienen, Belgium), Raftilose P95 (FOS; Orafti), and Oligomate 55 (galactooligosaccharide [GOS]; Yakult, Tokyo, Japan). Information about the chemical compositions of these NDO is given in Table 1. One culture was used as a control (no further additions), and the others were treated with clindamycin (20 μg/ml), a test oligosaccharide (10 g/liter), and both clindamycin and a test oligosaccharide. The clindamycin and oligosaccharides were added to the medium after it was autoclaved. Each vessel was sampled periodically for 48 h for immediate counting of C. difficile, while the predominant bifidobacteria were enumerated and identified to the species level. Samples for 16S rRNA and C. difficile cytotoxin assays were frozen at −80°C, and aliquots for fermentation product analysis were frozen at −20°C.
TABLE 1.
Chemical compositions and characteristics of NDO
| NDO | Chemical compositiona |
|---|---|
| FOS (Raftilose P95) | 95% oligosaccharide β(2-1)-fructan; 60% G-F, 40% F (DP, 2 to 8; average, DP, 4 to 5) |
| GOS (Oligomate 55) | Mainly 6′-galactosyllactose; DP, of oligosaccharide fraction, 2 to 5 (primarily DP 3); 55% pure |
| Inulin (Raftiline LS) | >99% oligosaccharide β(2-1)-fructan (inulin); average DP, 10 to 12 |
F, fructose; G, glucose; DP, degree of polymerization.
Bacterial enumeration.
Viable counts were determined as described previously (23, 25). Bacteria from the fermentation vessels were suspended and serially diluted (10-fold) by using anaerobic, half-strength peptone water (5 g of peptone per liter and 2.5 g of NaCl per liter in distilled water). Samples were then spread in duplicate onto prereduced agar plates and incubated in an anaerobic chamber (10% H2, 10% CO2, 80% N2) for 48 h before colonies were counted. Cycloserine-cefoxitin-fructose agar was used for isolation of C. difficile, and Beerens agar (4) was employed to select for bifidobacteria.
C. difficile cytotoxin assay.
The presence of C. difficile cytotoxin was detected by observing the cytopathic effects (CPE) of sample supernatants on Vero (African green monkey) cells and was confirmed by neutralization with Clostridium sordellii antitoxin (Public Health Laboratory Service, Cambridge, England). All samples taken from the batch cultures and both continuous culture vessels were tested. Two wells of a cell culture tray were inoculated with 0.1 ml of supernatant from each sample. C. sordellii antitoxin (0.1 ml) was added to one of the wells and incubated overnight at 37°C before microscopic examination for CPE. Culture supernatant from a highly toxigenic strain of C. difficile (NCTC 11207) was used as a positive control. Cytotoxin-positive samples were diluted in phosphate-buffered saline (pH 7.0), and the toxin titer was the dilution at which less than 50% of the monolayer showed evidence of CPE.
Fermentation product analysis.
A Pye model 204 gas chromatograph fitted with a flame ionization detector was used to measure short-chain fatty acids (SCFA) and other carboxylic acids. SCFA were extracted by using procedures described by Holdeman et al. (17), and 50 mM tert-butylacetic acid was included as an internal standard. The SCFA were separated by using Unicam 10% FFAP 100/120 mesh Chromosorb WAW-DMCS in a glass column (length, 1.8 m; inside diameter, 2 mm). The injector, detector, and column temperatures were 250, 300, and 155°C, respectively. The flow rates of the N2 carrier gas, H2, and air were 50, 30, and 370 ml/min. Lactate and succinate were measured by using the same analytical system after methylation of the samples (17). The injector, detector, and column temperatures were 175, 175, and 165°C, respectively.
16S rRNA analysis.
rRNA was extracted, blotted, and hybridized by using procedures described previously (18). Bacteria were disrupted mechanically by using a reciprocating shaker during the first phenol extraction, and this was repeated after the preparation was heated at 60°C for 2 min. This was followed by two phenol-chloroform-isoamyl alcohol extractions and one chloroform-isoamyl alcohol extraction. Total rRNA was precipitated overnight at −20°C with ammonium acetate, washed twice in ethanol, and resuspended in nuclease-free water. Nucleic acid samples were denatured and diluted prior to blotting in triplicate onto hybridization membranes along with reference RNA series for normalization. One oligonucleotide sequence directed against C. difficile (5′-ACTGAGAGTAGCTTTA-3′) (45), one oligonucleotide sequence directed against bifidobacteria (5′-CCGGTTTTMAGGGATCC-3′) (10), one oligonucleotide sequence directed against the Bacteroides-Porphyromonas-Prevotella group (5′-GCACTTAAGCCGACACCT-3′) (10), and one oligonucleotide sequence directed against total bacterial rRNA (5′-GCTGCCTCCCGTAGGAGT-3′) (1) were 5′ end labeled with 32P by using [γ-32P]ATP. The membranes were then prehybridized for 2 h before labeled probe was added in fresh buffer and they were hybridized for a further 16 h. Following this, the membranes were washed for 2 h in a sodium dodecyl sulfate-1× SSC solution (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and then twice at each of the experimentally determined wash temperatures (42, 46, 46, and 54°C). The hybridization signal was quantified by electronic autoradiography (Instant Imager; Canberra Packard, Panbourne, Berks, United Kingdom).
Cellular fatty acid analysis.
Bifidobacteria were identified to the species level on the basis of the fatty acid methyl esters extracted from 30 ml of an overnight culture grown in peptone-yeast extract-Tween broth (17). The Tween was removed prior to extraction by washing the cells in 0.7% MgSO4. Fatty acid methyl esters were separated by using 5% phenylmethyl silicone capillary columns and flame ionization detectors attached to a Hewlett-Packard model 6890 gas chromatograph under conditions described previously (18). The peaks were integrated, and isolates were identified by automatic comparison of fatty acid profiles by using the Moore library of the Microbial Identification System (MIDI, Newark, Del.) (28).
Continuous culture of C. difficile and bifidobacteria.
A two-stage continuous culture system was used to investigate interactions between C. difficile NCTC 11223 and fecal bifidobacteria isolated from the fermentation vessels. Bifidobacterium adolescentis, Bifidobacterium angulatum, Bifidobacterium bifidum, and Bifidobacterium catenulatum were used to inoculate a 0.10-liter (working volume) glass fermentation vessel containing peptone-yeast extract-glucose medium (17). A second chemostat inoculated with only C. difficile NCTC 11223 was added to create a two-stage continuous culture system in which the C. difficile culture was fed directly into the bifidobacteria chemostat. Anaerobic conditions were maintained by sparging the cultures with oxygen-free N2 gas, and the temperature (37°C) and pH (5.5) were automatically controlled as described above. The growth conditions for the two vessels were identical. Once the system had achieved a steady state, as determined by fermentation product analysis and bacterial cell counting, samples were taken for cytotoxin analysis and enumeration of bacteria at specific growth rates corresponding to dilution rates of 0.50 and 0.52 h−1.
Chemicals.
Unless stated otherwise, all chemicals were obtained from Sigma Chemical Co. (Poole, Dorset, United Kingdom). Bacteriologic culture media were purchased from Oxoid Ltd. (Basingstoke, Hamps, United Kingdom).
Statistical analyses.
The results obtained for the three different NDO with each of the three fecal samples were grouped together to form a treatment group (n = 9). These results were compared to the results for control vessels (n = 3) by using analysis of variance calculated with Data Desk software, version 6.0 (Data Description Inc., Ithaca, N.Y.). Viable counts and SCFA were compared at 12 h, while 16S rRNA was analyzed at 6 h.
RESULTS
Viable counts of C. difficile and bifidobacteria in fermentation vessels.
Table 2 shows that addition of NDO to fermentors containing fecal bacteria inhibited growth of C. difficile significantly in normal microbiotas (P < 0.05), although the variation between subjects and NDO treatment was high. GOS was initially more effective in reducing the number of the pathogen, and then inulin became more effective. However, C. difficile proliferated in cultures supplemented with clindamycin, and under these circumstances NDO did not inhibit its growth. Indeed, the viable counts of C. difficile in cultures containing FOS and clindamycin were reproducibly higher than those in fermentation vessels containing clindamycin alone.
TABLE 2.
Viable counts of C. difficile in fecal cultures supplemented with NDO and clindamycin
| Treatment |
C. difficile viable counts (log10CFU/ml)a |
||||
|---|---|---|---|---|---|
| 0 h | 6 h | 12 h | 24 h | 48 h | |
| Control | 5.8 ± 0.1 | 5.8 ± 0.1 | 5.8 ± 0.1 | 5.8 ± 0.4 | 5.3 ± 0.4 |
| NDO treatment group | |||||
| FOS | 5.7 ± 0.1 | 5.6 ± 0.1 | 5.4 ± 0.1 | 5.2 ± 0.1 | 4.4 ± 0.6 |
| GOS | 5.7 ± 0.1 | 5.2 ± 0.3 | 5.0 ± 0.2 | 4.7 ± 0.2 | 4.8 ± 0.2 |
| Inulin | 5.8 ± 0.1 | 5.7 ± 0.2 | 5.2 ± 0.4 | 4.1 ± 0.5 | 4.4 ± 0.3 |
| Mean | 5.7 ± 0.1 | 5.6 ± 0.2 | 5.3 ± 0.4b | 4.9 ± 0.5 | 4.5 ± 0.7 |
| Clindamycin control | 5.8 ± 0.1 | 6.7 ± 0.2 | 7.6 ± 0.4 | 7.2 ± 0.2 | 7.0 ± 0.7 |
| Clindamycin + NDO treatment group | |||||
| Clindamycin + FOS | 5.7 ± 0.1 | 7.2 ± 0.4 | 8.1 ± 0.2 | 8.3 ± 0.5 | 7.7 ± 0.1 |
| Clindamycin + GOS | 5.7 ± 0.1 | 6.6 ± 0.2 | 7.7 ± 0.2 | 7.4 ± 0.1 | 7.3 ± 0.4 |
| Clindamycin + inulin | 5.8 ± 0.1 | 6.9 ± 0.2 | 7.8 ± 0.2 | 8.0 ± 0.5 | 6.2 ± 0.2 |
| Mean | 5.7 ± 0.1 | 6.9 ± 0.4 | 7.7 ± 0.5 | 8.1 ± 0.4 | 7.0 ± 0.6 |
Mean ± standard deviation (n = 3).
There was a significant difference between the control and the NDO treatment group as determined by analysis of variance (P ≤ 0.05).
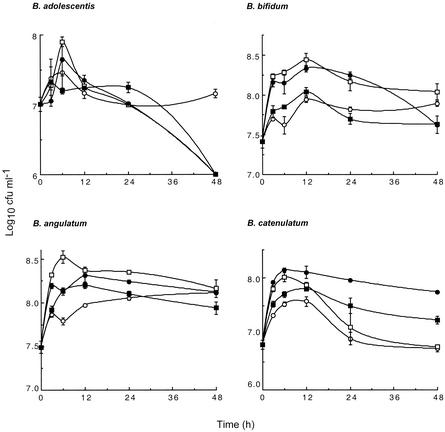
Addition of NDO to normal fecal cultures resulted in increased total bifidobacterial counts (P ≤ 0.001), and again, large variations were observed between volunteer fecal material and NDO treatment (Table 3). Clindamycin treatment resulted in marked reductions in the number of bifidobacteria, and when a combination of clindamycin with an NDO was used, these bacteria were further suppressed (P < 0.05). The enhanced inhibitory activity of clindamycin against bifidobacterial populations was observed with all three NDO tested, although the greatest antibifidobacterial activity was observed with clindamycin and FOS or GOS. In normal fecal cultures of one of the subjects studied in more detail, each NDO was found to stimulate growth of the four predominant bifidobacterial species to different degrees (Fig. 1). The highest cell population densities occurred with GOS for B. adolescentis, B. angulatum, and B. bifidum, while the preferred substrate for growth of B. catenulatum was FOS.
TABLE 3.
Viable counts of bifidobacteria in fecal cultures supplemented with NDO and clindamycin
| Treatment | Bifidobacterial viable counts (log10 CFU ml−1)a |
||||
|---|---|---|---|---|---|
| 0 h | 6 h | 12 h | 24 h | 48 h | |
| Control | 7.8 ± 0.6 | 8.2 ± 0.4 | 8.2 ± 0.5 | 8.1 ± 0.5 | 7.6 ± 0.6 |
| NDO treatment group | |||||
| FOS | 7.8 ± 0.6 | 8.7 ± 0.3 | 8.7 ± 0.4 | 8.6 ± 0.4 | 7.8 ± 0.7 |
| GOS | 7.8 ± 0.6 | 8.8 ± 0.3 | 8.7 ± 0.5 | 8.5 ± 0.4 | 6.8 ± 1.5 |
| Inulin | 7.9 ± 0.7 | 8.6 ± 0.4 | 8.6 ± 0.5 | 8.3 ± 0.2 | 7.6 ± 0.9 |
| Mean | 7.8 ± 0.6 | 8.7 ± 0.3 | 8.7 ± 0.4b | 8.5 ± 0.3 | 7.4 ± 1.0 |
| Clindamycin control | 8.0 ± 0.6 | 6.2 ± 1.3 | 6.0 ± 1.2 | 5.5 ± 1.1 | 5.1 ± 0.9 |
| Clindamycin + NDO treatment group | |||||
| Clindamycin + FOS | 7.8 ± 0.6 | 5.0 ± 0.3 | 4.3 ± 0.2 | 3.1 ± 0.8 | 3.3 ± 0.9 |
| Clindamycin + GOS | 7.8 ± 0.6 | 5.1 ± 0.5 | 4.3 ± 0.3 | 3.5 ± 0.7 | 3.4 ± 1.0 |
| Clindamycin + Inulin | 7.9 ± 0.7 | 5.5 ± 1.2 | 5.1 ± 1.3 | 4.4 ± 1.7 | 4.2 ± 1.3 |
| Mean | 7.8 ± 0.6 | 5.2 ± 0.7 | 4.5 ± 0.8c | 3.6 ± 1.2 | 3.6 ± 1.0 |
Mean ± standard deviation (n = 3).
There was a significant difference between the control and the NDO treatment group as determined by analysis of variance (P ≤ 0.001).
There was a significant difference between the clindamycin control and the group that was treated with clindamycin and NDO (P ≤ 0.05).
FIG. 1.
Effects of NDO on four predominant bifidobacterial populations in fecal cultures. The data points are means, and the error bars indicate standard deviations (n = 2). Symbols: ○, control; □, GOS; •, FOS; ▪, inulin.
16S rRNA analysis.
The total amount of bacterial rRNA in normal fecal cultures increased at least threefold during the first 6 h of incubation and then declined to approximately 50 μg/ml by 24 h (results not shown). Vessels containing clindamycin showed no such increase, and the rRNA concentrations remained relatively constant over the experimental time course. Addition of NDO resulted in minor changes in the total bacterial rRNA levels, but none of the changes were significant.
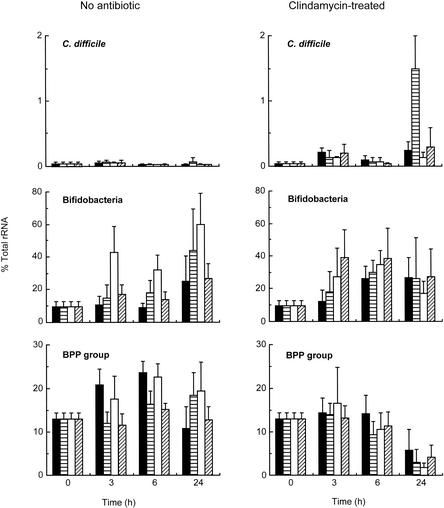
Figure 2 shows that the percentage of bacteroides-prevotella-porphyromonas (BPP group) rRNA in the fermentation vessels without the antibiotic ranged from 10 to 25%. Addition of NDO caused a reduction in the amount of the BPP group rRNA in normal fecal cultures supplemented with FOS and inulin. However, at 6 h, the decrease was not statistically significant.
FIG. 2.
Percentages of specific rRNA in fecal cultures supplemented with NDO with or without clindamycin. The bars indicate means, and the error bars indicate standard errors (n = 3). ▪, control; ▤, FOS; □, GOS; ▨, inulin.
The amount of bifidobacterial rRNA expressed as a percentage of the total bacterial rRNA increased as a result of addition of NDO to normal fecal cultures (P < 0.05). GOS rapidly induced bifidobacterial rRNA synthesis, and by 24 h FOS and GOS increased the amount of bifidobacterial rRNA to 40 and 60% of the total bacterial rRNA detected, respectively. In clindamycin-containing cultures, all three carbohydrates induced increases in the percentage of bifidobacterial rRNA within 3 h, and this rRNA accounted for almost 40% of the total bacterial rRNA in slurries supplemented with clindamycin and inulin. However, the values were not statistically significant by 6 h.
rRNA specific to C. difficile was detected only at very low levels in all fecal cultures, and no significant differences were found at 6 h. However, while the rRNA levels remained low in normal fecal incubation mixtures throughout the experiment, the cultures supplemented with clindamycin were markedly different after 24 h. The most pronounced changes were seen in fermentation vessels containing clindamycin and FOS, in which over 1% of the total bacterial rRNA could be ascribed to C. difficile.
Fermentation products.
The amounts of all SCFA increased over the course of the experiment, and acetate, propionate, and butyrate were the major fermentation products (Fig. 3). Changes in the levels of these SCFA were found to be significant at 12 h (P < 0.05) in normal fecal batch cultures supplemented with NDO, and a reduction in the level of isovalerate (P < 0.05) was also observed (results not shown). Addition of clindamycin to the fermentation vessels resulted in reductions in the formation of all SCFA except isobutyrate and isocaproate, which were detected only in cultures containing the antibiotic. In these antibiotic-treated cultures, NDO did not induce significant changes at 12 h.
FIG. 3.
Fermentation products in fecal cultures supplemented with NDO with or without clindamycin. (a) Acetate, propionate, and butyrate concentrations at 12 h. (b) Lactate and succinate concentrations at 3 and 12 h. The bars indicate means, and the error bars indicate standard errors (n = 3). C, control; Clind, clindamycin. ▪, acetate; ▤, propionate; □, butyrate; ▩, lactate; ▨, succinate.
Figure 3 shows that lactate and succinate were transitory fermentation products in normal fecal cultures. NDO supplementation resulted in lactate accumulation in the first 3 h, with the lactate concentration peaking at over 30 mM, and also increased the succinate concentrations. However, the levels of both these organic acids were markedly reduced after 12 h of incubation, and no significant differences were found at this time. In clindamycin-treated cultures, both of these fatty acids were detected throughout the experiments.
C. difficile cytotoxin production.
No cytotoxin was detected at any time in vessels containing a normal microbiota. The only time at which cytotoxic activity was found was 48 h, and this activity was limited to the fermentation vessels supplemented with both clindamycin and FOS (toxin titer, 27 ± 19). Neutralization of the CPE with C. sordellii antitoxin confirmed that C. difficile toxin was the cytotoxic agent. C. difficile cytotoxic activity was not detected in any chemostat during the continuous culture studies.
Bifidobacterial antagonism of C. difficile in continuous culture.
To test whether bifidobacteria were responsible for inhibiting the growth of C. difficile in the NDO experiments, a two-stage continuous culture system was constructed in which the pathogen was grown in a chemostat which fed a second vessel containing bifidobacteria. Table 4 shows the growth of C. difficile NCTC 11223 and bifidobacteria in this model system. We found that B. angulatum and B. bifidum became established at high levels when they were inoculated into the chemostat, although the growth of these organisms was inhibited in the presence of B. adolescentis and B. catenulatum. No inhibition of C. difficile was observed with a combination of B. angulatum and B. bifidum or with a combination of B. adolescentis and B. catenulatum at the specific growth rates tested.
TABLE 4.
Effect bifidobacteria on the growth of C. difficile in continuous culture
| Expt | Dilution rate (h−1) | Vessel | Organism | Viable count (log10 CFu/ml)a |
|---|---|---|---|---|
| 1 | 0.50 | 1 | C. difficile | 8.0 ± 0.1 |
| 2 | C. difficile | 8.4 ± 0.1 | ||
| B. adolescentis | 8.1 ± 0.3 | |||
| B. catenulatum | 8.6 ± 0.0 | |||
| 2 | 0.52 | 1 | C. difficile | 8.4 ± 0.0 |
| 2 | C. difficile | 8.6 ± 0.0 | ||
| B. angulatum | 8.2 ± 0.1 | |||
| B. bifidum | 7.8 ± 0.1 |
Mean ± standard deviation (n = 2).
DISCUSSION
The barrier effect of the intestinal microbiota is a fundamental factor in the pathophysiology of C. difficile-induced colitis (46). Attempts to increase colonization resistance by using probiotics and fecal enemas have yielded favorable results in some clinical trials (11, 31, 40), but these procedures have the disadvantage of introducing foreign microorganisms into the gut. Prebiotics in the form of NDO overcome this problem by stimulating growth of the autochthonous colonic microbiota. The abilities of FOS, GOS, and inulin to increase bifidobacterial populations in the colonic microbiota have been described previously (14, 19), and in this study we similarly showed that addition of NDO resulted in small but significant increases in the numbers of these bacteria (Table 3). RNA analyses demonstrated that these carbohydrates also enhanced the metabolic potential of bifidobacterial populations in the fecal cultures (Fig. 2). The ability of NDO to increase colonization resistance against invading pathogens is related to the fact that these compounds should be utilized only by a small group of nonpathogenic, protective organisms, such as bifidobacteria. Although viable counts showed that C. difficile was outcompeted by the normal microbiota in the presence of NDO, these results did not indicate whether the NDO were utilized selectively by the bifidobacteria.
Normally, the pH values in the distal colon range from 6 to 7, and this fact is often used in systems modeling of the colon (34). However, autopsy studies have shown that the cecal pH is around 5.5 (9, 24), and in situ radiometric techniques have demonstrated that carbohydrates such as lactulose can reduce the cecal pH to values as low as 4.5 (7, 12). This provided the rationale for performing fermentation experiments at pH 5.5 in this study.
The bacteroides group has been widely reported to be the major culturable group of organisms in the colon (14, 21, 26), and retrospective use of the BPP group probe demonstrated that there was a degree of selectivity in the organisms which fructan-based oligosaccharides, in particular, stimulated during the first 6 h of incubation, when the NDO were being rapidly fermented. One factor affecting this could have been the purity of the NDO preparations, because the GOS contained proportionally more free sugars than the FOS and inulin contained (Table 1). GOS did induce large changes in the proportion of bacteroides and bifidobacterial rRNA in these studies. The bacteroides and bifidobacterial rRNA accounted for approximately 60% of total rRNA during the early stages of fermentation. Probes designed for other major bacterial groups were not used in this investigation, but it was not entirely unexpected that these two probes accounted for a high percentage of the microbiota. Sghir et al. demonstrated that the bacteroides probe alone could account for between 7 and 74% of the total RNA in feces (37). Furthermore, the conditions in the batch cultures would initially have been more similar to those of the cecum, and comparisons of human cecal material and human fecal material indicate that the proportions of specific rRNA can vary markedly between the two (26).
The fact that the Northern hybridization technique does not distinguish between viable and nonviable cells meant that it was less suitable for assessing the inhibition of C. difficile. The C. difficile probe was originally developed in conjunction with PCR to allow detection of as few as 10 C. difficile cells (15), although our results suggest that without amplification, use of this probe would be limited to samples containing relatively high numbers of bacteria (ca. 108 cells). Viable counting showed that NDO inhibited C. difficile in normal fecal microbiotas (Table 2). This could be attributed to increased bifidobacterial populations that stimulated competition for nutrients, as well as the production of inhibitory substances, such as SCFA (27). However, changes in these two bacterial populations were not directly related, since variations in the degree of bifidogenicity of these carbohydrates could not be correlated with reductions in the number of C. difficile cells. For example, inulin resulted in a relatively small increase in the number of bifidobacteria, while the carbohydrate that showed the least suppression of C. difficile was found to be FOS. This could be explained by the presence of more fructose monosaccharide in the FOS preparation that could be utilized by C. difficile. Moreover, since NDO were found to stimulate different species of bifidobacteria to various degrees (Fig. 1), the ability of inulin to increase populations of strains present at very low levels cannot be discounted. The way in which inulin suppressed growth of C. difficile, despite its lower rate of fermentation, indicates that this carbohydrate with long-chain molecules should be the NDO of choice for individuals at risk of infection by this organism, since it is more likely to be available for fermentation in the distal gut, where pseudomembranous colitis develops (8).
Chemostat modeling studies (Table 4) demonstrated that bacterial interactions could result in inhibition of growth, since all four bifidobacteria could not become established in the chemostat at any one time. This interspecies competition further highlights the complex interactions among bacteria within the gut ecosystem. Filter-sterilized supernatants of the bifidobacteria grown in continuous cultures showed that suppression of C. difficile could be related to pH, although populations of this bacterium remained stable at pH 5.5 (results not shown). Many bacteria have increased sensitivity to SCFA as the pH becomes more acidic (35), and operating the culture vessels at pH 5.5 undoubtedly affected the growth of C. difficile. Acetate, propionate, and butyrate suppress both growth and toxin production by C. difficile at concentrations as low as 10 mM, and these effects are pH dependent (2, 27, 32). In this study, SCFA concentrations were lower in clindamycin-treated vessels, and C. difficile proliferated under these culture conditions. However, the rates of fermentation of the NDO could not be directly correlated with pathogen suppression.
Previous fecal culture studies showed that inhibition of C. difficile did not always correlate with low culture pH or the presence of specific SCFA (5). It was concluded that microbial competition was the mechanism of colonization resistance, instead of the production of soluble antimicrobial substances. The presence of a complex microbiota is still considered to be the major factor that suppresses C. difficile colonization, although the exact components remain unknown (6, 44). Interestingly, fecal cultures from breast-fed infants were more inhibitory and generally more acidic than fecal cultures from bottle-fed infants (5). Bifidobacteria are likely to predominate in the stools of breast-fed babies (16, 39). However, in the continuous culture experiments described here, bifidobacteria had little effect on C. difficile. Thus, although an NDO was a suppressive factor in fecal batch cultures, colonization resistance against C. difficile was not a bifidobacterium-specific response. Instead, the barrier effect of the intestinal microbiota involves multiple factors that affect pathogen growth (5), and in the case of C. difficile these processes can be enhanced by NDO in vitro.
Treatment of the anaerobic microbiota with an antibiotic resulted in NDO having no protective effect, while the antibacterial activity of clindamycin was enhanced. This antibiotic prevents peptide bond formation at the bacterial 50S ribosomal subunit by inhibiting peptidyl transferase, thereby arresting protein synthesis. Stimulation of bacterial growth by NDO results in a greater demand for protein synthesis, potentitating the antibiotic effects in actively growing populations. Interestingly, cultures inoculated simultaneously with clindamycin and FOS were the only cultures that exhibited cytotoxic activity. The severe breakdown in colonization resistance in these microbiotas raises questions concerning interactions of NDO and antibiotics in the gut. Gaskins et al. (13) reported a similar effect, and in their study C. difficile populations were found to be larger in mice treated with cefoxitin and a FOS supplement than in mice given the antibiotic alone. However, despite the increase in C. difficile counts, the titers of C. difficile toxin A were significantly reduced when FOS was given with the antibiotic.
In conclusion, NDO exhibited metabolic selectivity in fecal cultures since bifidobacterial growth was stimulated more than growth of bacteroides and related organisms. Utilization of these carbohydrates by the microbiota also resulted in reductions in C. difficile populations, although continuous culture studies and SCFA analyses indicated that this was not due to bifidobacteria. Clindamycin allowed overgrowth of C. difficile, and fermentable carbohydrates augmented its antibacterial effects. Thus, NDO could be useful for enhancing resistance to colonization by C. difficile prophylactically, but further work is needed to investigate these effects in human subjects, since they may be of little value in patients undergoing antibiotic therapy.
REFERENCES
- 1.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arzese, A., P. Corradino, and G. A. Botta. 1990. Effect of short chain fatty acids on growth and toxin production of pathogenic clostridia, p. 178. In S. P. Borriello (ed.), Clinical and molecular aspects of anaerobes. Wrightson Biomedical, Petersfield, England.
- 3.Balch, W. E., G. E. Fox, L. J. Magrum, C. R. Woese, and R. S. Wolfe. 1979. Methanogens: reevaluation of a unique biological group. Microbiol. Rev. 43:260-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beerens, H. 1991. Elective and selective isolation medium for Bifidobacterium spp. Lett. Appl. Microbiol. 11:155-157. [Google Scholar]
- 5.Borriello, S. P., and F. E. Barclay. 1986. An in-vitro model of colonisation resistance to Clostridium difficile infection. J. Med. Microbiol. 21:299-309. [DOI] [PubMed] [Google Scholar]
- 6.Borriello, S. P., and M. H. Wilcox. 1998. Clostridium difficile infections of the gut: the unanswered questions. J. Antimicrob. Chemother. 41S:C67-C69. [DOI] [PubMed]
- 7.Brown, R. L., J. A. Gibson, G. E. Sladen, B. Hicks, and A. M. Dawson. 1974. Effects of lactulose and other laxatives on ileal and colonic pH as measured by a radiotelemetry device. Gut 15:999-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen, M. B., and R. A. Giannella. 1991. Bacterial infections: pathophysiology, clinical features, and treatment, p. 395-428. In S. F. Phillips, J. H. Pemberton, and R. G. Shorter (ed.), The large intestine: physiology, pathophysiology and disease. Raven Press, New York, N.Y.
- 9.Cummings, J. H., E. W. Pomare, W. J. Branch, C. P. Naylor, and G. T. Macfarlane. 1987. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut 28:1221-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dore, J., A. Sghir, G. Hannequart-Gramet, G. Corthier, and P. Pochart. 1998. Design and evaluation of a 16S rRNA-targeted oligonucleotide probe for specific detection and quantitation of human faecal Bacteroides populations. Syst. Appl. Microbiol. 21:65-71. [DOI] [PubMed] [Google Scholar]
- 11.D'Souza, A. L., C. Rajkumar, J. Cooke, and C. J. Bulpitt. 2002. Probiotics in prevention of antibiotic associated diarrhoea: meta-analysis. BMJ 324:1361.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flourie, B., C. Florent, J. P. Jouany, P. Thivend, F. Etanchaud, and J. C. Rambaud. 1986. Colonic metabolism of wheat starch in healthy humans. Effects on fecal outputs and clinical symptoms. Gastroenterology 90:111-119. [DOI] [PubMed]
- 13.Gaskins, H. R., R. I. Mackie, T. May, and K. A. Garleb. 1996. Dietary fructo-oligosaccharide modulates large intestinal inflammatory responses to Clostridium difficile in antibiotic-compromised mice. Microb. Ecol. Health Dis. 9:157-166. [Google Scholar]
- 14.Gibson, G. R., E. R. Beatty, X. Wang, and J. H. Cummings. 1995. Selective stimulation of bifidobacteria in the human colon by oligofructose and inulin. Gastroenterology 108:975-982. [DOI] [PubMed] [Google Scholar]
- 15.Gumerlock, P. H., Y. J. Tang, F. J. Meyers, and J. Silva, Jr. 1991. Use of the polymerase chain reaction for the specific and direct detection of Clostridium difficile in human feces. Rev. Infect. Dis. 13:1053-1060. [DOI] [PubMed] [Google Scholar]
- 16.Harmsen, H. J., A. C. Wildeboer-Veloo, G. C. Raangs, A. A. Wagendorp, N. Klijn, J. G. Bindels, and G. W. Welling. 2000. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J. Pediatr. Gastroenterol. Nutr. 30:61-67. [DOI] [PubMed] [Google Scholar]
- 17.Holdeman, L., E. P. Cato, and W. E. C. Moore. 1977. Anaerobe laboratory manual, 4th ed. Virginia Polytechnic Institute and State University, Blacksburg.
- 18.Hopkins, M. J., R. Sharp, and G. T. Macfarlane. 2001. Age and disease related changes in intestinal bacterial populations assessed by cell culture, 16S rRNA abundance, and community cellular fatty acid profiles. Gut 48:198-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito, M., Y. Deguchi, A. Miyamori, K. Matsumoto, H. Kikuch, K. Matsumoto, T. Kobayashi, T. Yajima, and T. Kan. 1990. Effects of administration of galactooligosaccharides on the human faecal microflora, stool weight and abdominal sensation. Microb. Ecol. Health Dis. 3:285-292. [Google Scholar]
- 20.Ito, Y., H. Moriwaki, Y. Muto, N. Kato, K. Watanabe, and K. Ueno. 1997. Effect of lactulose on short-chain fatty acids and lactate production and on the growth of faecal flora, with special reference to Clostridium difficile. J. Med. Microbiol. 46:80-84. [DOI] [PubMed] [Google Scholar]
- 21.Kleessen, B., B. Sykura, H. J. Zunft, and M. Blaut. 1997. Effects of inulin and lactose on fecal microflora, microbial activity, and bowel habit in elderly constipated persons. Am. J. Clin. Nutr. 65:1397-1402. [DOI] [PubMed] [Google Scholar]
- 22.Kolida, S., K. Tuohy, and G. R. Gibson. 2002. Prebiotic effects of inulin and oligofructose. Br. J. Nutr. 87(Suppl. 2):193-217. [DOI] [PubMed]
- 23.Macfarlane, G. T., J. H. Cummings, and C. Allison. 1986. Protein degradation by human intestinal bacteria. J. Gen. Microbiol. 132:1647-1656. [DOI] [PubMed] [Google Scholar]
- 24.Macfarlane, G. T., G. R. Gibson, and J. H. Cummings. 1992. Comparison of fermentation reactions in different regions of the human colon. J. Appl. Bacteriol. 72:57-64. [DOI] [PubMed] [Google Scholar]
- 25.Macfarlane, S., M. E. Quigley, M. J. Hopkins, D. F. Newton, and G. T. Macfarlane. 1998. Polysaccharide degradation by human intestinal bacteria during growth under multi-substrate limiting conditions in a three-stage continuous culture system. FEMS Microbiol. Ecol. 26:231-243. [Google Scholar]
- 26.Marteau, P., P. Pochart, J. Dore, C. Bera-Maillet, A. Bernalier, and G. Corthier. 2001. Comparative study of bacterial groups within the human cecal and fecal microbiota. Appl. Environ. Microbiol. 67:4939-4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.May, T., R. I. Mackie, G. C. Fahey, Jr., J. C. Cremin, and K. A. Garleb. 1994. Effect of fiber source on short-chain fatty acid production and on the growth and toxin production by Clostridium difficile. Scand. J. Gastroenterol. 29:916-922. [DOI] [PubMed] [Google Scholar]
- 28.Microbial ID. 1992. Microbial identification system operational manual. Microbial ID Inc., Newark, Del.
- 29.Naaber, P., K. Klaus, E. Sepp, B. Bjorksten, and M. Mikelsaar. 1997. Colonization of infants and hospitalized patients with Clostridium difficile and lactobacilli. Clin. Infect. Dis. 25(Suppl. 2):S189-S190. [DOI] [PubMed]
- 30.Oli, M. W., B. W. Petschow, and R. K. Buddington. 1998. Evaluation of fructooligosaccharide supplementation of oral electrolyte solutions for treatment of diarrhea: recovery of the intestinal bacteria. Dig. Dis. Sci. 43:138-147. [DOI] [PubMed] [Google Scholar]
- 31.Roffe, C. 1996. Biotherapy for antibiotic-associated and other diarrhoeas. J. Infect. 32:1-10. [DOI] [PubMed] [Google Scholar]
- 32.Rolfe, R. D. 1984. Role of volatile fatty acids in colonization resistance to Clostridium difficile. Infect. Immun. 45:185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rolfe, R. D., S. Helebian, and S. M. Finegold. 1981. Bacterial interference between Clostridium difficile and normal fecal flora. J. Infect. Dis. 143:470-475. [DOI] [PubMed] [Google Scholar]
- 34.Rumney, C. J., and I. R. Rowland. 1992. In vivo and in vitro models of the human colonic flora. Crit. Rev. Food Sci. Nutr. 31:299-331. [DOI] [PubMed] [Google Scholar]
- 35.Russell, J. B., and F. Diez-Gonzalez. 1998. The effects of fermentation acids on bacterial growth. Adv. Microb. Physiol. 39:205-234. [DOI] [PubMed] [Google Scholar]
- 36.Sghir, A., J. M. Chow, and R. I. Mackie. 1998. Continuous culture selection of bifidobacteria and lactobacilli from human faecal samples using fructooligosaccharide as selective substrate. J. Appl. Microbiol. 85:769-777. [DOI] [PubMed] [Google Scholar]
- 37.Sghir, A., G. Gramet, A. Suau, V. Rochet, P. Pochart, and J. Dore. 2000. Quantification of bacterial groups within human fecal flora by oligonucleotide probe hybridization. Appl. Environ. Microbiol. 66:2263-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharp, R., S. Fishbain, and G. T. Macfarlane. 2001. Effect of short-chain carbohydrates on human intestinal bifidobacteria and Escherichia coli in vitro. J. Med. Microbiol. 50:152-160. [DOI] [PubMed] [Google Scholar]
- 39.Stark, P. L., and A. Lee. 1982. The microbial ecology of the large bowel of breast-fed and formula-fed infants during the first year of life. J. Med. Microbiol. 15:189-203. [DOI] [PubMed] [Google Scholar]
- 40.Tvede, M., and J. Rask-Madsen. 1989. Bacteriotherapy for chronic relapsing Clostridium difficile diarrhoea in six patients. Lancet i:1156-1160. [DOI] [PubMed]
- 41.Vanderhoof, J. A., and R. J. Young. 1998. Use of probiotics in childhood gastrointestinal disorders. J. Pediatr. Gastroenterol. Nutr. 27:323-332. [DOI] [PubMed] [Google Scholar]
- 42.van der Waaij, D. 1988. Evidence of immunoregulation of the composition of intestinal microflora and its practical consequences. Eur. J. Clin. Microbiol. Infect. Dis. 7:103-106. [DOI] [PubMed] [Google Scholar]
- 43.van der Waaij, D., J. M. Berghuis-de Vries, and L.-V. Lekkerkerk. 1971. Colonization resistance of the digestive tract in conventional and antibiotic-treated mice. J. Hyg. (London) 69:405-411. [DOI] [PMC free article] [PubMed]
- 44.Wilson, K. H. 1993. The microecology of Clostridium difficile. Clin. Infect. Dis. 16(Suppl. 4):S214-S218. [DOI] [PubMed]
- 45.Wilson, K. H., R. Blitchington, B. Hindenach, and R. C. Greene. 1988. Species-specific oligonucleotide probes for rRNA of Clostridium difficile and related species. J. Clin. Microbiol. 26:2484-2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilson, K. H., and R. Freter. 1986. Interaction of Clostridium difficile and Escherichia coli with microfloras in continuous-flow cultures and gnotobiotic mice. Infect. Immun. 54:354-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilson, K. H., and J. N. Sheagren. 1983. Antagonism of toxigenic Clostridium difficile by nontoxigenic C. difficile. J. Infect. Dis. 147:733-736. [DOI] [PubMed] [Google Scholar]
- 48.Wolf, B. W., J. A. Meulbroek, K. P. Jarvis, and K. A. Garleb. 1995. Effect of dietary supplementation with fructooligosaccharides on survival time in a hamster model of Clostridium difficile colitis, p. 23. In Proceedings of the First International Conference on the Molecular Genetics and Pathogenesis of the Clostridia. American Society for Microbiology, Washington, D.C.