Abstract
Pseudomonas aeruginosa JB2 can use 2-chlorobenzoate (2-CBa), 3-CBa, 2,3-dichlorobenzoate (2,3-DCBa), and 2,5-DCBa as sole carbon and energy sources, whereas strain 142 can only grow on 2-CBa and 2,4-DCBa. Both strains, however, harbor the same halobenzoate 1,2-dioxygenase (ohbAB) and chlorocatechol (clcABD) degradation genes necessary for the metabolism of ortho-CBas. In addition, the hybABCD operon, encoding a salicylate 5-hydroxylase, is also found in both strains. The expression of ohbAB, hybABCD, and clcABD operons was measured in cultures grown on different CBas as the sole carbon source and also in glucose-grown cells supplemented with CBas as inducers. A method to standardize real-time reverse transcription-PCR experimental data was used that allows the comparison of semiquantitative mRNA accumulation in different strains and culture conditions. In both strains, the ohb and hyb systems were induced in cells grown on 2-CBa or DCBas, whereas clc was induced only by DCBas. Repression by catabolite was observed both on ohb and clc systems in glucose-grown cells. Chlorocatechol 1,2-dioxygenase activity in JB2 was detected even in clc-repressed conditions, confirming the presence of additional isofunctional genes previously detected in P. aeruginosa 142. Although similar levels of induction of ohbAB were observed in strain JB2 grown on either benzoate, monochlorobenzoates, or DCBas, the ohbAB operon of strain 142 was only strongly induced by growth on 2-CBa and, to a lesser extent, on 2,4-DCBa. This observation suggests that regulation of the ohbAB operon may be different in both strains. The concomitant induction of ohb and hyb by CBas may allow the formation of hybrid halobenzoate dioxygenase(s) composed of Ohb/Hyb dioxygenase subunits and Hyb ferredoxin/ferredoxin reductase components.
Chlorobenzoates (CBas) are intermediates produced during the aerobic degradation of polychlorinated biphenyls. Although ortho-substituted CBas appear to be particularly recalcitrant to biodegradation, several strains have been reported to utilize 2-CBa and dichlorobenzoate (DCBa) derivatives as a sole carbon and energy source (8-10, 16, 33). Degradation of 2-CBa is initiated by the dihydroxylation of the substrate by a halobenzoate 1,2-dioxygenase, which, after decarboxylation and dehalogenation at C2, yields catechol. When DCBas are used, the same enzyme activity results in the production of chlorocatechols. Both catechol and chlorocatechols are then channeled through the ortho-cleavage metabolic pathway to the tricarboxylic acid (TCA) cycle intermediates (7, 14, 20, 31). The halobenzoate 1,2-dioxygenase genes have been characterized in Burkholderia cepacia 2CBS (13), Burkholderia sp. strain TH2 (33), Pseudomonas aeruginosa 142 (34), and P. aeruginosa JB2 (17): in Burkholderia the enzyme belongs to the two-component dioxygenase family, similar to the benzoate/toluate dioxygenase enzyme encoded by xylXYZ, whereas a three-component dioxygenase, displaying significant similarity to the biphenyl dioxygenase of Novosphingobium aromaticivorans and salicylate 5-hydroxylase of Ralstonia sp. strain U2, has been reported for Pseudomonas. Both the 142 and the JB2 strains of P. aeruginosa harbor the ohbAB operon, which encodes the small (OhbA) and large (OhbB) subunits of the halobenzoate 1,2-dioxygenase component (6, 17, 34). However, the ohbAB operon does not include the genes for the ferredoxin and ferredoxin reductase necessary to build the three-component dioxygenase (29). In addition, a DNA region including the clcABDE operon for chlorocatechol ortho-cleavage pathway and the hybABCDEF operon, encoding the three components of a salicylate 5-hydroxylase and putative transport proteins, were also identified in both strains (6, 18).
Little is known about the regulation of the halobenzoate metabolic pathway in bacteria. In Burkholderia sp. strain TH2, the 2-halobenzoate dioxygenase operon, cbdABC, has been shown to be under the control of cbdS an araC/xylS-type regulatory gene, which is induced by 2-CBa, 2-methylbenzoate, and benzoate (33). In P. aeruginosa 142, we have previously reported the induction of ohbAB and hybABCD by 2-CBa and several DCBas, as well as the catabolite repression of ohbAB and clcABDE in cells grown on glucose (6). The catechol and chlorocatechol operons in P. putida have been shown to be controlled by CatR and ClcR, respectively, which are LysR-type transcriptional regulators induced by cis,cis-muconate and 2-chloro-cis,cis-muconate (2CM), the products of the 1,2-dioxygenase activity on catechol and chlorocatechol, respectively (2, 3, 25; reviewed in reference 24). Similar observations have been made for the closely related cbnRAB operon of Ralstonia eutropha NH9 (27).
The range of CBas that support growth differs in P. aeruginosa 142 and JB2: strain 142 can use 2-CBa and 2,4-DCBa but not 3-CBa, 2,3-DCBa, or 2,5-DCBa as a carbon and energy source, whereas JB2 has a broader substrate range, which includes 2-CBa, 3-CBa, 2,3-DCBa, and 2,5-DCBa but not 2,4-DCBa. It has been shown that the dioxygenase component encoded by ohbAB purified from strain 142 may dihydroxylate a range of CBas that do not support growth (29). This indicates that other factors, in addition to enzyme specificity, play a role in the ability of these strains to use any given CBa. The effect of CBas, or their metabolic intermediates, on the induction of the catabolic operons, inefficient CBa uptake, or the presence of alternate genes for CBa metabolism may account for these discrepancies, and explain the differences found in the range of growth substrates between the two strains. We have made use here of real-time reverse transcription-PCR (RT-PCR) to further identify the CBas that trigger the expression of ohb, hyb, and clc operons in both strains. In addition, we present data suggesting the presence of an alternative (chloro)catechol 1,2-dioxygenase system which does not belong to the clc/tfd/tfc family.
MATERIALS AND METHODS
Bacterial strains, culture media, and growth conditions.
Cultures of P. aeruginosa 142 (30) and JB2 (16) were grown aerobically at 37°C either on 50 ml of mineral media (MM) (15) or the same medium containing 10 mM glucose (MMG). Both MM and MMG were supplemented when required with 2 mM (chloro)benzoates. All cultures were started by adding 0.25 ml of a previous culture, grown on MM supplemented with 2-CBa, to 100 ml of fresh medium in 500-ml Erlenmeyer flasks. Growth was monitored spectrophotometrically at 550 nm.
DNA isolation and PCR conditions.
Bacterial DNA was obtained according to the method described by Ausubel et al. (1) from 200-ml cultures. PCRs were carried out in reaction mixtures of 50 μl containing ca. 100 ng of total DNA. The PCR primers were designed to have a Tm in the 58 to 60°C range. Amplification was carried out by using Applied Biosystems AmpliTaq Gold DNA polymerase according to the manufacturer's instructions and a Gene-Amp PCR system 2700. The PCR program consisted of a 9-min hot start at 95°C, followed by 30 cycles of denaturation (30 s at 94°C), annealing (30 s at 58°C), and extension (1 min at 72°C), with final extension of 10 min at 72°C. Low-stringency PCR was carried out by lowering the annealing temperature to 45°C.
Nucleotide sequence determination.
The PCR products obtained were further purified by filtration through Chroma-Spin 100 columns (Clontech), and both strands of the amplified fragments were sequenced either directly by using the same oligonucleotides as primers or after being subcloned into a pGEM-T vector (Promega) by using Applied Biosystems sequencing kits and a model 377 automatic sequencer. The clcABD sequences found in strain JB2 was deposited at GenBank under accession number AF164958.
Assay for (chloro)catechol 1,2-dioxygenase activity.
The activity was monitored as the accumulation of cis,cis-muconate in the presence 10 mM EDTA. Cell extracts were prepared from 100-ml cultures grown to late exponential phase and processed as described previously (29).
RNA isolation and quantitation by real-time PCR.
Total RNA was purified from a 1.5-ml sample of cultures grown to late exponential phase by using an isolation column (Qiagen RNeasy). Residual DNA was removed by incubation with 50 U of RNase-free DNase (Roche) for 30 min at 25°C, followed by heat inactivation for 15 min at 95°C. Total RNA was quantified after being stained with SYBR green II (Molecular Probes) by using a Perkin-Elmer LS-50B fluorimeter, and its integrity was checked by electrophoresis in 1.5% agarose gels. RT was performed with 20 ng of total RNA incubated with 1 U of avian myeloblastosis virus reverse transcriptase (Promega)/μl in the presence of 250 μM concentrations of random hexamers and 1 U of RNase inhibitor (RNasin; Roche)/μl for 50 min at 42°C. RNA quantitation was performed by amplification of the cDNA obtained from the reverse transcriptase reaction in the real-time sequence detection system 7700 from Applied Biosystems. The PCR primers used for 16S rRNA measurements were 16SF (CAGGATTAGATACCCTGGTAGTCCAC) and 16SR (GACTTAACCCAACATCTCACGACAC), based on the sequence of P. aeruginosa 16S rRNA (AF237678). Transcripts of ohbA, hybA, and clcA were quantified with the following primer pairs, respectively: ohbAF (TAGTCCGCTGAAGGCGATACA) and ohbAR (TCTGGTAGCTGCCGTCTTCG), based on the ohbA sequence of strain 142 (AF121970); hybAF (TGACTATGTCCTGGCCTGCA) and hybAR (AACGGCTTGGCAGGTTTAAG), based on hybA from strain JB2 (AF087482); and clcAF (AAACTCGTGACGGATTCCCAG) and clcAR (GCAGTCATCGTCCACCCATTT), which match conserved internal clcA sequences found in plasmid pAC27 (11) and strains 142 (AF161263) and JB2 (AF164958 and AF087482).
Real-time PCRs were carried out in 96-well plates. The 25-μl reaction mixtures contained 12.5 μl of SYBR Green PCR master mix (which includes the heat-activated TaqGold enzyme [Applied Biosystems]), 200 μM concentrations of each specific oligonucleotide primer, and 2 μl of different dilutions of the reverse transcriptase product, corresponding to 0.2 to 2 ng of total RNA. The reaction conditions were as follows: 10 min at 95°C for enzyme activation and 40 cycles of 1 min at 95°C and 30 s at 60°C. Fluorescence due to the binding of the SYBRgreen fluorochrome to double-stranded DNA was measured twice per cycle. The increment in fluorescence versus reaction cycle was plotted, and the threshold cycle value (CT) (19) was obtained by positioning of the threshold baseline at the mid-exponential phase of the curve. CT can be defined as:
 |
(1) |
where E is the PCR efficiency, T0 is the initial amount of DNA, and K is the amount of DNA at the CT cycle. A plot of CT versus logT0 would yield a straight line in which the slope equals −1/logE.
For accurate comparison of real-time PCR results obtained with different primer pairs, it is necessary to assure that PCR efficiency is similar among the different reactions. Prior to the actual measurements, PCR efficiency for each primer pair used in this work was checked from the slopes obtained from real-time PCR runs by using serial dilutions of genomic DNA. The slopes obtained by using the 16SF-16SR, ohbAF-ohbAR, hybAF-hybAR, and clcAF-clcAFR primer pairs were similar: −1/log E = −3.44 ± 0.2, allowing the comparison of results.
The conversion of CT values to standardized mRNA equivalents was performed as follows. A reference curve was defined by using the equation:
 |
(2) |
from which an mRNA equivalent (R) was defined as the amount of mRNA that yielded a CT of 40 under our experimental conditions. To account for the variations in the positioning of the threshold baseline between different CT measurements, a 10-fold dilution series of 1 ng to 0.01 pg of total RNA μl−1 from a culture grown on 2-CBa was included in every PCR plate and amplified with the 16SF-16SR primers. For all of the reactions in the plate, a new set of corrected CTS were calculated as follows:
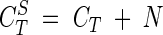 |
(3) |
where N is the difference between the experimental 16S rRNA CT and the 16S rRNA CT calculated from the reference line by using equation 2. CTS values were obtained for all transcripts analyzed in the same plate and, subsequently, used to calculate the corresponding mRNA equivalents extrapolated from the reference curve (RS). Finally, the standardized mRNA equivalents (R′) used in this work were obtained from the equation:
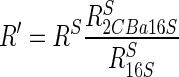 |
(4) |
where RS, RS16S, and RS2CBa16S are the values found by extrapolation from the CTS on the reference curve for the transcript analyzed, the 16S rRNA under the same growth conditions, and the 16S rRNA of the culture grown on 2-CBa, respectively. R′ represents a measure of the abundance of the target transcript normalized to the abundance of the 16S rRNA under the same growth conditions. Mean values were obtained from at least three experiments.
RESULTS AND DISCUSSION
Identification of ohbAB, hybABCDEF, and clcABDE in P. aeruginosa 142 and JB2.
Previous work had indicated the presence of the ohbAB gene cluster in JB2 (17), as well as hybABCDE and clcABD genes in P. aeruginosa 142 (6). The presence of the complete operons in our lab isolates was confirmed by PCR amplification and DNA sequencing of fragments of ca. 200 bp selected from each hybABCDE and ohbAB open reading frame (data not shown) and the complete sequencing of the clcABD genes of JB2. The nucleotide sequences determined for all three gene clusters differed <2% between both strains. The sequence of the clcABD genes found in JB2 was not identical to the one previously posted by others in the GenBank database (AF087482) and, therefore, was cataloged as a separate entry (AF164958). The physical genetic arrangement of the three operons, including the regions sequenced, is shown in Fig. 1. The oligonucleotide pairs ohbAF-ohbAR, hybAF-hybAR, and clcAF-clcAR were selected as primers for real-time RT-PCRs.
FIG. 1.
Map of the ohb (A), hyb (B), and clc (C) gene clusters in P. aeruginosa strains 142 and JB2. The arrows indicate gene orientations. Putative transposition, regulatory, and transport-related genes are represented by hatched, dotted, and empty arrows, respectively. The proteins encoded by ohbA and ohbB are the small and large subunits of the 2-halobenzoate 1,2-dioxygenase. ohbR is the gene for Lys-R type of regulatory protein. hybA, hybB, hybC, and hybD encode, respectively, the ferredoxin reductase, the α and β dioxygenase subunits, and the ferredoxin of the salicylate 5-hydroxylase. hybE, hybF, and hybG encode members of an ABC-type membrane transport system. The gene products of hybR and hybH show similarities to Lys-R type regulators. top and tnpA are the topoisomerase and transposase genes, respectively. The approximate positions of fragments amplified by RT-PCR are indicated as gray boxes.
Expression of 16SrRNA genes in cultures grown on glucose-supplemented media.
It is well established that the bacterial ribosome content, and therefore the amount of 16S rRNA, increases with the growth rate of the culture (5, 26). In order to use the expression of the 16S rRNA as a reference for the quantitation of target mRNA in cultures grown in the presence of glucose, we calculated experimentally the ratio of the average 16S rRNA produced in cultures grown on glucose plus CBa versus the average of 16S rRNA produced in cultures grown on CBa. A standard CT versus the log RNA curve was constructed by using as a template total RNA from eight different cultures of JB2 and 142, grown on MM plus CBa, and the 16SRNAF-16SRNAR primers. The curve was defined by the equation:
 |
(5) |
where T16S is the amount of total RNA (in picograms) used as a template.
The average CT obtained from 1 ng of total RNA from cultures grown on glucose plus CBa was 17.97 ± 1.40. This value, translated to the standard curve, corresponds to 2.8 ng of total RNA. This indicates that, for equal amounts of total RNA isolated from P. aeruginosa, there was a 2.8-fold increase in 16S rRNA in cells grown on glucose-supplemented medium. This factor was subsequently used for the correction of R′ mRNA data obtained from glucose-supplemented cultures.
Expression of ohbAB and hybABCDEF in P. aeruginosa 142 and JB2 grown on different CBas.
The expression of ohbAB and hybABCDEF mRNA was measured as the accumulation of the transcripts of the first gene for each operon, ohbA and hybA. Although, due to possible posttranscriptional processes, this would not necessarily reflect the actual amounts of the enzymes in the cell, it may be an accurate indication of the regulation of expression at the promoter level of these operons. Expression was measured in cultures of P. aeruginosa 142 grown on MM supplemented with 2-CBa and 2,4-DCBa, whereas in JB2 the cultures were grown on 2-CBa and 3-CBa, as well as on 2,3-DCBa and 2,5-DCBa (Fig. 2). The induction of the operons show several similarities in both strains: 2-CBa and DCBas can induce both ohb and hyb systems, whereas ohb (but not hyb) appears to be repressed in glucose-supplemented cultures. These results confirm our previous data obtained for P. aeruginosa 142 by using a different experimental approach (6) and extend the findings to a different CBa degrader strain. In spite of these similarities, several details point to the existence of differences in the patterns of induction between the strains. Expression of ohbA with 2-CBa in strain 142 reaches higher values than in strain JB2, whereas similar levels of induction are found for hybA with this substrate. In addition, benzoate is a clear inducer for ohb in JB2 but not in strain 142. Furthermore, we could not detect induction of either ohb or hyb in cultures growing on glucose supplemented with nonsubstrate CBas (3-CBa or 2,3-DCBa for strain 142; 2,4-DCBa for strain JB2 [data not shown]). These differences between P. aeruginosa JB2 and 142 suggest that the regulator proteins of these operons may not be identical (or behave identically) in both strains. As it has been recently shown, the oxygenase encoded by hybABCD is actually a salicylate hydroxylase, which does not display any activity on CBas (18). The induction of hybABCDE by these nongrowth substrates might be explained as a low-specificity interaction of the CBas with the salicylate regulator. However, the involvement of the hyb operon with CBa degradation cannot be ruled out: the flavoprotein, HybA, and the ferredoxin, HybD, components of the hydroxylase may combine with the large and small subunits and of the dioxygenase component encoded by ohbA and ohbB. It has been shown that the combination of OhbA or OhbB with unknown ferredoxin and ferredoxin reductase components of Escherichia coli yields a fully active ring-hydroxylating oxygenase (17, 34); it is therefore likely that the concomitant induction of ohbAB and hybABCD by the CBas in P. aeruginosa 142 and JB2 may favor the formation of OhbAB-HybAD hybrids. Moreover, two other combinations may arise, such as OhbA-HybABD or OhbB-HybACD. Changes in substrate specificity have been reported in similar component shuffling events involving elements of the toluene (tod) and biphenyl (bphA) operons for three-component dioxygenases (12). The implication of these possible oligomer rearrangements with the range of CBas that can be used as substrates remains to be elucidated.
FIG. 2.
Expression of ohb and hyb operons in P. aeruginosa strains 142 and JB2 grown in the presence of different CBas. Solid bars indicate cultures grown in the absence of glucose; shaded bars indicate cultures grown in MMG supplemented with CBas. The standardized mRNA equivalents R′ (see Materials and Methods) were obtained from RT-PCRs by using the ohbAF-ohbAR and hybAF-hybAR primer pairs. Cells were collected at late exponential stage (optical densities at 550 nm of 0.8 to 1.0 for glucose-grown cells and of 0.5 to 0.7 for CBa-grown cells).
The repression of ohbAB in the presence of glucose indicates that this operon is, at least, one of the catabolic repression systems proposed for Pseudomonas (35). The hybABCDE operon, on the contrary, appears to elude such repression. Since gene expression was measured in cells grown to the late exponential phase, we wondered whether another global regulation system, dependent on the growth phase (21), plays a role in the induction of ohb and hyb operons in glucose-supplemented media. To test this possibility, samples of cultures grown on MMG plus 2-CBa, 3-CBa, 2,3-DCBa, or 2,5-DCBa were collected both at mid-exponential phase (optical density at 550 nm of 0.6) and 24 h after the cultures reached saturation. The average ratio of mRNA equivalents accumulated in late log phase versus stationary phase was 1.17 ± 0.9 for ohbA and 0.07 ± 0.02 for hybA, indicating that the presence of hybA transcript is strongly reduced once the culture stops growing, whereas similar amounts of ohbA transcript were detected in both growth stages. This result suggests that ohb and hyb are mainly expressed during the exponential growth phase, and the induction of the hyb system in glucose-grown cells is due to the lack of catabolite repression, rather than to a stationary-phase-related regulation system.
Induction of the ortho-cleavage chlorocatechol metabolic pathway.
The expression of the clcABD operon in strains 142 and JB2 was measured by real-time PCR by using the primers clcAF-clcAR, which match identical internal sequences of the clcA gene found in plasmid pAC27 (11), and strains 142 (AF161263) and JB2 (AF164958 and AF087482). Strain JB2 again displayed less accumulation of clcABD mRNA than did strain 142 (Fig. 3). In addition, DCBas appear to be better inducers than 2-CBa. This was the expected result, since it has been shown that clcABDE is induced by 2CM in P. putida (22), and this intermediate results from the ortho-ring cleavage of chlorocatechols produced by a 2-halobenzoate dioxygenase activity on DCBas. However, 3-CBa, which also yields 2CM from a benzoate dioxygenase activity (16), is a poor inducer for the clc operon in strain JB2. Since strain JB2 can use 3-CBa as sole carbon source, either 3-CBa is channeled through a different pathway or there is an alternate ortho-cleavage system available for the degradation of chlorocatechols. We have previously proposed the existence of such an alternate system for P. aeruginosa 142, based on the detection of chlorocatechol 1,2-dioxygenase activity in glucose-grown cells (6). This activity, which is not under catabolite repression, was also detected in the strain JB2 growing on 3-CBa (Table 1), suggesting that P. aeruginosa may harbor a second ortho-cleavage system regulated by different control networks. Since no additional PCR amplification bands or unresolved base positions were found during the sequencing of clcABD, it seems likely that the gene sequence of this alternate (chloro)catechol 1,2-dioxygenase is quite divergent from clcA and would not be amplified or detected in the RT-PCRs. In addition, the alternate dioxygenase shows activity on both catechol and 3-chlorocatechol, whereas the activity induced by 2,3-DCBa appears to be specific for 4-chlorocatechol (Table 1). These results may be explained assuming that the pathway for 2,3-DCBa produces an intermediate (putatively 2CM), which acts as inducer of the clc system, whereas 3-CBa would be channeled through an alternate pathway that does not yield a good inducer for clc but, conversely, induces the expression of the alternate catechol 1,2-dioxygenase. Growth on 2,5-DCBa appears to share the characteristics of both mechanisms. To test the presence of the alternate chlorocatechol metabolic genes in strains JB2 and 142, we carried out PCRs under nonstringent conditions with primers designed on other genes of the clcA family: Rhodococcus opacus clcA, plasmid p51 tcbC, and plasmid pJP4 tfdCI and tfdCII. No amplification could be detected, suggesting that this putative alternate system may be structurally different from the clcA dioxygenase family.
FIG. 3.
Expression of the clc operons in P. aeruginosa 142 (A) and JB2 (B) strains grown in different CBas as the sole carbon source (solid bars) or in glucose supplemented media (shaded bars). R′ was calculated from RT-PCRs by using clcAF-clcAR primer pairs.
TABLE 1.
Catechol and chlorocatechol 1,2-dioxygenase activities in cell extracts of P. aeruginosa JB2
| Growth substrate | Catechol 1,2-dioxygenase activity (nmol min−1 mg−1) on substratea:
|
||
|---|---|---|---|
| Catechol | 3-Chlorocatechol | 4-Chlorocatechol | |
| 2-CBa | 35 | 2 | 4 |
| 3-CBa | 19 | 19 | 9 |
| 2,3-DCBa | ND | 3 | 54 |
| 2,5-DCBa | 12 | 14 | 26 |
| Benzoate | 23 | 2 | 3 |
| Glucose | ND | ND | ND |
| 2-CBa + glucose | 8 | 1 | 10 |
| 3-CBa + glucose | 40 | 57 | 23 |
| 2,3-DCBa + glucose | 5 | 8 | 13 |
| 2,5-DCBa + glucose | 6 | 3 | 2 |
| Benzoate + glucose | 5 | 2 | 2 |
Average activity of three independent determinations with a standard deviation of <20% of the given values.
The results shown in the Table 1 also indicate the presence of a third catechol 1,2-dioxygenase activity induced by 2-CBa and benzoate that does not display activity on chlorocatechols. This activity may correspond to the catABD operon detected in P. aeruginosa (32).
The mechanisms for catabolite repression in Pseudomonas sp. are not yet well understood. In P. putida and P. aeruginosa, the Crc protein is involved in the repression of genes implicated in the metabolism of some sugars and nitrogenated compounds (4), but it appears to be active in rich media or in MM supplemented with amino acids rather than in sugar-grown cells (35). In a previous report (23), it was shown that TCA cycle intermediates repressed the expression of clc in P. putida. Based on the comparatively high levels of induction on glucose, gluconate, or pyruvate, it was proposed that none of these three compounds was involved in catabolite repression. However, no data was provided on the clc expression in cells growing only on CBa and, consequently, a possible effect of glucose on repression could not be ruled out. In the same study, it was shown that fumarate behaves in vitro as an anti-inducer of the clc operon in P. putida, probably by binding to the regulator protein, ClcR, and preventing the interaction with the inducer molecule, 2CM. Compared to cells growing on 2 mM CBa, cells growing on 10 mM glucose-supplemented medium may have higher levels of TCA cycle intermediates, which would lead to repression of the clc system. The repression of ohb under the same conditions might also be mediated by TCA cycle intermediates.
The recruitment of closely related catabolic systems in a single strain by, most likely, horizontal gene transfer is a key feature for the utilization of novel substrates as carbon and energy sources. The three genetic clusters analyzed in this work are placed in the neighborhood of transposable elements, and direct evidence of the transfer of ohbAB and hybABCD has been reported (17, 28). The data presented here suggest that the transfer of these operons among different strains may lead to changes in their transcriptional regulation characteristics, and these changes may affect the ability to use a range of substrates for growth.
Acknowledgments
This work was supported in part by grants BIO94-0471 (from the Spanish Comisión Intrerministerial de Ciencia y Tecnología) and CAM 07 M/0450.
We thank A. Méndez for technical assistance. DNA sequencing and quantitative PCR were performed at the Genomics and Proteomics Core Service Facility of the UCM.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1991. Current protocols in molecular biology, vol. 1. John Wiley & Sons, Inc., New York, N.Y.
- 2.Chugani, S. A., M. R. Parsek, and A. M. Chakrabarty. 1998. Transcriptional repression mediated by LysR-type regulator CatR bound at multiple binding sites. J. Bacteriol. 180:2367-2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chugani, S. A., M. R. Parsek, C. D. Hershberger, K. Murakami, A. Ishihama, and A. M. Chakrabarty. 1997. Activation of the catBCA promoter: probing the interaction of CatR and RNA polymerase through in vitro transcription. J. Bacteriol. 179:2221-2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collier, D. N., P. W. Hager, and J. P. V. Phibbs. 1996. Catabolite repression control in pseudomonads. Res. Microbiol. 147:551-561. [DOI] [PubMed] [Google Scholar]
- 5.Condon, C., C. Squires, and C. L. Squires. 1995. Control of rRNA transcription in Escherichia coli. Microbiol. Rev. 59:623-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corbella, M. E., A. Garrido-Pertierra, and A. Puyet. 2001. Induction of the halobenzoate catabolic pathway and cometabolism of ortho-chlorobenzoates in Pseudomonas aeruginosa 142 grown on glucose-supplemented media. Biodegradation 12:149-157. [DOI] [PubMed] [Google Scholar]
- 7.Dorn, E., and H. J. Knackmuss. 1978. Chemical structure and biodegradabiity of halogenated aromatic compounds: two catechol-1,2-dioxygenases from a 3-chlorobenzoate-grown pseudomonad. Biochem. J. 174:73-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engesser, K. H., and P. Schulte. 1989. Degradation of 2-bromo-, 2-chloro-, and 2-fluorobenzoate by Pseudomonas putida CLB250. FEMS Microbiol. Lett. 60:143-148. [DOI] [PubMed] [Google Scholar]
- 9.Fava, F., F. Baldoni, and L. Marchetti. 1996. 2-Chlorobenzoic acid and 2,5-dichlorobenzoic acid metabolism by crude extracts of Pseudomonas sp. CPE2 strain. Lett. Appl. Microbiol. 22:275-279. [DOI] [PubMed] [Google Scholar]
- 10.Fetzner, S., R. Muller, and F. Lingens. 1989. Degradation of 2-chlorobenzoate by Pseudomonas cepacia 2CBS. Biol. Chem. Hoppe-Seyler 370:1173-1182. [DOI] [PubMed] [Google Scholar]
- 11.Frantz, B., and A. M. Chakrabarty. 1987. Organization and nucleotide sequence determination of a gene cluster involved in 3-chlorocatechol degradation. Proc. Natl. Acad. Sci. USA 84:4460-4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Furukawa, K., J. Hirose, S. Hayashida, and K. Nakamura. 1994. Efficient degradation of trichloroethylene by a hybrid aromatic ring dioxigenase. J. Bacteriol. 176:2121-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haak, B., S. Fetzner, and F. Lingens. 1995. Cloning, nucleotide sequence, and expression of the plasmid-encoded genes for the two-component 2-halobenzoate 1,2-dioxygenase from Pseudomonas cepacia 2CBS. J. Bacteriol. 177:667-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hardwood, C. S., and R. E. Parales. 1996. The β-ketoadipate pathway and the biology of self-identity. Annu. Rev. Microbiol. 50:553-590. [DOI] [PubMed] [Google Scholar]
- 15.Hareland, W. A., R. L. Crawford, P. J. Chapman, and S. Dagley. 1975. Metabolic function and properties of 4-hydroxyphenilacetic acid 1-hydroxylase from Pseudomonas acidovorans. J. Bacteriol. 121:272-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hickey, W., and D. D. Focht. 1990. Degradation of mono-, di-, and trihalogenated benzoic acids by Pseudomonas aeruginosa JB2. Appl. Environ. Microbiol. 56:3842-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hickey, W. J., and G. Sabat. 2001. Integration of matrix-assisted laser desorption ionization-time of flight mass spectrometry and molecular cloning for the identification and functional characterization of mobile Pseudomonas aeruginosa strain JB2. Appl. Environ. Microbiol. 67:5648-5655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hickey, W. J., G. Sabat, A. S. Yuroff, A. R. Arment, and J. Pérez-Lesher. 2001. Cloning, nucleotide sequencing, and funtional analysis of a novel, mobile cluster of biodegradation genes from Pseudomonas aeruginosa strain JB2. Appl. Environ. Microbiol. 67:4603-4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Higuchi, R., C. Fockler, G. Dollinger, and R. Watson. 1993. Kinetic PCR analysis: real-time monitoring DNA amplification reactions. Biotechnology 11:1026-1030. [DOI] [PubMed] [Google Scholar]
- 20.Kuhm, A. E., N. Schlömann, H. J. Knackmuss, and D. H. Pieper. 1990. Purification and characterization of dichloromuconate cycloisomerase from Alcaligenes eutrophus JMP134. Biochem. J. 266:877-883. [PMC free article] [PubMed] [Google Scholar]
- 21.Marqués, S., M. Manzanera, M. M. González-Pérez, M. T. Gallegos, and J. L. Ramos. 1999. The XylS-dependent Pm promoter is transcribed in vivo by RNA polymerase with σ32 or σ38 depending on the growth phase. Mol. Microbiol. 31:1105-1113. [DOI] [PubMed] [Google Scholar]
- 22.McFall, S. M. 1997. 2-Chloromuconate and ClcR-mediated activation of the clcABD operon: in vitro transcriptional and DNase I footprint analyses. J. Bacteriol. 179:3655-3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McFall, S. M., B. Abraham, C. G. Narsolis, and A. M. Chakrabarty. 1997. A tricarboxylic acid cycle intermediate regulating transcription of a chloroaromatic biodegradative pathway: fumarate-mediated repression of the clcABD operon. J. Bacteriol. 179:6729-6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McFall, S. M., S. A. Chugani, and A. M. Chakrabarty. 1998. Transcriptional activation of the catechol and chlorocatechol operons: variations on a theme. Gene 223:257-267. [DOI] [PubMed] [Google Scholar]
- 25.McFall, S. M., T. J. Klem, N. Fujita, A. Ishihama, and A. M. Chakrabarty. 1997. DNase I footprinting, DNA bending and in vitro transcription analyses of ClcR and CatR interactions with the clcABD promoter: evidence of a conserved transcriptional activation mechanism. Mol. Microbiol. 24:965-976. [DOI] [PubMed] [Google Scholar]
- 26.Nomura, M., R. Gourse, and G. Baughman. 1984. Regulation of the synthesis of ribosomes and ribosomal components. Annu. Rev. Biochem. 53:75-117. [DOI] [PubMed] [Google Scholar]
- 27.Ogawa, N., S. M. McFall, T. J. Klem, K. Miyashita, and A. M. Cakrabarty. 1999. Transcriptional activation of the chlorocatechol degradative genes of Ralstonia eutropha NH9. J. Bacteriol. 181:6697-6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pérez-Lesher, J., and W. J. Hickey. 1995. Use of an s-triazine nitrogen source to select for and isolate a recombinant chlorobenzoate-degrading Pseudomonas. FEMS Microbiol. Lett. 133:47-52. [Google Scholar]
- 29.Romanov, V., and R. T. Hausinger. 1994. Pseudomonas aeruginosa 142 uses a three-component ortho-halobenzoate 1,2-dioxygenase for metabolism of 2,4-dichloro- and 2-chlorobenzoate. J. Bacteriol. 176:3368-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Romanov, V. P., G. M. Grechkina, V. M. Adanin, and I. I. Starovoitov. 1993. Oxidative dehalogenation of 2-chlorobenzoate and 2,4-dichlorobenzoate by Pseudomonas aeruginosa. Microbiology 62:532-536. [PubMed] [Google Scholar]
- 31.Schmidt, E., and H.-J. Knackmuss. 1980. Chemical structure and biodegradability of halogenated aromatic compounds: conversion of chlorinated muconic acids into maleoylacetic acid. Biochem. J. 192:339-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, and I. T. Paulsen. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 33.Suzuki, K., N. Ogawa, and K. Miyashita. 2001. Expression of 2-halobenzoate dioxygenase genes (cbdSABC) involved in the degradation of benzoate and 2-halobenzoate in Burkholderia sp. TH2. Gene 262:137-145. [DOI] [PubMed] [Google Scholar]
- 34.Tsoi, T. V., E. G. Plotnikova, J. R. Cole, W. F. Guerin, M. Bagdasarian, and J. M. Tiedje. 1999. Cloning, expression, and nucleotide sequence of the Pseudomonas aeruginosa 142 ohb genes coding for oxygenolytic ortho dehalogenation of halobenzoates. Appl. Environ. Microbiol. 65:2151-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuste, L., and F. Rojo. 2001. Role of the crc gene in catabolic repression of the Pseudomonas putida GPo1 alkane degradation pathway. J. Bacteriol. 183:6197-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]





