Abstract
Several distinct naphthalene dioxygenases have been characterized to date, which provides the opportunity to investigate the ecological significance, relative distribution, and transmission modes of the different analogs. In this study, we showed that a group of naphthalene-degrading isolates from a polycyclic aromatic hydrocarbon (PAH)-contaminated hillside soil were phenotypically and genotypically distinct from naphthalene-degrading organisms isolated from adjacent, more highly contaminated seep sediments. Mineralization of 14C-labeled naphthalene by soil slurries suggested that the in situ seep community was more acclimated to PAHs than was the in situ hillside community. phnAc-like genes were present in diverse naphthalene-degrading isolates cultured from the hillside soil, while nahAc-like genes were found only among isolates cultured from the seep sediments. The presence of a highly conserved nahAc allele among gram-negative isolates from the coal tar-contaminated seep area provided evidence for in situ horizontal gene transfer and was reported previously (J. B. Herrick, K. G. Stuart-Keil, W. C. Ghiorse, and E. L. Madsen, Appl. Environ. Microbiol. 63:2330-2337, 1997). Natural horizontal transfer of the phnAc sequence was also suggested by a comparison of the phnAc and 16S ribosomal DNA sequences of the hillside isolates. Analysis of metabolites produced by cell suspensions and patterns of amplicons produced by PCR analysis suggested both genetic and metabolic diversity among the naphthalene-degrading isolates of the contaminated hillside. These results provide new insights into the distribution, diversity, and transfer of phnAc alleles and increase our understanding of the acclimation of microbial communities to pollutants.
The biochemical, enzymatic, and genetic details of microbial naphthalene degradation have been examined extensively since the early description in 1964 of a naphthalene metabolic pathway in Pseudomonas (9). Studies of naphthalene degradation are significant for at least four reasons: (i) naphthalene's aromatic character poses unique biochemical challenges for enzymatic attack (24, 67); (ii) naphthalene is a common pollutant that serves as a chemical model for the degradation of polycyclic aromatic hydrocarbons (PAHs) (56), which are often carcinogenic (49); (iii) insights are provided into the behavior of catabolic plasmids, the lateral transfer of genetic information among bacteria, and the evolution of oxygenase genes and enzymes (19, 22, 51, 59, 60, 63); and (iv) our abilities to effectively manage and treat polluted environments and to engineer novel enzymes for new technologies are increased (15, 48).
In all pure cultures of naphthalene-mineralizing bacteria that have been examined, the aerobic metabolism of naphthalene is initiated by a multicomponent enzyme system called naphthalene dioxygenase (NDO) (EC 1.14.12.12) (56). The NDO of Pseudomonas putida NCIB 9816-4 has been studied comprehensively and shown to be a three-component dioxygenase. Electrons from NAD(P)H are transferred via reductase (encoded by nahAa) and ferredoxin (encoded by nahAb) components to the α3β3 catalytic oxygenase. The large subunits of this oxygenase (encoded by nahAc) each contain a Rieske [2Fe-2S] center and mononuclear nonheme iron (26) and are thought to confer substrate specificity (46). The ability to degrade naphthalene has been demonstrated for a wide range of bacterial genera, and a number of distinct NDO large subunits have been characterized and shown to have various degrees of nucleotide and amino acid sequence similarities to the canonical genes from Pseudomonas spp. (1, 5, 10, 12, 13, 17, 20, 27, 28, 31, 32, 42, 53, 57, 58).
The characterization of these distinct NDO analogs allows researchers to address questions concerning the ecological significance, relative distribution, and transmission of the different dioxygenases. A relevant example is phnAc, which was first described in 1999 when Burkholderia sp. strain RP007 was isolated from a PAH-contaminated site in New Zealand by using phenanthrene as a substrate (32). The phnAc gene was subsequently amplified from DNA extracted from pristine and contaminated soils in New Zealand, but it was not detected in a group of PAH-degrading bacteria isolated from those same soils (34), which suggested that it may be difficult to culture the phnAc-carrying organisms. Quantitative studies using competitive PCR showed that phnAc was present at equivalent or greater concentrations than was nahAc in contaminated New Zealand soils and that the addition of naphthalene to a pristine soil enriched it for phnAc but not for nahAc (33). In that study, phnAc was also shown to be present in pristine soils from Siberia, the Antarctic, and New Zealand. Subsequently, a group of PAH-degrading bacteria isolated from pristine soils in Japan and petroleum-contaminated soil in Indonesia were shown to contain phnAc alleles (62).
In this study, our objective was to compare and contrast, at the organism and gene levels, naphthalene-degrading bacterial guilds from adjacent but physically distinct areas of the same field site in upstate New York. Surprisingly, we found that culturable naphthalene-degrading bacteria from the adjacent sites were phenotypically and taxonomically distinct and that the nahAc allele was distributed only among isolates from the highly contaminated seep sediments while the phnAc allele was distributed only among taxonomically divergent hosts isolated from the less-contaminated hillside soil.
MATERIALS AND METHODS
Site description, sampling, and sample characterization.
Samples were obtained from a coal tar-contaminated area located in South Glens Falls in upstate New York. This site, known as site 24, has been studied intensively, and details, including the site history and sample characteristics, have been published previously (2, 22, 35, 65, 66). Sampling was done at four locations near the seep area, which was located at the foot of a hillside approximately 400 m from the original coal tar deposit. The seep sample was taken aseptically from saturated naphthalene-contaminated sediments immediately within the contaminated portion of the seep. The hillside soil sample was obtained within 3 m of the seep on an adjacent hillside down gradient from the contaminated source that was approximately 1.5 m higher in elevation than the area where the seep sample was taken. Vegetation on the hillside included pine trees. The upstream sample was obtained from saturated sediments 25 m upstream in a northerly direction from the seep in a shallow drainage that emptied into the seep area. The adjacent sample was taken from saturated sediments in a drainage that was expected to contain no or low contamination and was parallel to, and 50 m east of, the seep. Sediment samples were obtained aseptically 10 cm below the surface, while the hillside soil sample was taken at a depth of 2 cm. All samples (25 to 30 g [wet weight]) were placed on ice in the field and maintained at 4°C until subsampling on the following day for mineralization, plating, and characterization.
Sample characterization, strain isolation, and reference cultures.
All chemicals and reagents were of the highest available quality and were purchased from Sigma Chemical Co. (St. Louis, Mo.) unless otherwise noted. Soil and sediment samples were tested for mineralization of 14C-labeled naphthalene in sealed flasks as previously described (35). Preparation and counting of total bacteria by acridine orange fluorescent-direct counting were also done as described previously (41) but with results reported as the means for two smears, 15 fields counted per smear. For dilution plating, 10 g of each sample was suspended in 90 ml of 0.1% sodium pyrophosphate (pH 7.0), further diluted in a 10-fold series in phosphate-buffered solution (0.07% K2HPO4, 0.03% KH2PO4 [pH 7.0]), plated in three replicates per dilution on Difco (Detroit, Mich.) R2A agar medium (for total counts) and on Stanier's mineral salts medium (54) solidified with 1.5% Noble agar (Difco), and incubated in the presence of naphthalene vapor. For each sample, 20 to 30 colonies with diameters greater than 1 mm were selected randomly from replicate naphthalene dilution plates that had between 20 and 200 well-defined colonies. Presumptive growth on naphthalene as a sole carbon source was confirmed by plating on mineral salts medium with and without naphthalene vapor. Isolation and characterization of strains were done as previously described (22).
P. putida G7 was a gift of G. Sayler, University of Tennessee. P. putida NCIB 9816-4 and Comamonas testosteroni GZ42 were gifts from G. Zylstra, Rutgers University.
PCR assays.
Genomic DNA was obtained from bacterial isolates as previously reported (64). The presence of an amplifiable NDO large-subunit gene was determined by using PCR assays with a range of different PCR primers, primer combinations, and protocols. These included primers targeting nahAc from P. putida G7 (primers nahAc1 and nahAc3 andprotocol as described previously [21]); degenerate primers targeting a range of NDO sequences, including nahAc from P. putida G7 and nahAc1 from C. testosteroni GZ42 (forward primers Ab248F, Ac114F, and Ac307F, reverse primers Ac596R, Ac893R, and Ac1095R, and protocol as described previously [65]); and primers targeting phnAc of Burkholderia sp. strain RP007 (forward primers 6897F and 8073F, reverse primers 8420R and 9047R, and protocol as described previously [32] except that the temperature cycling program was modified to 35 cycles of 94°C for 30 s, 52°C for 30 s, and 72°C for 60 s). As positive controls, PCR amplifications using 16S rRNA primers 16SP-5 and 16SP-3 (21) were performed with each isolate. Amplification of the 16S rRNA genes from the hillside isolates was performed as previously described by a modification of the method described by Herrick et al. (22), using the eubacterial primers 27f and 1492r (30). PCR amplification was performed with a 50-μl volume with a primer concentration of 1 μM and a cycling regime of 94°C for 5 min (1 cycle); 94°C for 1 min, 56°C for 45 s, and 72°C for 45 s (35 cycles); and 72°C for 10 min (1 cycle). The amplicons were analyzed by electrophoresis on a 1% agarose gel and ligated into the pCR2.1 vector (Invitrogen, Carlsbad, Calif.). The constructed plasmids were introduced into Escherichia coli INV-F competent cells (One Shot; Invitrogen), and sequencing of the resulting white clones was completed on an ABI Prism 377XL instrument (Applied Biosystem Instruments, Foster, Calif.) by using M13 forward and/or reverse primers.
DNA hybridization.
Digoxigenin-labeled probes were prepared by using PCR as described previously (21). Two probes were used: a 702-bp nahAc probe was prepared from P. putida G7 by using primers nahAc1 and nahAc3, and a 482-bp nahAc probe was prepared from C. testosteroni GZ42 by using primers Ac114F and Ac596R. Five micrograms of DNA from each isolate was denatured in 0.1 volume of a solution containing 4 M NaOH plus 0.1 M Na EDTA and blotted onto a Magna Graph (Micron Separations Inc., Westborough, Mass.) nylon membrane by using a vacuum-blotter (Bio-Rad, Hercules, Calif.) and baked at 65°C according to the manufacturer's (MSI) instructions. Prehybridization and hybridization were carried out in accordance with the instructions from the manufacturer for the Genius system (Boehringer-Mannheim, Indianapolis, Ind.). Hybridization and prehybridization were carried out at 65°C for the G7 probe and at 55°C for the GZ42 probe. Percent mismatch for a given wash temperature (Tm) and SSC concentration ([Na+]; 1×SSC is 0.15 M NaCl plus 0.015 M sodium citrate) was calculated by using the formula Tm = [81.5 + 16.6 (log concentration of Na+) + 0.41(%G+C) − 600/l] − % mismatch, where l is the length of the hybrid in base pairs (38, 50). The bound probe was detected by using the Genius kit and Lumiphos with chemiluminescent exposure of X-ray film (G7) or by using Nitro Blue Tetrazolium and X-phosphate (GZ42) according to the manufacturer's instructions (Boehringer-Mannheim).
Cloning, sequencing, and analysis of PCR amplicons from hillside isolates.
Cloning of the PCR products was performed with the pGEM-T Easy Vector System I (Promega Corporation, Madison, Wis.) according to the manufacturer's instructions. Plasmid preparations were performed with the Promega Wizard-Plus SV Minipreps kit. Sequencing reactions were performed with the Thermo Sequenase fluorescence-labeled primer cycle sequencing kit with 7-deaza-dGTP (Amersham Life Science, Piscataway, N.J.) and analyzed on an IR2 Long ReadIR2 automated DNA sequencer (Li-Cor, Inc, Lincoln, Nebr.). The nucleotide sequences were determined in both directions. The phnAc sequences were aligned and the percent identities were calculated by using the CLUSTALW and CLUSTALDIST programs available at San Diego Supercomputing Center's The Biology Workbench (http://workbench.sdsc.edu). 16S rRNA sequences were aligned and analyzed by using sequence match and similarity matrix programs available at the Ribosomal Database Project II (37).
Metabolite production by pure cultures.
Cultures were grown on liquid Stanier's mineral salts medium supplemented with naphthalene at 24°C in the dark with shaking at 225 rpm, harvested by centrifugation, washed twice in an equal volume of 50 mM KH2PO4 (pH 7.25) at 4°C, and resuspended in 10 ml of the KH2PO4 buffer. Fifty microliters of 200 mM substrate (naphthalene or naphthalene which was uniformly labeled with deuterium) in N,N-dimethylformamide was added to 5 ml of the concentrated cell suspension, and metabolism was allowed to proceed for 5 min. Suspensions were extracted twice with 7.5 ml of neutralized ethyl acetate (64), which was then dried by using anhydrous Na2SO4 and concentrated under an atmosphere of N2 to a volume of 100 μl. Extracts were derivatized with 10 μl of bis(trimethylsilyl)trifluoroacetimide (Supelco, Bellefonte, Pa.) for 2 to 5 min prior to gas chromatography-mass spectrometry (GC-MS) analysis. Negative controls included washed cell suspensions with no added substrate, KH2PO4 buffer (no cells) plus naphthalene, and in some cases uninduced cell suspensions (grown on yeast extract-peptone rather than on naphthalene). Studies were also performed with E. coli JM109(pDTG601), which carries the cloned toluene dioxygenase genes from P. putida F1 (68). Studies designed to determine whether a strain used a dioxygenase-mediated attack in the initial oxidation of naphthalene were performed with 10-ml volumes of cell suspensions in 25-ml serum bottles with crimp tops. To remove ambient dissolved 16O2 from the cell suspension, N2 was passed through a 0.22-μm-pore-size filter and gently bubbled through the liquid for 5 min. Stripped gases were allowed to exit via an 18-gauge syringe needle piercing the seal. A measured amount of 18O2 (Cambridge Isotope Laboratories, Andover, Mass.) or a mixture of 18O2 and 16O2 was introduced with a syringe, after which the bottle was inverted and shaken vigorously to dissolve the headspace gas. The isotope composition of the oxygen in the headspace was measured by sampling with a gas-tight syringe and analysis via GC-MS. The ratio of 18O2 to 16O2 was adjusted by the addition of further gas, if necessary, and reanalyzed prior to the addition of naphthalene and subsequent assay of metabolite production. The cell suspension assays were performed three times, and the identity of detected compounds was verified by comparison with the retention times and mass spectra of authentic standards.
GC-MS analysis.
To determine the amount of naphthalene in sediment and soil samples, 4 g (wet weight) of soil or 5 g (wet weight) of sediment was mixed with 3 ml of a 9:1 mixture of hexane-butanol and shaken for 5 days in Teflon-sealed glass vials as previously described (36). After the mixtures settled, 0.9 μl of the organic phase was analyzed as outlined below. Metabolites produced by washed-cell suspensions were analyzed by injecting 9 μl of the derivatizeded ethyl acetate extract into a Hewlett-Packard (H-P) model 5890 Series II gas chromatograph equipped with an H-P 5 (5% phenyl methyl silicone) fused-silica capillary column (30 m by 0.25 mm with a 0.25-mm film thickness) connected to an H-P model 5971A quadrupole mass selective detector operated at an electron energy of 70 eV and a detector voltage of 1,600 to 3,000 V. A splitless injection was used, with a 1-min delay before purge. Helium was the carrier gas at a linear gas velocity of 25 cm/s. The injector and transfer line were maintained at 250 and 300°C, respectively. The ion source pressure was maintained at 1.0 × 10−5 torr. The GC temperature profile was 40°C for 1 min followed by increases of 10°C/min to 250°C.
Nucleotide sequence accession numbers.
The phnAc and 16S rRNA gene sequences are available under GenBank accession numbers AY154358 to AY154379.
RESULTS
Characterization of samples.
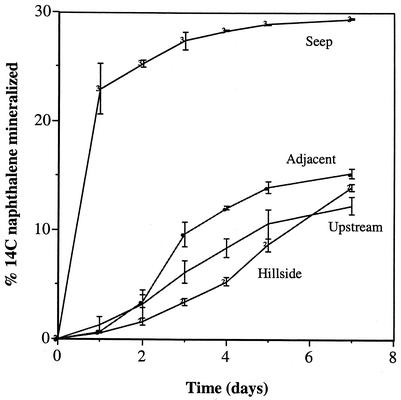
The chemical and microbiological characteristics of the four samples are presented in Table 1. The seep area lies in a direct line of the main flow of contaminated groundwater at the site. As expected, the naphthalene concentration was near saturation in the seep sediments but considerably lower in the other three samples. All samples were equivalent with regard to total microscopic counts and viable counts of bacteria. The seep sediment rapidly mineralized naphthalene with no apparent lag phase (Fig. 1). By comparison, the mineralization patterns by the nonseep (less-contaminated) samples displayed a clear lag phase, lower rates, and lesser extents of [14C]naphthalene respiration. In contrast to the naphthalene mineralization results, mineralization of 14C-labeled p-hydroxybenzoate was rapid and showed no noticeable lag in any of the samples (data not shown). Para-hydroxybenzoate serves as a useful positive control for aromatic hydrocarbon degradation because it is easily metabolized by many heterotrophic bacteria and because it is structurally similar to many naturally occurring phenolic compounds (14, 35).
TABLE 1.
Chemical and microbiological characteristics of samplesa
| Sample designationb | Type of sample | Water content (%) | Naphthalene conc (μg · cm3−1) | Acridine orange microscopic count of total bacteria [(±SD) · 109 · gdw−1] | Total no. of cultured heterotrophsc [(±SD) · 106 · gdw−1] | Total no. of cultured naphthalene-degrading bacteriad [(±SD) · 104 · gdw−1] |
|---|---|---|---|---|---|---|
| Seep | Sediment | 61.7 | 22.00 | 4.98 (±0.87) | 1.6 (±0.3) | 4.6 (±1.6) |
| Hillside | Soil | 36.3 | 0.80 | 3.37 (±0.10) | 1.8 (±0.5) | 3.2 (±0.7) |
| Adjacent | Sediment | 51.1 | 0.17 | 4.01 (±0.39) | 1.1 (±0.6) | Below detection |
| Upstream | Sediment | 61.3 | 0.70 | 5.23 (±0.46) | 0.54 (±0.25) | Below detection |
gdw, gram (dry weight) of cells.
See Materials and Methods for a description of sampling locations.
Grown on R2A medium.
Only large colonies (∼3 mm in diameter) that responded to naphthalene vapor were counted. See Materials and Methods for details.
FIG. 1.
Cumulative 14CO2 production (mineralization) from 14C-labeled naphthalene by samples from the study site. Points represent the means of results for triplicate samples relative to values for poisoned controls. Error bars indicate 1 standard deviation greater than and less than the mean.
Isolation and enumeration of naphthalene-degrading bacterial strains.
Naphthalene-degrading bacteria were readily isolated from both the seep and hillside samples without prior enrichment. Dilution plates containing bacteria from the contaminated seep and hillside samples were incubated in the presence of naphthalene vapor and yielded robust colonies (>3 mm in diameter after 3 days of incubation at 20°C) at a population density of ca. 104 cells/g (Table 1). By using relative growth with and without naphthalene vapor as a presumptive criterion for naphthalene catabolic capability, 18 presumptive naphthalene-degrading bacterial isolates were picked and purified from the contaminated seep plates and 23 were picked from the hillside sample plates. Of these, 17 seep and 22 hillside isolates (Table 2) mineralized [14C]naphthalene to 14CO2 after 2 days of incubation at 25°C.
TABLE 2.
Characteristics of naphthalene-mineralizing bacterial isolates from adjacent site habitats, seep sediment and hillside soil
| Habitat/strain | Gram stain/morphologya | Oxidase | Closest taxon identified byb:
|
16S rRNA (% identity) | |
|---|---|---|---|---|---|
| API-NFT | BIOLOG | ||||
| Seep | |||||
| Cg1 | Neg./rod | + | Pseudomonas putida | Pseudomonas putida A1 | Pseudomonas gramini (100) |
| Cg2 | Neg./rod | + | Pseudomonas fluorescens | Pseudomonas fluorescens G | Pseudomonas azotoformans (99.7) |
| Cg3 | Pos./rod | − | ND | Micrococcus diversus | NDc |
| Cg4 | Neg./rod | + | No ID | No ID (Pseudomonas fulva) | Pseudomonas amygdali (99.4) |
| Cg5 | Neg./rod | + | (Pseudomonas fluorescens) | Pseudomonas fluorescens G | Pseudomonas gramini (100) |
| Cg7 | Neg./rod | + | No ID | (Pseudomonas fluorescens C) | Pseudomonas azotoformans (99.7) |
| Cg8 | Neg./rod | + | Pseudomonas fluorescens | No ID (Pseudomonas asplenii) | ND |
| Cg9 | Neg./rod | + | No ID | Pseudomonas fluorescens G | ND |
| Cg11 | Neg./rod | + | Ralstonia picketii | Pseudomonas mendocina | Pseudomonas amygdali (99.4) |
| Cg12 | Neg./rod | + | Pseudomonas fluorescens | (Pseudomonas fluorescens G) | ND |
| Cg13 | Pos./rod | − | ND | ND | ND |
| Cg14 | Pos./rod | − | ND | ND (Arthrobacter)d | ND |
| Cg15 | Neg./rod | + | Ralstonia picketii | No ID (Pseudomonas cichorii) | Pseudomonas amygdali (99.4) |
| Cg16 | Neg./rod | + | (Pseudomonas aureofaciens) | (Pseudomonas corrugata) | ND |
| Cg18 | Pos./rod | − | ND | Micrococcus diversus | ND |
| Cg20 | Pos./rod | − | ND | ND | ND |
| Cg21 | Neg./rod | − | Sphingomonas paucimobilis | Sphingobacterium mizutaii | Pseudomonas azotoformans (100) |
| Hillside | |||||
| Hg1 | Neg./rod | − | No ID | (Chryseomonas luteola) | Herbaspirillum seropedicae (96.6) |
| Hg2 | Neg./rod | − | No ID | (Burkholderia glathei) | Burkholderia glatheii (97.7) |
| Hg3 | Neg./rod | − | No ID | Burkholderia glathei | Burkholderia glatheii (97.8) |
| Hg4 | Neg./rod | − | No ID | Burkholderia glathei | Burkholderia glatheii (97.9) |
| Hg5 | Neg./rod | − | No ID | Burkholderia glathei | Burkholderia glatheii (98.0) |
| Hg6 | Neg./rod | − | Burkholderia cepacia | (Burkholderia glathei) | ND |
| Hg7 | Neg./coccus | − | No ID | (Burkholderia glathei) | Burkholderia glatheii (97.3) |
| Hg8 | Neg./rod | − | Burkholderia cepacia | Burkholderia gladioli | Burkholderia phenazinium (97.7) |
| Hg9 | Neg./rod | − | No ID | Burkholderia sp. (B. gladioli) | ND |
| Hg10 | Neg./rod | − | Burkholderia cepacia | Burkholderia gladioli | Burkholderia phenazinium (97.8) |
| Hg11 | Neg./rod | − | No ID | No ID (B. glathei) | Burkholderia glathei (98.1) |
| Hg12 | Neg./rod | − | Burkholderia cepacia | No ID (B. gladioli) | ND |
| Hg13 | Neg./rod | − | No ID | Burkholderia sp. (B. gladioli) | ND |
| Hg14 | Neg./rod | − | No ID | (Pseudomonas fluorescens G) | Burkholderia phenazinium (97.9) |
| Hg15 | Neg./rod | − | No ID | Burkholderia glathei | ND |
| Hg16 | Neg./rod | − | No ID | Burkholderia glathei | Burkholderia phenazinium (97.5) |
| Hg17 | Neg./rod | − | Burkholderia cepacia | Burkholderia gladtoli | Burkholderia glathei (98.1) |
| Hg18 | Neg./rod | + | No ID | Burkholderia glathei | Burkholderia glathei (98.0) |
| Hg19 | Neg./rod | − | No ID | No ID (B. glathei) | Burkholderia glathei (98.1) |
| Hg20 | Neg./rod | − | No ID | Xanthomonas maltophilia | ND |
| Hg21 | Neg./rod | − | No ID | No ID (Burkholderia glathei) | ND |
| Hg22 | Neg./rod | − | Burkholderia cepacia | Burkholderia gladioli | ND |
Neg., negative; Pos., positive.
Names in parentheses indicate the database taxon closest to the unidentified genus or species. ND, not determined; ID, identification. The API-NFT test kit is designed for gram-negative bacteria only.
ND, not done
Based on Biolog characterization and microscopic observation of coccus/irregular rod cycling
Identical enumeration and isolation procedures were applied to samples adjacent to and upstream from the seep area. For these samples, however, colony sizes were not larger in the presence of naphthalene vapor; thus, initial counts were deemed to be below the level of detection (Table 1). Naphthalene-degrading bacterial strains were obtained from these samples after seven or more days of incubation in a minimal broth medium plus naphthalene, indicating that naphthalene-catabolizing bacteria were present and could be enriched (data not shown).
Characterization of naphthalene-degrading isolates and comparison of hillside and seep isolates.
All isolates were characterized and, when possible, identified by using standard microbiological methods and the API Rapid-NFT and Biolog test kits (Table 2). 16S rRNA gene sequences (1,454 nucleotides, corresponding to nucleotides 28 through 1491 of rRNA from E. coli) for 14 of the hillside isolates were compared to rRNA sequences of characterized organisms. Partial 16S rRNA gene sequences (305 nucleotides, corresponding to nucleotides 23 through 328 of rRNA from E. coli) were obtained for a subset of the seep isolates as part of a previous study (22). The two sets of naphthalene-degrading isolates, one from the contaminated seep and the other from the hillside, were taxonomically and phenotypically distinct. Cluster analysis of the gram-negative isolates based on 95 carbon source utilization tests in the Biolog system revealed seven well-defined phenetic groups (data not show). Members of each of the phenetic groups were homogeneous with respect to the source of the sample from which they were isolated. Three of the groups consisted entirely of seep isolates, while four groups were composed entirely of isolates from the hillside sample. Isolates from the hillside sample were all gram-negative, oxidase-negative, mainly rod-shaped organisms. Most of these isolates were identified by substrate utilization profiles as members of the genus Burkholderia (Table 2). Partial 16S rRNA gene sequences supported this taxonomic affiliation, with most of the sequences matching (ca. 98% identity) that of one of three Burkholderia species: Burkholderia glathei, Burkholderia gladioli, or Burkholderia phenazinium. Strain Hg20 was identified by Biolog as Xanthomonas maltophilia, a member of the gamma-proteobacteria. No matches were found in the Biolog database for Hg1, a strain whose 16S rRNA gene sequence was found to share more than 96% identity with that of Herbaspirillum seropedicae. In contrast, the naphthalene-degrading bacteria from the contaminated seep samples were phenotypically and taxonomically more diverse. The seep guild was comprised of both gram-positive and -negative and oxidase-positive and -negative isolates (Table 2). The gram-negative isolates had substrate utilization profiles and 16S rRNA gene sequences that placed them within the rRNA group 1 (45) of the genus Pseudomonas. None of the hillside isolates fell within this genus.
Detection of nahAc homologs by PCR analysis and Southern hybridization.
As previously reported, all of the gram-negative seep isolates were found to possess a highly conserved allele of nahAc located on a pDTG1-like plasmid (22, 55). Previously optimized PCR primers and protocols (21, 65) and Southern hybridization were used to screen the hillside isolates for the presence of nahAc homologs. nahAc amplification products were not produced from the hillside strains nor from the gram-positive strains from the seep, and these strains did not hybridize with a 702-bp nahAc probe under nonstringent (∼13% mismatch) conditions. Under even less stringent conditions (∼25% mismatch), 11 of the 22 strains hybridized with the nahAc probe (Hg4, Hg7, Hg10, Hg11, Hg14, and Hg16 to Hg22). None of the seep gram-positive strains hybridized, even at conditions approximating 40% mismatch. A second PCR screen that targeted a wider array of genes encoding dioxygenase iron-sulfur protein large subunits was performed. This assay used a suite of degenerate primers shown to amplify a range of diverse large-subunit sequences, including nahAc2 from C. testosteroni GZ42, which is ∼80% identical at the nucleotide level to nahAc from P. putida G7 and to ndoB from P. putida NCIB 9816-4 (65). However, none of the hillside isolates produced a PCR product by this alternate PCR assay. Lack of amplification was probably not due to the unavailability of template DNA, since PCR amplification of the 16S rRNA gene for all of the isolates was successful (data not shown).
Amplification, sequencing, and analysis of phnAc homologs.
The hillside isolates were then screened for the presence of homologs of phnAc from Burkholderia sp. strain RP007 by using PCR primers that had been developed for studies of that gene (32, 33). Strain RP007 was isolated on phenanthrene from a PAH-contaminated soil from New Zealand and shown to possess a set of naphthalene-catabolic genes that were highly divergent from those that had previously been described. Using primers phn8073F and phn9047R, we were able to amplify a product of the expected size from all 14 of the Hg strains tested (Hg1 to Hg5, Hg7, Hg8, Hg10, Hg11, Hg14, and Hg16 to Hg19) (data not shown). These strains represented five of the six phenotypic clusters identified from carbon-substrate-utilization profiles. Hg20, the only member of the sixth cluster, could not be recovered from frozen stocks and therefore was not included in this analysis. The PCR results indicated that these strains contained phnAc homologs. This result was verified by cloning and sequencing the phn8073F-phn9047R amplicons from eight of the isolates and comparing the resulting sequences (992 nucleotides) to the sequence from Burkholderia sp. strain RP007. The sequences were aligned and the uncorrected percentages of dissimilarity were calculated by using ClustalW and ClustalDist programs at San Diego SuperComputing Center's The Biology Workbench (http://workbench.sdsc.edu) (Table 3). The phnAc nucleotide sequences from the hillside isolates were ca. 98% identical to that from Burkholderia sp. strain RP007. The new sequences were even more closely related to one another; in all but one case, they were more than 99% identical to one another. The sequences from strains Hg8 and Hg11 were 100% identical.
TABLE 3.
Distance matrix of phnAc and 16S rRNA gene sequences from hillside isolates
| Strain | Dissimilaritya to:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Hg2 | Hg4 | Hg8 | Hg10 | Hg11 | Hg14 | Hg16 | RP007 | |
| Hg1 | 0.002a | 0.001 | 0.005 | 0.006 | 0.005 | 0.008 | 0.003 | 0.099 |
| 0.104 | 0.102 | 0.101 | 0.102 | 0.097 | 0.103 | 0.107 | 0.097 | |
| Hg2 | 0.003 | 0.003 | 0.008 | 0.003 | 0.005 | 0.005 | 0.038 | |
| 0.006 | 0.040 | 0.037 | 0.008 | 0.039 | 0.043 | 0.039 | ||
| Hg4 | 0.006 | 0.005 | 0.006 | 0.008 | 0.002 | 0.036 | ||
| 0.038 | 0.035 | 0.006 | 0.036 | 0.041 | 0.036 | |||
| Hg8 | 0.005 | 0.000 | 0.009 | 0.008 | 0.030 | |||
| 0.003 | 0.034 | 0.004 | 0.008 | 0.031 | ||||
| Hg10 | 0.005 | 0.013 | 0.007 | 0.029 | ||||
| 0.032 | 0.002 | 0.006 | 0.029 | |||||
| Hg11 | 0.009 | 0.008 | 0.034 | |||||
| 0.033 | 0.037 | 0.034 | ||||||
| Hg14 | 0.005 | 0.027 | ||||||
| 0.006 | 0.028 | |||||||
| Hg16 | 0.033 | |||||||
| 0.033 | ||||||||
Dissimilarity of phnAc sequences is indicated in bold, white dissimilarity of 16S rRNA sequences is indicated in italics.
The natural horizontal transfer of genes has been inferred by showing that a highly conserved gene is shared by a group of taxonomically diverse hosts (22, 29). To examine whether lateral transfer of the phnAc allele might be taking place among the hillside microorganisms, we compared the phnAc sequence divergence with the16S rRNA gene sequence divergence of the same strains. rRNA gene sequences (1,454 nucleotides) were aligned and uncorrected percentages of dissimilarity were calculated by using the Similarity Matrix program at the Ribosomal Database Project II (37), excluding gaps and regions that could not be unambiguously aligned (Table 3). For several of the strains, the 16S rRNA gene sequences were markedly different while the phnAc sequences were identical or nearly identical. For example, the phnAc sequences of Hg1 and Hg4 differed by a single nucleotide in the region sequenced, but the 16S rRNA sequences of these strains shared only 90% identity. Similarly, the phnAc sequences of Hg8 and Hg11 were identical, but the 16S rRNA gene sequences of these strains shared only 97% identity.
The sequencing results suggested that diverse phnAc alleles may be present in the different hillside isolates. To investigate whether these isolates possessed a similar gene arrangement to that found in Burkholderia sp. strain RP007, other PCR primers targeted at regions of DNA flanking phnAc (32) were used to screen the isolates. The four possible combinations of two forward PCR primers (phn6897F and phn8073F) and two reverse primers (phn8420R and phn9047R) were used to screen for the presence of diverse phnAc homologs in several of the hillside isolates. All strains tested produced amplicons of the expected size with each of the four primer combinations, except Hg14, which produced amplicons of the expected size with all the primer combinations except phn6897F-phn9047R.
Metabolite production by pure cultures of phnAc-containing strains.
The cloned phn genes of Burkholderia sp. strain RP007 have been shown to produce 1,2-dihydroxynaphthalene and salicylate from naphthalene, and two distinct catechol 2,3-dioxygenase genes have been cloned from this strain and characterized (32). To examine the metabolic diversity of naphthalene catabolism in the phnAc-containing hillside isolates, metabolite production by pure cultures was examined by using GC-MS analysis. Seven of the hillside strains were chosen to represent the subgroups that were identified by carbon substrate utilization profiles. Characterized strains P. putida G7 and C. testosteroni GZ42 were included as controls. These control strains metabolize naphthalene to salicylate; P. putida G7 oxidizes salicylate to catechol by using a salicylate-1-hydroxylase (56), while it is thought that C. testosteroni GZ42 converts salicylate to gentisate, presumably via a salicylate-5-hydroxylase (16).
Metabolites produced from washed cell suspensions that were incubated briefly with naphthalene are included in Table 4. Parallel experiments in which deuterium-labeled naphthalene was substituted for the unlabeled substrate were performed to verify that these compounds originated from the added naphthalene substrate. Some variability was detected among the pathways for naphthalene catabolism in the hillside (Hg) strains (Table 4). When P. putida G7 cells were assayed, all of the expected metabolites from the catechol pathway were detected. Strains Hg4, Hg6, and Hg8 also produced all of the compounds expected from the catechol pathway, while Hg2 and Hg12 produced all of the compounds expected from the catechol pathway except for salicylaldehyde. C. testosteroni GZ42 produced all of the metabolites present in the gentisate pathway, as expected. Hg11 cell suspensions also produced all of the metabolites of the gentisate pathway. In Hg1 cell suspensions, only three of the previously reported metabolites were detected: 1,2-dihydroxy-1,2-dihydronaphthalene (1,2-DHDN); 1,2-dihydroxynaphthalene, and salicylate.
TABLE 4.
Potential metabolites detected in extracts of cell suspensions incubated with naphthalene
| Strain | Potential metabolite detecteda
|
% of 1,2-DHDN labeled with the indicated no. of 18O atomsb
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Catechol | Gentisate | Salicylate | Salicylald. | 1,2-DHN | 1,2-DHDN | 0c | 1d | 2e | |
| P. putida G7 | + | BD | + | + | + | + | 66 | 11 | 22 |
| C. testosteroni GZ42 | BD | + | + | + | + | + | — | ||
| Hg1 | BD | BD | + | BD | + | + | 46 | 9 | 45 |
| Hg2 | + | BD | + | BD | + | + | 55 | 9 | 36 |
| Hg3 | + | BD | + | BD | + | + | — | ||
| Hg4 | + | BD | + | + | + | + | 45 | 9 | 46 |
| Hg6 | + | BD | + | + | + | + | — | ||
| Hg8 | + | BD | + | + | + | + | 55 | 10 | 35 |
| Hg11 | BD | + | + | + | + | + | — | ||
| Hg12 | + | BD | + | BD | + | + | 62 | 10 | 28 |
+, detection; BD, below detection. Salicylald., salicyladehyde; 1,2-DHN, 1,2-dihydroxynaphthalene.
Mass spectrometry determined the distribution of 18O within 1,2-DHDN parent ions produced under an atmosphere of 50% 18O2-50% 16O2. —, not determined.
1,2-DHDN mass contained two 16O atoms. Molecular ion = M.
1,2-DHDN mass contained one 16O atoms and one 18O atom. Molecular ion = M + 2.
1,2-DHDN mass contained zero 16O atoms and two 18O atoms. Molecular ion = M + 4.
Three possible mechanisms can be envisioned for the enzymatic formation of 1,2-DHDN from naphthalene, and these three mechanisms can be distinguished by examining the patterns of 18O-labeled products formed under atmospheres containing 50/50 mixtures of 18O2 and 16O2. The first mechanism is the dioxygenase-mediated single-step addition of both atoms of molecular oxygen (O2) to adjacent carbons of a naphthalene molecule (56). Alternatively, a two-step attack — initial mono-oxygenase followed by the action of an epoxide hydrolase — would result in the addition of one atom of O that originated from O2 and one atom of O from water (6). Finally, two mono-oxygenations, a mechanism that we could find no reports of in the literature, would proceed via the sequential addition of two O atoms that each originated from the pool of O2 molecules. Thus, the predicted distributions of labeled products formed by the different mechanisms under atmospheres containing 50/50 mixtures of 18O2 and 16O2 areas follows: for a dioxygenase, two equal pools of 1,2-DHDN differing by 4 atomic mass units; for a mono-oxygenase followed by an epoxide hydrolase, two equal pools of 1,2-DHDN differing by 2 atomic mass units; and for sequential mono-oxygenations, three pools of 1,2-DHDN, in ratios of 1:2:1, each differing by 2 atomic mass units. Cell suspensions of all strains, when incubated in the presence of a mixture of 18O2 and 16O2, produced labeling patterns in 1,2-DHDN that deviated from theoretical ratios (discussed above) but conformed most closely with that of an initial dioxygenase attack (Table 4).
DISCUSSION
Several distinct NDOs have been characterized to date, but the ecological significance, relative distribution, and transmission of the different NDO analogs are poorly investigated. The phn genes have been described only recently, and it has been suggested several times that these genes were not discovered earlier because organisms with phn genotypes are more difficult to culture than are organisms with nah genotypes (32, 34). However, in this study, only phn-containing organisms were isolated from a hillside soil, which appears to conflict with the suggestion that culturing these organisms is problematic. Additionally, a recent study of PAH-degrading bacteria isolated from pristine and contaminated soils in Kuwait, Indonesia, Thailand, and Japan found that 6 out of 19 isolates hybridized to a phnAc probe (62). A likely explanation is that phnAc-utilizing organisms have been cultured routinely in the past but were not recognized as such because the phn genes had not been described. Several studies have reported the isolation of naphthalene-degrading organisms that did not hybridize to probes derived from nahAc of P. putida G7 and ndoB of P. putida NCIB 9816-4 (4, 7, 18, 39, 42, 47, 62). Studies of PAH dioxygenase gene distribution need to be updated by using the suite of recently described PAH dioxygenase genes as hybridization targets.
It also appears that the restriction of nahAc alleles to seep isolates and that of phnAc alleles to hillside isolates in this study are consistent with natural selective enrichment of the different genotypes in the two environments. While retrospective identification of selective factors is necessarily speculative, possible selective factors can be divided into two groups: those that relate to the PAH-degrading phenotype and those that are independent of this phenotype. With regard to the first group, there may have been selection for the nah genotype in the seep because the concentration of PAHs was higher and the phn genotype was inhibited, the mixture of PAHs in the seep was more susceptible to attack by the nah genotype, lateral transfer of the nah genotype occurred to a greater extent in the seep environment, or the nah genotype allowed for more rapid growth in the seep environment. Selective enrichment of microbial populations in response to pollutant contamination was observed by Ka et al. (25), who found that certain groups of 2,4-dichlorophenoxyacetic acid degraders which predominated after long exposure were found only at low frequencies initially, while others were dominant at first but isolated only rarely after prolonged contamination. Also, Ogunseitan et al. (43) found that seeding soils with salicylate, an inducer of naphthalene degradation, caused an increase in the proportion of nahAB-hybridizing strains in soil microcosms, while Laurie and Lloyd-Jones (33) found enrichment of phnAc but not nahAc after the addition of naphthalene to an uncontaminated soil.
Alternatively, a selective factor could be independent of the PAH-degrading phenotype. That is, there could have been selection for different taxonomic groups in the two environments, and the different NDO genotypes are coincidentally associated with those different taxonomic groups. Environmental factors that might selectively enrich for different taxonomic groups are diverse (e.g., moisture content or anaerobiosis), but an examination of the phylogenetic identity of the hillside isolates suggests one particular factor, vegetation. Hillside organisms primarily belonged to several species of Burkholderia, including B. glathei, B. gladioli, and B. phenazinium. All of these species are common soil inhabitants that have been found to reach much higher densities in the rhizosphere, representing half the culturable bacteria in some rhizosphere studies (61). Hg1 and Hg20 were the only hillside isolates that were not among these Burkholderia species. Hg1 was most closely related to H. seropedicae, which has been found only intimately associated with plant tissues and roots (3, 44), and Hg20 was most closely related to X. maltophilia, a common plant pathogen. Thus, it may be that the hillside community represented naphthalene degraders that are selected for in the rhizosphere environment. This interpretation leads to the suggestion that difficulties reported by other investigators in culturing phn-containing strains reflect problems with culturing Burkholderia on nonselective media from nonrhizosphere soils and sediments. However, this interpretation is tentative because we did not actively attempt to sample the rhizosphere, for example, by washing and crushing roots to sample endophytic bacteria. Furthermore, even efforts to plate rhizosphere samples onto selective media are not always particularly effective at recovering Burkholderia species (40). In addition, Pseudomonas species, including those that hybridize with nahAc probes, have been isolated from rhizosphere (7) and endophyte (52) samples.
An alternative explanation for the irregular distribution of NDO genes among site isolates is that the communities were limited by the genetic material available to them. This possibility seems unlikely given the widespread distribution of both alleles in pristine as well as contaminated soils (33, 34) and the likelihood of cotransport of PAHs and PAH-degrading bacteria (36). It would not be surprising if additional sampling produced some nahAc alleles in hillside isolates or phnAc alleles in seep isolates. However, it is unlikely that the trend described here would disappear with further sampling.
We previously reported evidence indicating that natural horizontal transfer of diverse nahAc-containing plasmids had taken place among the microbial inhabitants of the seep. An identical nahAc allele was shared by seven different seep isolates whose 16S rRNA gene sequences differed by as much as 7.9% (22). The sequence divergence of the highly conserved 16S rRNA gene, coupled with the lack of divergence of the nahAc allele, was taken as evidence that the nahAc allele had been recently transferred among the seep isolates. Large plasmids were identified in all of these isolates, and a 407-bp nahAc probe hybridized to plasmids in all but one (22). Filter matings between naphthalene-degrading bacterial isolates and their cured progeny revealed that the naphthalene-catabolic plasmids were self-transmissible, and distinct naphthalene-catabolic plasmids were retrieved directly from the microbial community indigenous to the contaminated site in filter matings by using a cured, rifampin-resistant, site-derived isolate as the recipient (23, 55). We initiated similar studies to investigate whether lateral transfer of phnAc alleles might be taking place among the members of the hillside community.
Partial phnAc sequences (992 nucleotides) and 16S sequences (1,454 nucleotides) were obtained from eight isolates in five of the six phenotypic clusters identified from carbon substrate-utilization profiles. Comparison of the phnAc sequences revealed greater diversity than was seen with the nahAc allele in the seep study (22), but the phnAc sequences were all closely related. Although only two strains had identical phnAc alleles (Hg8 and Hg11), the most dissimilar strains still shared 98.7% identity (Hg10 and Hg14). Comparison of the 16S rRNA gene sequences, however, revealed much greater divergence than the comparison of phnAc sequences. For example, the 16S rRNA gene sequences of Hg1 and Hg4 are only 90% identical, but the phnAc sequences differ by merely a single nucleotide in the 992-nucleotide region sequenced. Similarly, the 16S rRNA gene sequences of Hg8 and Hg11 share identical phnAc sequences but have 16S sequences that are only 97% identical. We interpret these results, which show that a highly conserved gene is shared by a group of taxonomically diverse hosts, as evidence that horizontal transfer of the phnAc allele has taken place among the members of the hillside community. That the hosts are in fact taxonomically diverse is further supported by the substrate-utilization profiles (Table 4).
There were two important differences between the results of this study and those obtained in the nahAc study (22). In this study, there were multiple alleles of phnAc present and in some pair-wise comparisons the phnAc sequences of the isolates were more highly diverged than were the 16S rRNA gene sequences. The divergence observed among the phnAc alleles of the hillside isolates, although slight, is interesting in light of the fact that a single nahAc allele was recovered from the seep isolates. This divergence may reflect lateral transfer that took place prior to, or over a longer period of time than, that of the nahAc allele. Alternatively, it may indicate transfer of multiple, dissimilar phn-carrying mobile elements among bacteria in the hillside environment. Genetic diversity within the phn genes was also suggested when Hg14 did not produce an amplification product with primers phn6897F and phn9047R. Although this is a negative result and therefore problematic to interpret, it may indicate a different gene order in Hg14 or it may reflect sequence divergence of the phnC gene at the site where phn6897F binds. Further studies, including the characterization of large plasmids (if present), should provide greater understanding of the means by which the phn genes are disseminated at the site.
We also examined whether the hillside isolates were metabolically diverse. By using GC-MS analysis of culture extracts, these isolates were tested to determine what metabolites they produced, their mechanism of initial ring attack, and the source of oxygen atoms added to the ring. Both the metabolites themselves and their deuterated and 18O-labeled analogs indicated uniformly that the mechanism of initial ring attack was dioxygenase mediated. Two distinct pathways for subsequent naphthalene catabolism have been characterized, one in which salicylate is oxidized to catechol and one in which salicylate is converted to gentisate, and evidence for each of these pathways was found in different subsets of the hillside isolates. Interestingly, evidence for the catechol pathway was demonstrated in one strain (Hg8) that had a phnAc allele identical to that of an isolate that may be using the gentisate pathway (Hg11). In addition to being the first indication that a phn-utilizing organism may catabolize naphthalene through the gentisate pathway, this result further supports the notion that multiple phn mobile elements may have been transferred at the site. In Hg1 cell suspensions, only three of the previously reported metabolites were detected. The inability of this assay to detect additional compounds in extracts from this strain could reflect differences in the rate at which it uses certain metabolites, or it could reflect the existence of further pathway variation. The detection of a compound in supernatant fluids of naphthalene-grown cells, or the demonstration that it is produced from naphthalene by induced cells, does not prove that the compound is an intermediary metabolite of a naphthalene-catabolic process. Compounds might be produced as dead-end metabolites, as side reactions, or as artifacts of the extraction and analysis procedure (8). A better approach for designating a compound as an intermediary metabolite can result from studies of enzymes isolated from an organism. Genetic approaches, such as that utilized by Eaton and Chapman to study the ring cleavage of 1,2-dihydroxynaphthalene (11), are more stringent and less likely to suffer from these difficulties. However, the methods used in this study are helpful for screening a large number of isolates to identify which organisms should be more intensively studied.
These results provide further evidence that lateral transfer is important in the biodegradation of contaminants in polluted field sites. They extend the known geographic distribution of phnAc alleles, expand the range of microbial genera known to contain phnAc alleles, and suggest previously unidentified metabolic diversity among phnAc-containing bacteria. Genetic investigations of the site-derived strains that were examined in this study and field studies targeting rhizosphere versus nonrhizosphere communities may be useful in furthering the understanding of PAH-degradative processes at this and other contaminated field sites. Greater understanding of these processes can assist with the development of new technologies and management strategies for use in contaminated field sites and can improve the knowledge of the evolution of natural bacterial populations.
Acknowledgments
Agencies that supported this research, via grants to E.L.M., included the National Science Foundation (grant MCB-0084175) and the Air Force Office of Scientific Research (grant 93-NL-073). This work was also supported in part by funds from the Howard Hughes Medical Institute (grant 52002680).
We are grateful to Karen G. Stuart-Keil, Erik C. Osborn, Alexandra T. Kameda, Sergio Manodori, and Lesley M. Jones for laboratory assistance.
REFERENCES
- 1.Andreoni, V., S. Bernasconi, M. Colombo, J. B. van Beilen, and L. Cavalca. 2000. Detection of genes for alkane and naphthalene catabolism in Rhodococcus sp. strain 1BN. Environ. Microbiol. 2:572-577. [DOI] [PubMed] [Google Scholar]
- 2.Bakermans, C., A. M. Hohnstock-Ashe, S. Padmanabhan, P. Padmanabhan, and E. L. Madsen. 2002. Geochemical and physiological evidence for mixed aerobic and anaerobic field biodegradation of coal tar waste by subsurface microbial communities. Microb. Ecol. 44:95-106. [DOI] [PubMed] [Google Scholar]
- 3.Baldani, J. I., V. L. D. Baldani, L. Seldin, and J. Döbereiner. 1986. Characterization of Herbaspirillum seropedicae gen. nov., sp. nov., a root-associated nitrogen-fixing bacterium. Int. J. Syst. Bacteriol. 36:86-93. [Google Scholar]
- 4.Berardesco, G., S. Dyhrman, E. Gallagher, and M. P. Shiaris. 1998. Spatial and temporal variation of phenanthrene-degrading bacteria in intertidal sediments. Appl. Environ. Microbiol. 64:2560-2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosch, R., E. Garcia-Valdes, and E. R. Moore. 1999. Genetic characterization and evolutionary implications of a chromosomally encoded naphthalene-degradation upper pathway from Pseudomonas stutzeri AN10. Gene 236:149-157. [DOI] [PubMed] [Google Scholar]
- 6.Cerniglia, C. E., J. R. Althaus, F. E. Evans, J. P. Freeman, R. K. Mitchum, and S. K. Yang. 1983. Stereochemistry and evidence for an arene oxide-NIH shift pathway in the fungal metabolism of naphthalene. Chem. Biol. Interact. 44:119-132. [DOI] [PubMed] [Google Scholar]
- 7.Daane, L. L., I. Harjono, G. J. Zylstra, and M. M. Häggblom. 2001. Isolation and characterization of polycyclic aromatic hydrocarbon-degrading bacteria associated with the rhizosphere of salt marsh plants. Appl. Environ. Microbiol. 67:2683-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dagley, S., and P. J. Chapman (ed.). 1973. Evaluation of methods used to determine metabolic pathways. Methods Microbiol. 6A:217-268.
- 9.Davies, J. I., and W. C. Evans. 1964. Oxidative metabolism of phenanthrene and anthracene by soil pseudomonads. The ring fission mechanism. Biochem. J. 91:251-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denome, S. A., D. C. Stanley, E. S. Olson, and K. D. Young. 1993. Metabolism of dibenzothiophene and naphthalene in Pseudomonas strains: complete DNA sequence of an upper naphthalene catabolic pathway. J. Bacteriol. 175:6890-6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eaton, R. W., and P. J. Chapman. 1992. Bacterial metabolism of naphthalene: construction and use of recombinant bacteria to study ring cleavage of 1,2-dihydroxynaphthalene and subsequent reactions. J. Bacteriol. 174:7542-7554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuenmayor, S. L., M. Wild, A. L. Boyes, and P. A. Williams. 1998. A gene cluster encoding steps in conversion of naphthalene to gentisate in Pseudomonas sp. strain U2. J. Bacteriol. 180:2522-2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geiselbrecht, A. D., B. P. Hedlund, M. A. Tichi, and J. T. Staley. 1998. Isolation of marine polycyclic aromatic hydrocarbon (PAH)-degrading Cycloclasticus strains from the Gulf of Mexico and comparison of their PAH degradation ability with that of Puget Sound Cycloclasticus strains. Appl. Environ. Microbiol. 64:4703-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghiorse, W. C., and J. T. Wilson. 1988. Microbial ecology of the terrestrial subsurface. Adv. Appl. Microbiol 33:107-172. [DOI] [PubMed] [Google Scholar]
- 15.Gibson, D. T., and R. E. Parales. 2000. Aromatic hydrocarbon dioxygenases in environmental biotechnology. Curr. Opin. Biotechnol. 11:236-243. [DOI] [PubMed] [Google Scholar]
- 16.Goyal, A. K., and G. J. Zylstra. 1997. Genetics of naphthalene and phenanthrene degradation by Comamonas testosteroni. J. Ind. Microbiol. Biotechnol. 19:401-407. [DOI] [PubMed] [Google Scholar]
- 17.Goyal, A. K., and G. J. Zylstra. 1996. Molecular cloning of novel genes for polycyclic aromatic hydrocarbon degradation from Comamonas testosteroni GZ39. Appl. Environ. Microbiol. 62:230-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamann, C., J. Hegemann, and A. Hildebrandt. 1999. Detection of polycyclic aromatic hydrocarbon degradation genes in different soil bacteria by polymerase chain reaction and DNA hybridization. FEMS Microbiol. Lett. 173:255-263. [DOI] [PubMed] [Google Scholar]
- 19.Harayama, S., M. Kok, and E. L. Neidle. 1992. Functional and evolutionary relationships among diverse oxygenases. Annu. Rev. Microbiol. 46:565-601. [DOI] [PubMed] [Google Scholar]
- 20.Hedlund, B. P., A. D. Geiselbrecht, T. J. Bair, and J. T. Staley. 1999. Polycyclic aromatic hydrocarbon degradation by a new marine bacterium, Neptunomonas naphthovorans gen. nov., sp. nov. Appl. Environ. Microbiol. 65:251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herrick, J. B., E. L. Madsen, C. A. Batt, and W. C. Ghiorse. 1993. Polymerase chain reaction amplification of naphthalene-catabolic and 16S rRNA gene sequences from indigenous sediment bacteria. Appl. Environ. Microbiol. 59:687-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herrick, J. B., K. G. Stuart-Keil, W. C. Ghiorse, and E. L. Madsen. 1997. Natural horizontal transfer of a naphthalene dioxygenase gene between bacteria native to a coal tar-contaminated field site. Appl. Environ. Microbiol. 63:2330-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hohnstock, A. M., K. G. Stuart-Keil, E. E. Kull, and E. L. Madsen. 2000. Naphthalene and donor cell density influence field conjugation of naphthalene catabolism plasmids. Appl. Environ. Microbiol. 66:3088-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeffrey, A. M., H. J. C. Yeh, D. M. Jerina, T. R. Patel, J. F. Davey, and D. T. Gibson. 1975. Initial reactions in the oxidation of naphthalene by Pseudomonas putida. Biochemistry 14:566-584. [DOI] [PubMed] [Google Scholar]
- 25.Ka, J. O., W. E. Holben, and J. M. Tiedje. 1994. Genetic and phenotypic diversity of 2,4-dichlorophenoxyacetic acid (2,4-D)-degrading bacteria isolated from 2,4-D-treated field soils. Appl. Environ. Microbiol. 60:1106-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kauppi, B., K. Lee, E. Carredano, R. E. Parales, D. T. Gibson, H. Eklund, and S. Ramaswamy. 1998. Structure of an aromatic-ring-hydroxylating dioxygenase-naphthalene 1,2-dioxygenase. Structure 6:571-586. [DOI] [PubMed] [Google Scholar]
- 27.Khan, A. A., R.-F. Wang, W.-W. Cao, D. R. Doerge, D. Wennerstrom, and C. E. Cerniglia. 2001. Molecular cloning, nucleotide sequence, and expression of genes encoding a polycyclic aromatic ring dioxygenase from Mycobacterium sp. strain PYR-1. Appl. Environ. Microbiol. 67:3577-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kitagawa, W., A. Suzuki, T. Hoaki, E. Masai, and M. Fukuda. 2001. Multiplicity of aromatic ring hydroxylation dioxygenase genes in a strong PCB degrader, Rhodococcus sp. strain RHA1 demonstrated by denaturing gradient gel electrophoresis. Biosci. Biotechnol. Biochem. 65:1907-1911. [DOI] [PubMed] [Google Scholar]
- 29.Koonin, E. V., K. S. Makarova, and L. Aravind. 2001. Horizontal gene transfer in prokaryotes: quantification and classification. Annu. Rev. Microbiol. 55:709-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-148. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Inc., New York, N.Y.
- 31.Larkin, M. J., C. C. R. Allen, L. A. Kulakov, and D. A. Lipscomb. 1999. Purification and characterization of a novel naphthalene dioxygenase from Rhodococcus sp. strain NCIMB12038. J. Bacteriol. 181:6200-6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laurie, A. D., and G. Lloyd-Jones. 1999. The phn genes of Burkholderia sp. strain RP007 constitute a divergent gene cluster for polycyclic aromatic hydrocarbon catabolism. J. Bacteriol. 181:531-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laurie, A. D., and G. Lloyd-Jones. 2000. Quantification of phnAc and nahAc in contaminated New Zealand soils by competitive PCR. Appl. Environ. Microbiol. 66:1814-1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lloyd-Jones, G., A. D. Laurie, D. W. F. Hunter, and R. Fraser. 1999. Analysis of catabolic genes for naphthalene and phenanthrene degradation in contaminated New Zealand soils. FEMS Microbiol. Ecol. 29:69-79. [Google Scholar]
- 35.Madsen, E. L., J. L. Sinclair, and W. C. Ghiorse. 1991. In situ biodegradation: microbiological patterns in a contaminated aquifer. Science 252:830-833. [DOI] [PubMed] [Google Scholar]
- 36.Madsen, E. L., C. T. Thomas, M. S. Wilson, R. L. Sandoli, and S. E. Bilotta. 1996. In situ dynamics of aromatic hydrocarbons (AHs) and bacteria capable of AH metabolism in a coal tar waste-contaminated field site. Environ. Sci. Technol. 30:2412-2416. [Google Scholar]
- 37.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. J. Parker, P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meinkoth, J., and G. Wahl. 1984. Hybridization of nucleic acids immobilized on solid supports. Anal. Biochem. 138:267-272. [DOI] [PubMed] [Google Scholar]
- 39.Meyer, S., R. Moser, A. Neef, U. Stahl, and P. Kampfer. 1999. Differential detection of key enzymes of polyaromatic-hydrocarbon-degrading bacteria using PCR and gene probes. Microbiology 145:1731-1741. [DOI] [PubMed] [Google Scholar]
- 40.Miller, S. C. M., J. J. LiPuma, and J. L. Parke. 2002. Culture-based and non-growth-dependent detection of the Burkholderia cepacia complex in soil environments. Appl. Environ. Microbiol. 68:3750-3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moré, M. I., J. B. Herrick, M. C. Silva, W. C. Ghiorse, and E. L. Madsen. 1994. Quantitative cell lysis of indigenous microorganisms and rapid extraction of microbial DNA from sediment. Appl. Environ. Microbiol. 60:1572-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moser, R., and U. Stahl. 2001. Insights into the genetic diversity of initial dioxygenases from PAH-degrading bacteria. Appl. Microbiol. Biotechnol. 55:609-618. [DOI] [PubMed] [Google Scholar]
- 43.Ogunseitan, O. A., I. L. Delgado, Y.-L. Tsai, and B. H. Olson. 1991. Effect of 2-hydroxybenzoate on the maintenance of naphthalene-degrading pseudomonads in seeded and unseeded soil. Appl. Environ. Microbiol. 57:2873-2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olivares, F. L., V. L. D. Baldani, V. M. Reis, J. I. Baldani, and J. Döbereiner. 1996. Occurrence of the endophytic diazotrophs Herbaspirillum spp. in roots, stems, and leaves, predominantly of Gramineae. Biol. Fertil. Soil 21:197-200. [Google Scholar]
- 45.Palleroni, N. J. 1993. Pseudomonas classification. A new case history in the taxonomy of gram-negative bacteria. Antonie Leeuwenhoek 64:231-251. [DOI] [PubMed] [Google Scholar]
- 46.Parales, R. E., M. D. Emig, N. A. Lynch, and D. T. Gibson. 1998. Substrate specificities of hybrid naphthalene and 2,4-dinitrotoluene dioxygenase enzyme systems. J. Bacteriol. 180:2337-2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pellizari, V. H., S. Bezborodnikov, J. F. Quensen III, and J. M. Tiedje. 1996. Evaluation of strains isolated by growth on naphthalene and biphenyl for hybridization of genes to dioxygenase probes and polychlorinated biphenyl-degrading ability. Appl. Environ. Microbiol. 62:2053-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pieper, D. H., and W. Reineke. 2000. Engineering bacteria for bioremediation. Curr. Opin. Biotechnol. 11:262-270. [DOI] [PubMed] [Google Scholar]
- 49.Pitot, H. C., III, and Y. P. Dragan. 1995. Chemical carcinogenesis. In L. J. Cassarett, M. O. Amdur, and C. D. Klaassen (ed.), Cassarett and Doull's toxicology: the basic science of poisons, 5th ed. Macmillan Publishing Co., New York, N.Y.
- 50.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 51.Sayler, G. S., S. W. Hooper, A. C. Layton, and J. M. H. King. 1990. Catabolic plasmids of environmental and ecological significance. Microb. Ecol. 19:1-20. [DOI] [PubMed] [Google Scholar]
- 52.Siciliano, S. D., N. Fortin, A. Mihoc, G. Wisse, S. Labelle, D. Beaumier, D. Ouellette, R. Roy, L. G. Whyte, M. K. Banks, P. Schwab, K. Lee, and C. W. Greer. 2001. Selection of specific endophytic bacterial genotypes by plants in response to soil contamination. Appl. Environ. Microbiol. 67:2469-2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simon, M. J., T. D. Osslund, R. Saunders, B. D. Ensley, S. Suggs, A. Harcourt, W. C. Suen, D. L. Cruden, D. T. Gibson, and G. J. Zylstra. 1993. Sequences of genes encoding naphthalene dioxygenase in Pseudomonas putida strains G7 and NCIB 9816-4. Gene 127:31-37. [DOI] [PubMed] [Google Scholar]
- 54.Stanier, R. Y., N. J. Palleroni, and M. Douderoff. 1966. The aerobic pseudomonads: a taxonomic study. J. Gen. Microbiol. 53:1010-1019. [DOI] [PubMed] [Google Scholar]
- 55.Stuart-Keil, K. G., A. M. Hohnstock, K. P. Drees, J. B. Herrick, and E. L. Madsen. 1998. Plasmids responsible for horizontal transfer of naphthalene catabolism genes between bacteria at a coal tar-contaminated site are homologous to pDTG1 from Pseudomonas putida NCIB 9816-4. Appl. Environ. Microbiol. 64:3633-3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sutherland, J. B., F. Rafii, A. A. Khan, and C. E. Cerniglia. 1995. Mechanisms of polycyclic aromatic hydrocarbon degradation, p. 269-306. In L. Y. Young and C. E. Cerniglia (ed.), Microbial transformation and degradation of toxic organic chemicals. John Wiley & Sons, Inc., New York, N.Y.
- 57.Takizawa, N., N. Kaida, S. Torigoe, T. Moritani, T. Sawada, S. Satoh, and H. Kiyohara. 1994. Identification and characterization of genes encoding polycyclic aromatic hydrocarbon dioxygenase and polycyclic aromatic hydrocarbon dihydrodiol dehydrogenase in Pseudomonas putida OUS82. J. Bacteriol. 176:2444-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Treadway, S. L., K. S. Yanagimachi, E. Lankenau, P. A. Lessard, G. Stephanopoulos, and A. J. Sinskey. 1999. Isolation and characterization of indene bioconversion genes from Rhodococcus strain I24. Appl. Microbiol. Biotechnol. 51:786-793. [DOI] [PubMed] [Google Scholar]
- 59.van der Meer, J. R., W. M. DeVos, S. Harayama, and A. J. B. Zehnder. 1992. Molecular mechanisms of genetic adaptation to xenobiotic compounds. Microbiol. Rev. 56:677-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van der Meer, J. R., C. Werlen, S. F. Nishino, and J. C. Spain. 1998. Evolution of a pathway for chlorobenzene metabolism leads to natural attenuation in contaminated groundwater. Appl. Environ. Microbiol. 64:4185-4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Viallard, V., I. Poirier, B. Cournoyer, J. Haurat, S. Wiebkin, K. Ophel-Keller, and J. Balandreau. 1998. Burkholderia graminis sp. nov., a rhizospheric Burkholderia species, and reassessment of [Pseudomonas] phenazinium, [Pseudomonas] pyrrocinia, and [Pseudomonas] glathei into Burkholderia. Int. J. Syst. Bacteriol. 48:549-563. [DOI] [PubMed] [Google Scholar]
- 62.Widada, J., H. Nojiri, K. Kasuga, T. Yoshida, H. Habe, and T. Omori. 2002. Molecular detection and diversity of polycyclic aromatic hydrocarbon-degrading bacteria isolated from geographically diverse sites. Appl. Microbiol. Biotechnol. 58:202-209. [DOI] [PubMed] [Google Scholar]
- 63.Williams, P. A., and J. R. Sayers. 1994. The evolution of pathways for aromatic hydrocarbon oxidation in Pseudomonas. Biodegradation 5:195-217. [DOI] [PubMed] [Google Scholar]
- 64.Wilson, K. 1994. Preparation of genomic DNA from bacteria, p. 2.4.1-2.4.5. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, and K. Struhl (ed.), Current protocols in molecular biology, vol. 1. John Wiley & Sons, New York, N.Y. [DOI] [PubMed]
- 65.Wilson, M. S., C. Bakermans, and E. L. Madsen. 1999. In situ, real-time catabolic gene expression: extraction and characterization of naphthalene dioxygenase mRNA transcripts from groundwater. Appl. Environ. Microbiol. 65:80-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wilson, M. S., and E. L. Madsen. 1996. Field extraction of a transient intermediary metabolite indicative of real time in situ naphthalene biodegradation. Environ. Sci. Technol. 30:2099-2103. [Google Scholar]
- 67.Zhang, X., E. R. Sullivan, and L. Y. Young. 2000. Evidence for aromatic ring reduction in the biodegradation pathway of carboxylated naphthalene by a sulfate reducing consortium. Biodegradation 11:117-124. [DOI] [PubMed] [Google Scholar]
- 68.Zylstra, G. J., L. P. Wackett, and D. T. Gibson. 1989. Trichloroethylene degradation by Escherichia coli containing the cloned Pseudomonas putida F1 toluene dioxygenase genes. Appl. Environ. Microbiol. 55:3162-3166. [DOI] [PMC free article] [PubMed] [Google Scholar]