Abstract
The deacetoxycephalosporin C synthase (DAOCS) from Streptomyces clavuligerus was engineered with the aim of enhancing the conversion of penicillin G into phenylacetyl-7-aminodeacetoxycephalosporanic acid, a precursor of 7-aminodeacetoxycephalosporanic acid, for industrial application. A single round of random mutagenesis followed by the screening of 5,500 clones identified three mutants, G79E, V275I, and C281Y, that showed a two- to sixfold increase in the kcat/Km ratio compared to the wild-type enzyme. Site-directed mutagenesis to modify residues surrounding the substrate resulted in three mutants, N304K, I305L, and I305M, with 6- to 14-fold-increased kcat/Km values. When mutants containing all possible combinations of these six sites were generated to optimize the ring expansion activity for penicillin G, the double mutant, YS67 (V275I, I305M), showed a significant 32-fold increase in the kcat/Km ratio and a 5-fold increase in relative activity for penicillin G, while the triple mutant, YS81 (V275I, C281Y, I305M), showed an even greater 13-fold increase in relative activity toward penicillin G. Our results demonstrate that this is a robust approach to the modification of DAOCS for an optimized DAOCS-penicillin G reaction.
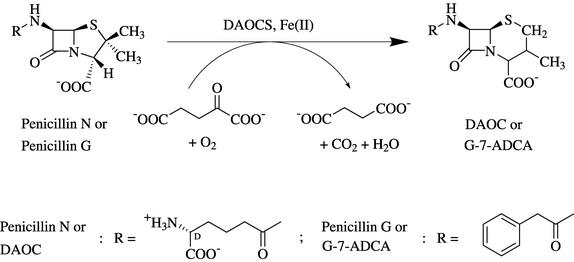
Cephalosporins are β-lactam antibiotic agents widely used for the clinical treatment of bacterial infection. Three pharmaceutically important oral cephalosporins, cephradine, cephalexin, and cephadroxil, are derived from 7-aminodeacetoxycephalosporanic acid (7-ADCA) by the addition of different side chains. The committed step in 7-ADCA synthesis in industry is the chemical expansion of the five-membered thiazolidine ring of penicillin G into the six-membered dihydrothiazine ring of phenylacetyl-7-ADCA (G-7-ADCA), followed by enzymatic side chain cleavage of G-7-ADCA. Given the strong negative environmental impact of the chemical process currently used for ring expansion, the development of an alternative enzymatic biosynthetic method for the production of the cephalosporin nucleus is vital. A lead enzyme, deacetoxycephalosporin C synthase (DAOCS or expandase), an Fe(II)- and α-ketoglutarate-dependent dioxygenase that catalyzes the expansion of penicillin N into deacetoxycephalosporin C (DAOC) (Fig. 1), has been chosen as a Green target in the industrial production of 7-ADCA. DAOCS is encoded by the cefE gene in the cephalosporin-producing bacteria, Streptomyces clavuligerus (21), Nocardia lactamdurans (9), and Lysobacter lactamgenus (20), and by the cefEF gene, encoding a bifunctional DAOCS/deacetylcephalosporin C synthase (or hydroxylase) activity, in the fungus, Cephalosporium acremonium (31). When the S. clavuligerus DAOCS gene was introduced into Penicillium chrysogenum, the recombinant fungus was able to expand adipoyl-6-APA into adipoyl-7-ADCA (10) or penicillin G into G-7-ADCA (5). The side chains of these cephalosporins can then be easily removed with a penicillin acylase to yield 7-ADCA (5, 10). In the wild-type C. acremonium strain, DAOC, expanded from penicillin N by DAOCS/expandase activity of the cefEF gene product, is subsequently hydroxylated to deacetylcephalosporin C (DAC) by DAC synthase/hydrolase activity of the same gene product. Therefore, in the C. acremonium ΔcefEF-cefE strain, the penicillin N could be expanded into DAOC by DAOCS (encoded by the introduced S. clavuligerus cefE), but DAOC could not be hydroxylated further. After purification of DAOC from the fermentation broth of the strain, the d-α-aminoadipic acid side chain of DAOC was then removed to generate 7-ADCA using a d-amino acid oxidase-glutaryl acylase enzymatic reaction (37).
FIG. 1.
Ring expansion of penicillin N into DAOC and penicillin G into G-7-ADCA catalyzed by DAOCS.
DAOCS was initially thought to have limited substrate activity in that it could convert penicillin N, but not other penicillin analogues, including penicillin G (13, 14, 29) (Fig. 1). However, the finding that resting cells and extracts of S. clavuligerus can expand the ring of penicillin G by a modified reaction condition (8) enabled further development in the process of biotransformation, since penicillin G not only is more stable than penicillin N but also is commercially available at a relatively low cost. Attempts have been made to increase the expansion activity towards penicillin G using resting cells grown in the presence of methanol or ethanol (16) or by further modifying the reaction conditions (2, 17, 18, 32). Moreover, penicillin G has been successfully expanded using immobilized S. clavuligerus cells (12) or immobilized DAOCS, purified from a transformant yeast expressing S. clavuligerus DAOCS (1), albeit with a low efficiency. On the other hand, modification of DAOCS for increased activity towards penicillin G has also been attempted after the characterization of S. clavuligerus DAOCS purified from an overexpression Escherichia coli transformant on ring expansion of penicillin G (28). The availability of the atomic structure of S. clavuligerus DAOCS and a proposed catalytic model (24, 28, 36) has facilitated a rational design approach to the engineering of DAOCS (6, 7, 23-27, 33, 34). However, despite enormous efforts, the mutated DAOCSs produced have only shown an approximately twofold increase in penicillin G conversion activity (6, 7, 24, 27).
The primary object of this study was therefore to engineer DAOCS to enhance the conversion rate in penicillin G ring expansion because of its importance in the pharmaceutical industry. In addition to the rational design approach, mutants were also generated by random mutagenesis, and a screening method was developed to select desirable mutants. Mutation sites giving an increased kcat/Km ratio were combined manually by site-directed mutagenesis to attempt to obtain mutants with even higher enzymatic activities. Mutants with a 32-fold-increased kcat/Km ratio and a 13-fold-increased activity compared to that of the wild-type (WT) enzyme were obtained.
MATERIALS AND METHODS
Materials.
Unless otherwise stated, all chemicals were purchased from Merck and Sigma. S. clavuligerus was obtained from the Culture Collection and Research Center. Strain E. coli ESS was kindly provided by Arnold L. Demain (Drew University, Madison, N.J.). Oligonucleotides were synthesized by Genset (Singapore Biotech). DNA sequencing was performed by Mission Biotech using an ABI PRISM dye terminator cycle sequencing kit and an ABI 377 DNA sequencer. Penicillin G was obtained from Harbin Pharmaceutical Co., and [14C]penicillin G was from Moravek. Penicillin N, DAOC, and G-7-ADCA were synthesized and characterized in this laboratory as described by Baldwin et al. (3, 4) and de Koning et al. (11). Electrospray ionization spectrometry was performed by Protech Laboratory. Other materials and suppliers were as follows: enzymes for DNA manipulations (Promega); DNA gel extraction kit, GFX micro plasmid prep kit, and fast-performance liquid chromatography apparatus and columns (Amersham Pharmacia Biotech); PCR clean up-M kit (Viogene); QuikChange site-directed mutagenesis kit (Stratagene); pET24a, pET30a, BL21(DE3), and Tuner(DE3) cells (Novagen); Bradford reagent, 30% acrylamide-bis-acrylamide solution, and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) apparatus (Bio-Rad); Pefabloc SC, isopropyl-β-d-thiogalactopyranoside (IPTG), lysozyme, and leupeptin (Roche); paper disks (Toyo Roshi Kaisha); penicillinase (Bacto Penase concentrate; Difco); UF 0.5 and 15 devices (nominal molecular weight limit, 5,000; Millipore); high-performance liquid chromatography (HPLC) set (LC-10AT, Sil-10AD, SPD-10A; Shimadzu); C18 column (250 ×4.6 mm, 5 μ; Hypersil); and HPLC data analysis software (Scientific Information Service Corporation).
Plasmid.
The cefE gene was cloned from S. clavuligerus using a Zero Blunt TOPO PCR cloning kit and inserted into the BamHI-HindIII of pET24a downstream of a T7 · Tag sequence to produce pYB4.
Random mutagenesis and screening.
The cefE gene was cut with BamHI and HindIII from plasmid pYB4 and treated at 65°C for 2 h with 0.8 M hydroxylamine in 0.1 M phosphate buffer (pH 6.0) containing 1 mM EDTA (35). The mutagen-treated DNA was cleaned up using a PCR clean up-M kit and then ligated into the BamHI-HindIII site of pET24a, and the ligation mixture was used to transform BL21(DE3) cells. Transformants selected on Luria-Bertani (LB) plates containing kanamycin at 50 μg/ml were grown in 96-well plates, in which each transformant was cultured in one well containing 56.7 μl of LB medium-50 mM 3-(N-morpholino)propanesulfonic acid (MOPS) buffer (pH 7.0) supplemented with kanamycin (50 μg/ml) for 20 h at 30°C. Then, 6.3 μl of IPTG (5 mM in LB medium/50 mM MOPS buffer [pH 7.0]) was added for 2 h to indirectly induce DAOCS expression, followed by the addition of 7 μl of lysozyme (100 mg/ml in 50 mM MOPS buffer [pH 6.5]) for 1 h to disrupt the cell. DAOCS activity was assayed for 1 h at 30°C after addition of 30 μl of 1 to 5 mM penicillin G in assay buffer A (50 mM MOPS [pH 6.5], 18 mM FeSO4, 40 mM ascorbate, and 25.6 mM α-ketoglutarate [16]). Paper disks (diameter, 8 mm) were loaded with 50 μl of the above assay mixture and then allowed to dry before being placed on LB plates containing 50,000 IU of penicillinase per ml seeded with E. coli ESS (a β-lactam-supersensitive mutant [8]). The plates were incubated overnight at 37°C, and clones with a clear zone larger than that of the WT control strain were selected and subjected to further screening.
For the second screening, the selected clones were grown at 30°C in 10 ml of LB medium containing kanamycin (50 μg/ml) to an A600 of 0.5 to 0.7, and then 0.5 mM IPTG was added for 2 h to indirectly induce DAOCS expression. The cells were then harvested, washed, suspended in 600 μl of 50 mM MOPS buffer (pH 6.5), disrupted by sonication (XL-2010; Misonix Inc.), and centrifuged at 14,000 × g for 20 min at 4°C, and the supernatants were collected as cell extracts. The protein concentration was determined using the Bradford reagent, and then a sample of cell extract containing 20 μg of protein was assayed for DAOCS activity using a thin-layer chromatography assay as follows: (i) incubation lasted for 1 h at 30°C in assay buffer B (50 mM MOPS [pH 6.5], 1.8 mM FeSO4, 4 mM ascorbate, 2.56 mM α-ketoglutarate, 0.1 mM [14C]penicillin G; final volume, 10 μl); (ii) the reaction was stopped by addition of 10 μl of ethanol; and (iii) 2.5 μl of the mixture was analyzed by spotting on a silica plate (60 F254), followed by development of the plate in a tank with a solvent system of chloroform:acetone:acetic acid (6:5:0.5, vol/vol) to separate [14C]penicillin G and [14C]G-7-ADCA. The plate was removed from the tank, allowed to dry in air, and subjected to autoradiography.
Mutant generation by site-directed mutagenesis.
A QuikChange site-directed mutagenesis kit was used to prepare mutated cefE genes using various mutagenic primers designed individually according to the manufacturer's manual, and then the mutated genes were cloned into the NdeI-HindIII site of pET30a and used to transform Tuner(DE3) cells. All mutations were confirmed by DNA sequencing. The mutated transformants were grown overnight at 30°C in 5 ml of LB medium containing kanamycin (50 μg/ml) and used to inoculate 150 ml of the same medium, and then the cells were grown for 1 h at 30°C. For expression of DAOCS, 0.1 mM IPTG was added to the cells for 4 h, and then the cells were harvested; washed; suspended in 5 ml of a solution containing 50 mM MOPS (pH 7.5), 1 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol (DTT), and leupeptin (0.5 μg/ml); disrupted by sonication (VCX 750; Sonics and Materials); and centrifuged at 15,000 × g for 15 min at 4°C. The supernatants were collected as cell extracts, and their protein concentrations were determined as described previously. The crude mutant enzymes were then assayed for DAOCS activity by HPLC assay as described below.
Purification of DAOCSs.
WT cell extract (2 ml) was applied to a HiTrap Q column (5 ml) preequilibrated with column buffer (50 mM MOPS [pH 7.5], 1 mM DTT, 0.4 mM Pefabloc SC, leupeptin [0.5 μg/ml]), and then the column was washed with 20 ml of column buffer, which was followed by a further wash with 25 ml of column buffer containing 120 mM NaCl before elution of DAOCS using 20 ml of column buffer containing 150 mM NaCl. Fractions shown by SDS-PAGE to contain DAOCS were pooled (10 ml), concentrated on a UF 15 device (Millipore), and further purified on a Hiload Superdex 75 (16 by 60 mm) column using column buffer at a flow rate of 1 ml/min. Fractions containing DAOCS were again collected and concentrated on a UF 15 device and then were immediately stored at −80°C in a solution of 1 mM DTT and bovine serum albumin (2 mg/ml). Mutated DAOCSs were purified using similar procedures.
DAOCS activity assayed by HPLC.
Cell extract (20 μl) was added to a standard assay reaction mixture to give a final volume of 200 μl and final concentrations of 50 mM MOPS (pH 7.5), 1 mM FeSO4, 4 mM ascorbate, 4 mM α-ketoglutarate, and 7 mM penicillin G. After 20 min at 30°C, 200 μl of ethanol was added, the sample was centrifuged at 15,000 × g for 5 min at 4°C, and then penicillin G and G-7-ADCA in the supernatants (40 μl) were separated by HPLC using a mobile phase of 25 mM potassium phosphate (pH 6.5) containing 19% (vol/vol) acetonitrile at a flow rate of 1 ml/min with detection at 215 nm. The retention times for G-7-ADCA and penicillin G were 8.5 and 12.5 min, respectively. The product at 8.5 min was confirmed by electrospray ionization spectrometry to have a mass of 332 kDa.
For kinetic assays, two different substrates, penicillin G and penicillin N, were used. For penicillin G, the assay procedures were identical to those described above except that 20 μg of purified DAOCS was used, the concentration of α-ketoglutarate was reduced to 1 mM, and the concentration of penicillin G varied from 0.1 to 10 mM. For penicillin N, 0.5 μg of purified DAOCS was incubated for 10 min at 30°C with various concentrations of penicillin N (15 to 100 μM) in 240 μl of assay mixture (50 mM HEPES [pH 7.5], 0.1 mM FeSO4, 0.4 mM ascorbate, 0.1 mM α-ketoglutarate), and then the reaction was stopped by addition of the same volume of cold 10 mM EDTA (pH 7.5) and the mixture was subjected to ultrafiltration on a UF 0.5 device. The filtrate was analyzed by HPLC using 25 mM potassium phosphate, pH 6.5, as the mobile phase, the retention times of DAOC and penicillin N being 12 min and 13.5 min, respectively. The kinetic parameters were calculated using a Hanes-Woolf plot, as described by Hanes (19).
RESULTS
Isolation of mutant DAOCS with improved penicillin G conversion generated by random mutagenesis.
In this study, a chemical mutagen, hydroxylamine, was first used to generate a mutant library for the screening of mutants with increased DAOCS activity using a simple paper disk-based visual screening method for ring expansion activity. Among approximately 5,500 clones, 85 were selected on the basis of the formation of larger clear zones than those produced by the WT control strain. These clones were subjected to a second screening using a thin-layer chromatography method that required only a small amount of crude enzyme due to the high radiosensitivity provided by the use of [14C]penicillin G. After this two-step screening, six mutants were found to have increased conversion activity. When their DNA was sequenced, three point mutations, G79E, V275I, and C281Y, were found. To eliminate interference from the T7 · Tag at the N terminus of the enzyme, the mutated cefE genes alone was further cloned into the NdeI-HindIII site of pET30a and the plasmids generated were used to transform cells of E. coli strain Tuner(DE3), a lacYZ deletion mutant of E. coli BL21(DE3) that allows more tightly controlled expression of DAOCS by IPTG than BL21(DE3) does. The resultant mutants, YS59 (G79E), YS5 (V275I), and YS53 (C281Y), were confirmed by DNA sequencing, and crude cell extracts assayed by HPLC were found to have higher ring expansion activity than the WT enzyme.
Isolation of DAOCS with improved penicillin G conversion by site-directed mutagenesis of residues near the substrate-binding site.
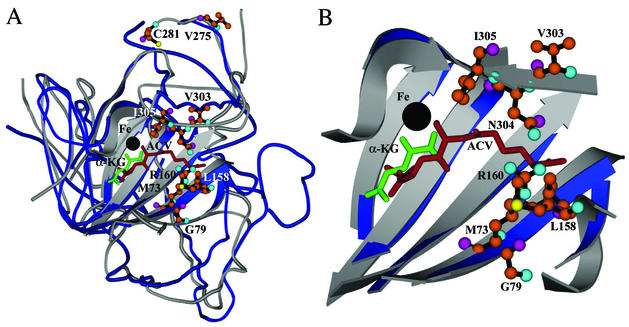
Although the structure of the DAOCS-Fe(II)-α-ketoglutarate-penicillin N complex is not available, that for the isopenicillin N synthase (IPNS)-Fe(II)-l-δ-(α-aminoadipoyl)-l-cysteinyl-d-valine (ACV) complex provides insights into the binding sites of DAOCS, since IPNS not only is a closely related enzyme but also shares a structurally conserved active center with DAOCS (30, 36). In a recent study (6), an approach involving the superimposition of the DAOCS and IPNS structures was used to identify DAOCS active-site residues that could be replaced by Leu in an attempt to generate a higher affinity for the hydrophobic substrate, penicillin G. The finding that the N304L mutant showed a substantial improvement in penicillin G conversion supports the feasibility of this rationally based approach. Using a similar approach, we identified DAOCS residues M73, L158, R160, V303, N304, and I305 surrounding the aminoadipoyl moiety of ACV (Fig. 2). Each of these six sites was changed first to an Ala residue and then to a positively charged residue (Lys), a negatively charged residue (Asp), a hydrophobic residue (Leu), and a sulfur-containing residue (Met) by site-directed mutagenesis. After confirming the substitution by sequencing, crude mutant enzymes were prepared for HPLC assay of penicillin G expansion activity as described in Materials and Methods. Using this method, three single mutants, YS12 (N304K), YS8 (I305L), and YS11 (I305M), with increased expansion activity were found.
FIG. 2.
(A) Superimposed structures of DAOCS and IPNS. DAOCS is shown in gray, and IPNS is shown in blue; ACV, the IPNS substrate, is shown in brown, and α-ketoglutarate, the DAOCS cosubstrate, is shown in green. The ferrous ions in IPNS and DAOCS are shown in black. The residues mutated in this study are shown in the ball-and-stick structure. (B) Close-up view of the catalytic site shown in panel A. This figure was prepared using MOLSCRIPT (22).
Purification of mutant DAOCSs and kinetic analysis.
The six DAOCS mutants with increased activity toward penicillin G produced by either random- or site-directed mutagenesis were purified by anion-exchange and gel filtration chromatography. Each of the purified DAOCSs was >90% pure as judged by SDS-PAGE analysis (data not shown). The kinetic parameters of the purified mutants and WT DAOCS toward penicillins G and N, determined by HPLC analysis, are shown in Tables 1 and 2, respectively.
TABLE 1.
Kinetic parameters for penicillin G conversion by WT and mutant DAOCSsa
| Strains | Amino acid substitution(s) | Km (mM) | kcat (s−1) | kcat/Km (M−1s−1) | Relative activity (%)b with penicillin G
|
|
|---|---|---|---|---|---|---|
| 1 mM | 10 mM | |||||
| YS16 | WT | 2.58 ± 0.13 | 0.0453 ± 0.0004 | 18 | 100 | 160 |
| YS59 | G79E | 0.75 ± 0.01 | 0.0315 ± 0.0001 | 42 | 90 | 130 |
| YS5 | V275I | 1.68 ± 0.11 | 0.0502 ± 0.0005 | 30 | 100 | 190 |
| YS53 | C281Y | 0.68 ± 0.20 | 0.0744 ± 0.0013 | 109 | 200 | 300 |
| YS12 | N304K | 0.22 ± 0.02 | 0.0564 ± 0.0001 | 256 | 220 | 240 |
| YS8 | I305L | 0.66 ± 0.04 | 0.0759 ± 0.0006 | 115 | 230 | 310 |
| YS11 | I305M | 0.75 ± 0.02 | 0.1452 ± 0.0008 | 194 | 380 | 580 |
| YS67 | V275I, I305M | 0.25 ± 0.05 | 0.1458 ± 0.0014 | 583 | 500 | 610 |
| YS81 | V275I, C281Y, I305M | NDc | ND | ND | 1,290 | 690 |
| YS96 | G79E, V275I, I305M | ND | ND | ND | 1,110 | 540 |
| YS94 | C281Y, N304K, I305M | ND | ND | ND | 650 | 230 |
| YS100 | G79E, V275I, I305L | ND | ND | ND | 470 | 240 |
| YS88 | G79E, V275I, C281Y | ND | ND | ND | 430 | 200 |
| YS74 | V275I, N304K, I305L | ND | ND | ND | 300 | 130 |
| YS76 | G79E, V275I, C281Y, I305L | ND | ND | ND | 250 | 90 |
All of the DAOCSs listed here were purified as described in Materials and Methods. All kinetic parameters are shown as the mean ± standard error of three independent experiments.
The relative activity of each mutant DAOCS was determined using 1 and 10 mM penicillin G, with the activity of the WT enzyme using 1 mM penicillin G being set as 100%.
ND, not determined.
TABLE 2.
Kinetic parameters for penicillin N conversion by WT and mutant DAOCSsa
| Strains | Amino acid substitution(s) | Km (mM) | kcat (s−1) | kcat/Km (M−1s−1) |
|---|---|---|---|---|
| YS16 | WT | 0.014 ± 0.003 | 0.307 ± 0.019 | 22,000 |
| YS59 | G79E | 0.009 ± 0.002 | 0.178 ± 0.010 | 20,000 |
| YS5 | V275I | 0.012 ± 0.002 | 0.252 ± 0.010 | 20,000 |
| YS53 | C281Y | 0.006 ± 0.0005 | 0.273 ± 0.007 | 47,000 |
| YS12 | N304K | 0.004 ± 0.0005 | 0.366 ± 0.022 | 92,000 |
| YS8 | I305L | 0.006 ± 0.001 | 0.284 ± 0.015 | 44,000 |
| YS11 | I305M | 0.012 ± 0.0005 | 0.310 ± 0.002 | 26,000 |
| YS67 | V275I, I305M | 0.013 ± 0.002 | 0.316 ± 0.016 | 24,000 |
All of the DAOCSs listed here were purified as described in Materials and Methods. All kinetic parameters are shown as the mean ± standard error of three independent experiments.
In the mutants generated by random mutagenesis, substitution by a negatively charged side chain, as in G79E (YS59), increased the binding of penicillin G and penicillin N by three and twofold, respectively, but reduced the rate of oxidation to 70% for penicillin G and to 60% for penicillin N. In V275I (YS5), the increase in hydrophobicity due to the Ile residue improved both the binding of penicillin G (2-fold) and its rate of conversion (1.1-fold) but had little effect on the activity toward penicillin N. Another mutant, C281Y (YS53), showed increased binding of penicillin G and penicillin N (4- and 2-fold, respectively) and an increased rate of conversion of penicillin G (1.6-fold), with little effect on penicillin N conversion rate.
The three mutants selected using the rationally based approach showed an even greater improvement. N304K (YS12) showed not only significantly increased binding of penicillin G and penicillin N (12-fold and 4-fold, respectively), but also a slight increase in the rate of conversion of both substrates (1.2-fold). Replacement of Ile305 with Leu (YS8) resulted in a 4-fold increase in penicillin G binding activity and a 1.7-fold increase in ring expansion activity and moderately improved binding of penicillin N (2-fold), with little effect on the rate of ring expansion. The other mutant, I305M (YS11), showed a marked improvement in both the Km (3-fold) and kcat (3.2-fold) toward penicillin G but little difference in activity towards penicillin N. The two mutants with the highest kcat/Km ratio for penicillin G were both active-site mutations, with N304K showing a 14-fold increase and I305M showing an 11-fold increase.
Construction of combined mutants.
In order to see whether these six mutants could have an additive effect on activity, mutants containing all 41 possible combinations of these substitutions (two to five substitutions) were generated manually by site-directed mutagenesis; then, after confirmation of the mutations by DNA sequencing, crude extracts of the various mutants were assayed for ring expansion activity on penicillin G using the HPLC method. Of the 41 mutants generated, 8 (shown in Table 1) were found to have a higher conversion rate for penicillin G. These eight mutants were further purified for kinetic analysis, but because of substrate inhibition, it was only possible to determine the kcat and Km for YS67 (V275I, I305M). As shown in Table 1, this mutant had a very low apparent Km (0.25 mM) for penicillin G, compared to the values of 1.68, 0.75, and 2.58 mM for mutants V275I and I305M and the WT enzyme, respectively, indicating that the double mutation increased the penicillin G binding activity. The kcat of YS67 for penicillin G was similar to that of YS11 (I305M), with a 3.2-fold increase compared to the WT enzyme. As a result, YS67 showed an ∼32-fold increase in the kcat/Km ratio, compared to an ∼2-fold increase for V275I and an ∼11-fold increase for I305M, suggesting a cooperative effect of these two point mutations. No significant change in kinetic parameters for the natural substrate, penicillin N, was found, but this is not surprising, since neither V275I nor I305M alone showed any significant change in activity towards penicillin N.
The activities of the eight mutants were determined using 1 and 10 mM penicillin G and expressed as relative activities compared to the activity of the WT using 1 mM penicillin G (Table 1). Using 1 mM penicillin G in the assay mixture, all had much higher activities than the WT enzyme, with mutant YS81 (V275I, C281Y, I305M) showing an interesting increase of about 13-fold. Although this mutant only showed a sevenfold increase in the presence of 10 mM penicillin G, indicating the effect of substrate inhibition, it still showed the highest activity of all mutants tested.
DISCUSSION
In this study, we used a random mutagenesis approach to engineer DAOCS for enhanced activity toward penicillin G. A disk-based screening method using a two-step screening procedure was developed which proved to be very efficient because of its high sensitivity and simplicity. Three mutants, G79E (YS59), V275I (YS5), and C281Y (YS53), with a two- to sixfold-increased kcat/Km ratio were obtained from 5,500 clones (Table 1). The lower Km values for these three mutants, particularly when penicillin G was used as a substrate, suggest that they have a better binding affinity. Based on the crystal structure of DAOCS (28, 36), these residues are distant from the binding pocket, with residue G79 lying on a β strand near the edge of the central sheet and residues V275 and C281 in a helix connecting to the C-terminal lid (Fig. 2). Since they are unlikely to be directly involved in enzymatic catalysis, the observed increased binding affinity may be due to a global effect of these mutations on the active-site conformation. The C-terminal lid has been proposed to play a role in binding, selecting, and/or orienting the penicillin substrate during catalysis (7, 24, 27, 28). Substitution of V275I or C281Y may thus indirectly affect the conformation of the nearby C-terminal lid, thereby resulting in a better conformation for binding penicillin G. In addition, C281Y may also play a role in the affinity binding of penicillin N (2-fold increase) and oxidation of penicillin G (1.6-fold increase). On the other hand, although G79E (YS59) had more than two times kcat/Km compared to the WT DAOCS, it also had the lowest relative activity among the enzymes tested, which should be due to its lowest kcat. This phenomenon seems controversial to the fact that YS59 was selected from enhanced activity in cell extract. However, it is possible that this mutant had more expression so that its cell extract was more active than the WT.
We also directly engineered the active-site pocket of DAOCS by site-directed mutagenesis to search for mutants with high activity towards the hydrophobic penicillin substrate, penicillin G. In a previous study (6) in which the hydrophilic residues R74, R160, R266, and N304, surrounding the presumed substrate in DAOCS were replaced with Leu to enhance hydrophobic enzyme-substrate interactions, only N304L showed increased (1.2-fold) transformation of penicillin G. In a recent study, substitution of the same residue, Asn304, with Ala resulted in increased kcat values for both penicillin N (1.6-fold) and penicillin G (1.8-fold), while the Km was unchanged (penicillin N) or showed a 2-fold increase (penicillin G) (26). The importance of this residue was confirmed in the present study, since our N304L mutant was 1.4-fold more active using 1 mM penicillin G than the WT enzyme (data not shown). In addition to testing these hydrophilic-to-hydrophobic substitutions, we also tested the effect of replacing these residues with a positively charged residue, a negatively charged residue, a hydrophobic residue, or a sulfur-containing residue. A striking finding of this work was that we were able to generate a remarkable point mutant, N304K (YS12), with significantly increased binding affinities for both penicillin G (12-fold increase) and penicillin N (4-fold increase) and a slight increase in the conversion rates for both substrates (1.2-fold) compared to the WT enzyme. These results support the idea that residue 304 plays an important role in both substrate binding and catalytic activity (6, 26). However N304K had significantly improved activities for both penicillin G and penicillin N, whereas N304A (26) and N304L (6) did not. In addition to N304, other point mutations at the nearby residue 305 also showed improved activity. I305L (YS8) and I305M (YS11) showed a 3- to 4-fold increased binding affinity for penicillin G (Table 1), but only the former had a 2-fold increased binding affinity for penicillin N (Table 2), while, for penicillin G, I305M showed the greatest increase in the kcat (3.2-fold), whereas I305L showed only a 1.7-fold increase. Thus, residue 305 may play a critical role both in substrate binding for proper catalysis and in modulating penicillin G ring expansion activity. Taken together, these results suggest that modification of the C-terminal residues surrounding the binding pocket of DAOCS, particularly residues 304 and 305, is a feasible and robust approach to optimizing the desired enzyme-substrate reaction.
In an attempt to obtain DAOCSs that were even more efficient in the conversion of penicillin G, mutants containing all 41 possible combinations of these substitutions (two- to five substitutions per mutant) were generated using site-directed mutagenesis and the double mutant, YS67 (275I, I305M), was found to show a great improvement (32-fold increase) in the kcat/Km ratio (Table 1). Using 1 mM penicillin G in the assay mixture, the activity of YS67 was 5-fold greater than that of the WT enzyme. Of the 41 mutants, the triple-substituted mutants, YS81 (V275I, C281Y, I305M) and YS96 (G79E, V275I, I305M), gave the greatest improvement, with a marked 13-fold increase with YS81 and an 11-fold increase with YS96 (Table 1). When the concentration of penicillin G was increased from 1 to 10 mM, although YS81 was still the most active mutant, YS81 and YS96 showed reduced activities (Table 1). In addition, YS81 was more active when 0.2 mM penicillin G was used than when 1 mM penicillin G was used (data not shown), indicating a severe substrate inhibition effect. Similar substrate inhibition was found for all other combined mutants, except YS67 (Table 1). This substrate inhibition might be due to the tight binding of penicillin G to, and/or slow release of G-7-ADCA from, the active site. Recently, the expansion activity of DAOCS toward penicillin G was also found to be reduced if the amount of α-ketoglutarate, a cosubstrate of DAOCS, was more than 0.2 mM (15). But we had also titrated the α-ketoglutarate amount ranging from 0.5 to 2 mM, with the penicillin G concentration fixed at 0.5 mM in the standard assay, and found that the optimal concentration of α-ketoglutarate was 1 mM. Thus, the substrate inhibition may not be due to the concentration of α-ketoglutarate being higher than 0.2 mM. None of the other combinations, including YS94, YS100, YS88, YS74, and YS76 (Table 1), showed better activity than YS81 and YS96.
In conclusion, S. clavuligerus DAOCS can be modified for higher catalytic activity towards an unnatural substrate, penicillin G. Both random mutagenesis and rational design approaches have been successfully used to produce point mutants with higher activities towards penicillin G. Moreover, several mutations can be combined to produce even more-active mutants, with the double mutant, YS67 (V275I, I305M), and the triple mutant, YS81 (V275I, C281Y, I305M), showing, respectively, a significant 5-fold and a marked 13-fold increase in relative activity towards penicillin G.
Acknowledgments
We thank Arnold L. Demain for kindly providing E. coli strain ESS and Jai-Shin Liu for the preparation of Fig. 2.
This work was supported by grants from the Ministry of Economic Affairs of the Republic of China (90J027), by an intramural fund from the National Yang-Ming University of the Republic of China, and by Synmax Biochemical Co. Ltd.
REFERENCES
- 1.Adrio, J. L., J. Velasco, G. Soler, M. Rodriguez Saiz, J. L. Barredo, and M. A. Moreno. 2001. Extracellular production of biologically active deacetoxycephalosporin C synthase from Streptomyces clavuligerus in Pichia pastoris. Biotechnol. Bioeng. 75:485-491. [DOI] [PubMed] [Google Scholar]
- 2.Baez-Vasquez, M. A., J. L. Adrio, J. M. Piret, and A. L. Demain. 1999. Further studies on the bioconversion of penicillin G into deacetoxycephalosporin G by resting cells of Streptomyces clavuligerus NP-1. Appl. Biochem. Biotechnol. 81:145-152. [DOI] [PubMed] [Google Scholar]
- 3.Baldwin, J. E., R. M. J. Crabbe, G. Knight, T. Nomoto, C. J. Schofield, and H.-H. Ting. 1987. The enzymatic ring expansion of penicillins to cephalosporins: site chain specificity. Tetrahedron. 43:3009-3014. [Google Scholar]
- 4.Baldwin, J. E., S. R. Herchen, and P. D. Singh. 1980. Syntheses of penicillin N, [6α-3H]penicillin N and [10-14C,6α-3H]penicillin N. Biochem J. 186:881-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bovenberg, R. A. L., B. P. Koekman, D. Schipper, and A. W. Vollebregt. March 1998. Process for the production of 7-ADCA via expandase activity on penicillin G. U.S. patent 5,731,165.
- 6.Chin, H. S., J. Sim, and T. S. Sim. 2001. Mutation of N304 to leucine in Streptomyces clavuligerus deacetoxycephalosporin C synthase creates an enzyme with increased penicillin analogue conversion. Biochem. Biophys. Res. Commun. 287:507-513. [DOI] [PubMed] [Google Scholar]
- 7.Chin, H. S., and T. S. Sim. 2002. C-terminus modification of Streptomyces clavuligerus deacetoxycephalosporin C synthase improves catalysis with an expanded substrate specificity. Biochem. Biophys. Res. Commun. 295:55-61. [DOI] [PubMed] [Google Scholar]
- 8.Cho, H., J. L. Adrio, J. M. Luengo, S. Wolfe, S. Ocran, G. Hintermann, J. M. Piret, and A. L. Demain. 1998. Elucidation of conditions allowing conversion of penicillin G and other penicillins to deacetoxycephalosporins by resting cells and extracts of Streptomyces clavuligerus NP1. Proc. Natl. Acad. Sci. USA 95:11544-11548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coque, J. J., J. F. Martin, and P. Liras. 1993. Characterization and expression in Streptomyces lividans of cefD and cefE genes from Nocardia lactamdurans: the organization of the cephamycin gene cluster differs from that in Streptomyces clavuligerus. Mol. Gen. Genet. 236:453-458. [DOI] [PubMed] [Google Scholar]
- 10.Crawford, L., A. M. Stepan, P. C. McAda, J. A. Rambosek, M. J. Conder, V. A. Vinci, and C. D. Reeves. 1995. Production of cephalosporin intermediates by feeding adipic acid to recombinant Penicillium chrysogenum strains expressing ring expansion activity. Bio/Technology 13:58-62. [DOI] [PubMed] [Google Scholar]
- 11.de Koning, J. J., H. J. Kooreman, H. S. Tan, and J. Verwij. 1975. One-step, high yield conversion of penicillin sulfoxides to deacetoxycephalosporins. J. Org. Chem. 40:1346-1347. [DOI] [PubMed] [Google Scholar]
- 12.Demain, A. L., and M. A. Baez-Vasquez. 2000. Immobilized Streptomyces clavuligerus NP1 cells for biotransformation of penicillin G into deacetoxycephalosporin G. Appl. Biochem. Biotechnol. 87:135-140. [DOI] [PubMed] [Google Scholar]
- 13.Dotzlaf, J. E., and W.-K. Yeh. 1987. Copurification and characterization of deacetoxycephalosporin C synthetase/hydroxylase from Cephalosporium acremonium. J. Bacteriol. 169:1611-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dotzlaf, J. E., and W.-K. Yeh. 1989. Purification and properties of deacetoxycephalosporin C synthase from recombinant Escherichia coli and its comparison with the native enzyme purified from Streptomyces clavuligerus. J. Biol. Chem. 264:10219-10227. [PubMed] [Google Scholar]
- 15.Dubus, A., M. D. Lloyd, H.-J. Lee, C. J. Schofield, J. E. Baldwin, and J. M. Frere. 2001. Probing the penicillin sidechain selectivity of recombinant deacetoxycephalosporin C synthase. Cell. Mol. Life Sci. 58:835-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernandez, M.-J., J. L. Adrio, J. M. Piret, S. Wolfe, S. Ro, and A. L. Demain. 1999. Stimulatory effect of growth in the presence of alcohols on biotransformation of penicillin G into cephalosporin-type antibiotics by resting cells of Streptomyces clavuligerus NP1. Appl. Microbiol. Biotechnol. 52:484-488. [DOI] [PubMed] [Google Scholar]
- 17.Gao, Q., and A. L. Demain. 2001. Improvement in the bioconversion of penicillin G to deacetoxycephalosporin G by elimination of agitation and addition of decane. Appl. Microbiol. Biotechnol. 57:511-513. [DOI] [PubMed] [Google Scholar]
- 18.Gao, Q., and A. L. Demain. 2002. Improvement in the resting-cell bioconversion of penicillin G to deacetoxycephalosporin G by addition of catalase. Lett. Appl. Microbiol. 34:290-292. [DOI] [PubMed] [Google Scholar]
- 19.Hanes, C. S. 1932. CLXVII. Studies on plant amylases. I. The effect of starch concentration upon the velocity of hydrolysis by the amylase of germinated barley. Biochem. J. 26:1406-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kimura, H., H. Miyashita, and Y. Sumino. 1996. Organization and expression in Pseudomonas putida of the gene cluster involved in cephalosporin biosynthesis from Lysobacter lactamgenus YK90. Appl. Microbiol. Biotechnol. 45:490-501. [DOI] [PubMed] [Google Scholar]
- 21.Kovacevic, S., B. J. Weigel, M. B. Tobin, T. D. Ingolia, and J. R. Miller. 1989. Cloning, characterization, and expression in Escherichia coli of the Streptomyces clavuligerus gene encoding deacetoxycephalosporin C synthetase. J. Bacteriol. 171:754-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kraulis, P. J. 1991. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24:946-950. [Google Scholar]
- 23.Lee, H.-J., M. D. Lloyd, I. J. Clifton, K. Harlos, A. Dubus, J. E. Baldwin, J.-M. Frere, and C. J. Schofield. 2001. Alteration of the co-substrate selectivity of deacetoxycephalosporin C synthase. The role of arginine 258. J. Biol. Chem. 276:18290-18295. [DOI] [PubMed] [Google Scholar]
- 24.Lee, H.-J., M. D. Lloyd, K. Harlos, I. J. Clifton, J. E. Baldwin, and C. J. Schofield. 2001. Kinetic and crystallographic studies on deacetoxycephalosporin C synthase (DAOCS). J. Mol. Biol. 308:937-948. [DOI] [PubMed] [Google Scholar]
- 25.Lee, H.-J., M. D. Lloyd, K. Harlos, and C. J. Schofield. 2000. The effect of cysteine mutations on recombinant deacetoxycephalosporin C synthase from S. clavuligerus. Biochem. Biophys. Res. Commun. 267:445-448. [DOI] [PubMed] [Google Scholar]
- 26.Lee, H.-J., C. J. Schofield, and M. D. Lloyd. 2002. Active site mutations of recombinant deacetoxycephalosporin C synthase. Biochem. Biophys. Res. Commun. 292:66-70. [DOI] [PubMed] [Google Scholar]
- 27.Lipscomb, S. J., H.-J. Lee, M. Mukherji, J. E. Baldwin, C. J. Schofield, and M. D. Lloyd. 2002. The role of arginine residues in substrate binding and catalysis by deacetoxycephalosporin C synthase. Eur. J. Biochem. 269:2735-2739. [DOI] [PubMed] [Google Scholar]
- 28.Lloyd, M. D., H.-J. Lee, K. Harlos, Z.-H. Zhang, J. E. Baldwin, C. J. Schofield, J. M. Charnock, C. D. Garner, T. Hara, A. C. Terwisscha van Scheltinga, K. Valegard, J. A. C. Viklund, J. Hajdu, I. Andersson, A. Danielsson, and R. Bhikhabhai. 1999. Studies on the active site of deacetoxycephalosporin C synthase. J. Mol. Biol. 287:943-960. [DOI] [PubMed] [Google Scholar]
- 29.Maeda, K., J. M. Luengo, O. Ferrero, S. Wolfe, M. Y. Lebedev, A. Fang, and A. L. Demain. 1995. The substrate specificity of deacetoxycephalosporin C synthetase (“expandase”) of Streptomyces clavuligerus is extremely narrow. Enzyme Microb. Technol. 17:231-234. [Google Scholar]
- 30.Roach, P. L., I. J. Clifton, C. M. H. Hensgens, N. Shibata, C. J. Schofield, J. Hajdu, and J. E. Baldwin. 1997. Structure of isopenicillin N synthase complexed with substrate and the mechanism of penicillin formation. Nature 387:827-830. [DOI] [PubMed] [Google Scholar]
- 31.Samson, S. M., J. E. Dotzlaf, M. L. Slisz, G. W. Becker, R. M. van Frank, L. E. Veal, W.-K. Yeh, J. R. Miller, S. W. Queener, and T. D. Ingolia. 1987. Cloning and expression of the fungal expandase/hydroxylase gene involved in cephalosporin biosynthesis. Bio/Techniques 5:1207-1214. [Google Scholar]
- 32.Sim, J., and T.-S. Sim. 2001. In vitro conversion of penicillin G and ampicillin by recombinant Streptomyces clavuligerus NRRL 3585 deacetoxycephalosporin C synthase. Enzyme Microbiol. Technol. 29:240-245. [Google Scholar]
- 33.Sim, J., and T.-S. Sim. 2000. Mutational evidence supporting the involvement of tripartite residues His183, Asp185, and His243 in Streptomyces clavuligerus deacetoxycephalosporin C synthase for catalysis. Biosci. Biotechnol. Biochem. 64:828-832. [DOI] [PubMed] [Google Scholar]
- 34.Sutherland, J. D., R. A. L. Bovenberg, and J. M. van der Laan. July 1999. Process for the production of SSC's via expandase activity on penicillin G. U.S. patent 5,919,680.
- 35.Tange, T., S. Taguchi, S. Kojima, K. Miura, and H. Momose. 1994. Improvement of a useful enzyme (subtilisin BPN′) by an experimental evolution system. Appl. Microbiol. Biotechnol. 41:239-244. [DOI] [PubMed] [Google Scholar]
- 36.Valegard, K., A. C. Terwisscha van Scheltinga, M. D. Lloyd, T. Hara, S. Ramaswamy, A. Perrakis, A. Thompson, H.-J. Lee, J. E. Baldwin, C. J. Schofield, J. Hajdu, and I. Andersson. 1998. Structure of a cephalosporin synthase. Nature 394:805-809. [DOI] [PubMed] [Google Scholar]
- 37.Velasco, J., J. L. Adrio, M. A. Moreno, B. Diez, G. Soler, and J. L. Barredo. 2000. Environmentally safe production of 7-aminodeacetoxycephalosporanic acid (7-ADCA) using recombinant strains of Acremonium chrysogenum. Nat. Biotechnol. 18:857-861. [DOI] [PubMed] [Google Scholar]