Abstract
Strains of the sweet potato soil rot pathogen Streptomyces ipomoeae had previously been divided into three groups based on their ability to inhibit one another during pairwise cocultivation. While group I strains are not antagonistic to members of the other groups, group II and group III strains produce separate substances that are inhibitory to strains outside their respective cognate groups. Here, we purified the group III inhibitory substance from the culture supernatant of a representative strain and found that it consists of a single 10-kDa cationic protein which is bacteriolytic for S. ipomoeae group I and II strains but which showed no inhibitory function against other streptomycetes or other bacterial genera tested. The structural gene for the inhibitor was cloned from a chromosomal library of the producing strain, and while the gene sequence revealed that the inhibitor is initially made in a larger precursor form, the deduced mature protein showed no significant homology to other known proteins. Our results demonstrate that S. ipomoeae group III inhibitory activity is manifested in the form of a highly specific, potentially novel bacteriocin, which we have designated ipomicin.
The gram-positive bacterial genus Streptomyces is comprised of over 500 mainly soil-dwelling saprophytic species that display a differentiating life cycle (19). Vegetative growth of a filamentous substrate mycelium precedes depletion of nutrients, with the latter signaling a requisite switch in growth involving the vertical projection of individual aerial hyphae, which ultimately become spore chains (7). This directional growth switch is associated with the production and release of secondary metabolites, including antibiotics. Such molecules are derived from complex biosynthetic pathways and probably offer a competitive advantage to producing strains as nutrients become scarce (6).
A few Streptomyces species cause parasitic plant diseases. Streptomyces scabiei, the most thoroughly studied and economically important Streptomyces pathogen, infects the underground tubers of potatoes (causing common scab disease) as well as similarly infecting underground structures of other plant species (20). The ability of streptomycetes to cause disease is correlated with the production of thaxtomins, a family of low-molecular-weight phytotoxic compounds (20).
Another Streptomyces plant pathogen, Streptomyces ipomoeae, causes soil rot, which is a destructive scab disease of the sweet potato plant Ipomoea batatas (L.) Lam (12). Hallmarks of the disease include decay (rotting) of fibrous feeder roots as well as the development of necrotic lesions on the fleshy storage roots (11). S. ipomoeae also infects other members of the plant family Convolvulaceae (13) but does not naturally infect potato. Besides differences in host range and disease etiology, S. ipomoeae differs from S. scabiei and other plant-pathogenic Streptomyces species in the spectrum of thaxtomin compounds produced and potentially in the biosynthetic pathway used for thaxtomin production (14, 18) as well as in the presence of certain other pathogenicity-associated genetic loci (5).
Prevention of soil rot has relied primarily on the development of resistant plant cultivars; however, resistance has shown some variability (9), an effect that may be influenced by as-yet-undetermined environmental and genetic factors. With the goal of developing alternative strategies for disease management, we previously began a study of genetic variation in S. ipomoeae by focusing on several genotypic and phenotypic characteristics (8). One phenotype characterized by inhibitory interactions seen during pairwise cocultivation of strains on agar plates led to the organization of 36 S. ipomoeae strains into three “inhibition groups.” Specifically, while group I strains are unable to inhibit any other strains, group II strains can inhibit members of both group I and group III, and similarly, group III strains can inhibit both group I and group II strains.
Here, we further examined the inhibition phenomenon by purifying an interstrain inhibitory substance from an S. ipomoeae group III strain. We found that the group III inhibitor consists of a highly stable cationic 10-kDa protein which is released into the culture supernatant and which is bacteriolytic for only sensitive S. ipomoeae strains and not other streptomycete species or other bacterial genera examined. A degenerate oligonucleotide derived from a partial amino acid sequence of this protein was used to isolate its structural gene from a genomic cosmid library. The nucleotide sequence of the gene revealed that the protein is initially made in a 13-kDa precursor form that includes an N-terminal signal sequence, which is then apparently removed prior to release of the 10-kDa inhibitor from the cell. This antimicrobial protein, which can be classified as a bacteriocin due to its proteinaceous ribosomally derived nature, shows no significant homology to other known proteins and represents just the second example (the other being from Streptomyces virginiae [24]) of a bacteriocin produced by a streptomycete.
MATERIALS AND METHODS
Bacterial strains, bacteriological methods, and plasmids.
The pathogenic S. ipomoeae strains 78-61 (group I), 88-35 (group II), 91-03, and 88-03 (both group III) have been described (8) and were preserved on silica gel crystals (22) and revived on S. ipomoeae growth agar (SIGA) (8, 10). Escherichia coli hosts for cloning were DH10B (Life Technologies Inc.) and XL1-Blue MR (Stratagene). S. scabiei strains were provided by D. Lambert (University of Maine, Orono) and were preserved and revived as indicated for S. ipomoeae. “Streptomyces cyanogenus” strain NRRL B-12354 was obtained as a lyophilized stock from the Agricultural Research Service Culture Collection (Peoria, Ill.). Streptomyces rochei strain 7434-AN4 (15), Streptomyces lividans TK23, Streptomyces coelicolor A3(2) (17), and Mycobacterium smegmatis mc2155 (26) have been described. All other bacteria were obtained as plate or slant cultures from the Louisiana State University undergraduate microbiology teaching laboratory. Transformation of E. coli was performed as previously described (25). Streptomyces species were cultured on SIGA or grown in tryptic soy broth (TSB) (Becton, Dickinson, and Company) at 30°C with shaking (300 rpm) unless otherwise noted. Tryptic soy agar (Becton, Dickinson, and Company) was used to culture Alcaligenes faecalis, Staphylococcus aureus, Enterobacter aerogenes, and Proteus vulgaris (all at 37°C), as well as Pseudomonas aeruginosa (at 30°C) and Serratia marcescens (at 25°C). E. coli and Enterococcus faecalis were cultured on Luria-Bertani (LB) agar (25) at 37°C, while Bacillus subtilis was grown at 37°C on nutrient agar (Becton, Dickinson, and Company), and Streptococcus strain B was cultured on Trypticase soy agar containing 5% sheep's blood (Becton, Dickinson, and Company) in a candle jar at 37°C.
The recombinant cosmid pSIP1 consists of an approximately 35-kb Sau3AI genomic fragment from S. ipomoeae strain 91-03 cloned into the cosmid vector pOJ446 (3). Subcloning of a 5.5-kb PvuII fragment from the pSIP1 insert into the same site of the E. coli vector pSP72 (Promega) created pSIP7. Further subcloning of a 0.85-kb Sau3AI fragment from the pSIP7 insert into the BamHI site of pSP72 created plasmid pSIP8.
Detection of bacteriolytic activity of group III inhibitor.
Spores of S. ipomoeae strain 91-03 were inoculated into 200 ml of TSB, and after growth for 48 h, mycelia were removed by centrifugation at 4,000 × g for 15 min at 4°C. The supernatant was passed through a 0.2-μm-pore-size filter to remove any remaining cells and then kept at 4°C for immediate use or frozen at −20°C for long-term storage. S. ipomoeae strains 78-61, 88-35, and 88-03 were grown in 100-ml TSB cultures overnight following inoculation with 100 μl of a relevant dense spore suspension. Mycelia were harvested from exponentially growing cultures by centrifugation at 4,000 × g for 15 min at 20°C and then resuspended in 10 ml of fresh TSB. Aliquots (1 ml) of the resuspended cells were used to inoculate flasks containing a 50-ml solution consisting of 40 ml of fresh TSB to which 10 ml of strain 91-03 culture supernatant had been added to give a final concentration (of added supernatant) of 20% (vol/vol). Controls using 50 ml of fresh TSB containing no added culture supernatant were also performed. Flasks were shaken at 30°C, and at various times, samples were taken for spectrophotometric measurement of absorbance at 600 nm and for viable counts, determined by plating appropriate dilutions on SIGA.
Plate bioassays to detect inhibition phenotype.
Plate bioassays were performed essentially as described previously (8) except that 5-μl aliquots of strain 91-03 culture supernatant (prepared as described above) were spotted onto streptomycete test lawns, and plates were incubated at 30°C for 3 days prior to examination. For specificity tests involving nonstreptomycete bacteria, the appropriate agar medium (indicated above) was spread with a bacterial test strain, and 10 μl of strain 91-03 culture supernatant was then overspotted. Plates were incubated at the relevant temperatures (see above) and examined for up to 3 days for zones of inhibition. For stability tests, 1-ml samples of strain 91-03 culture supernatant were adjusted to pH values ranging from 2.0 to 12.0 with HCl or NaOH and then kept at 40, 50, or 60°C for 1 h or at 80°C for 20 min. Following this treatment, samples were rapidly cooled on ice and brought back to the initial pH, and 5-μl aliquots were then tested against S. ipomoeae strain 88-35 by plate bioassay.
The location of group III inhibitor during each stage of purification (see below) was determined by testing for the presence of inhibitory activity using plate bioassays involving test strain 88-35. The original supernatant or fractions known to contain inhibitor (i.e., fractions I to V; see below) were quantified for inhibitor activity by spotting 5-μl drops of 10-fold stepwise dilutions of the supernatant or fraction onto test lawns.
Purification of S. ipomoeae group III inhibitor.
Purification of the group III inhibitor protein was based on protocols used previously to purify bacteriocins from other bacterial sources (16). Four 250-ml TSB cultures of S. ipomoeae strain 91-03 (each in a 1-liter flask) were grown to stationary phase, mycelia were removed by centrifugation at 4,000 × g for 15 min at 4°C, and 400 g of ammonium sulfate was then added to the combined supernatant. The protein precipitate was pelleted by centrifugation at 7,000 × g for 20 min and solubilized in 250 ml of 20 mM sodium acetate, pH 4.8 (buffer A). This solution (fraction I) was filtered through a 0.45-μm-pore-size filter and applied to a 5-ml High-S cation-exchange chromatography column (Bio-Rad) which had been preequilibrated with 25 ml of buffer A. The column was washed with 25 ml of buffer A, and proteins were eluted with a 100-ml linear gradient of 0 to 1.0 M sodium chloride (using distilled H2O [dH2O] against 1.0 M sodium chloride in 20 mM sodium phosphate buffer, pH 6.8) at a flow rate of 2.0 ml/min. Fractions with inhibitory activity were pooled and adjusted to 2.4 M ammonium sulfate in 250 ml of 100 mM sodium phosphate buffer pH 6.8 (fraction II). Fraction II was applied to a 5-ml methyl hydrophobic interaction chromatography column (Bio-Rad) preequilibrated with 25 ml of 2.4 M ammonium sulfate in 100 mM sodium phosphate pH 6.8 (buffer B). The column was washed with 25 ml of buffer B, and proteins were eluted with a 60-ml linear gradient from 2.4 M ammonium sulfate to no ammonium sulfate (using 2.4 M ammonium sulfate in buffer B against dH2O) at a flow rate of 2.0 ml/min. Fractions containing inhibitor were pooled (fraction III), passed through a 0.2-μm-pore-size filter, and then concentrated to 400 μl with a Centriprep YM-10 centrifugal filter (Millipore). The retentate was brought up to 15 ml with dH2O and reconcentrated to 400 μl, and this cycle was repeated once more to yield a final 400-μl retentate (fraction IV). The filtrates from all of these concentration cycles were pooled and concentrated to 200 μl with a Centriprep YM-3 centrifugal filter (fraction V). Fractions IV and V were kept at 4°C for immediate use or frozen at −20°C for long-term storage. Protein concentrations for fractions I to V were determined by using the protein dye reagent from Bio-Rad.
Gel electrophoresis and protein sequencing.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out with 18% polyacrylamide gels (25). Molecular weight markers (low range) were from Life Technologies Inc. Nondenaturing gel electrophoresis was performed in the same way except that SDS was omitted from the gels and electrophoresis buffer and proteins were resuspended at room temperature in sample buffer lacking SDS and 2-mercaptoethanol and were not boiled prior to loading. Also, in an attempt to avoid possible protein denaturation during electrophoresis, nondenaturing gels were placed at 4°C and electrophoresed at 50 V.
Following SDS-PAGE of 8.5 μg of fraction IV, proteins were electroblotted to an Immobilon-P transfer membrane (Millipore) as described previously (28). After blotting, the membrane was wetted in 100% methanol, washed with three changes of dH2O to remove glycine, and then stained with a fresh solution of 0.1% Coomassie brilliant blue R-250 in 50% methanol and 7% acetic acid for 2 min. The blot was then destained in 50% methanol for 15 min, incubated in 100% methanol for 10 min to completely destain the background, and dried, and the stained 10-kDa protein band was cut out and subjected to N-terminal sequencing on an Applied Biosystems Procise sequencer located at the Protein Chemistry Core Laboratory at Baylor College of Medicine (Houston, Tex.).
Determination of inhibitor function for the 10-kDa protein species.
Following electrophoresis of fraction IV samples on a nondenaturing gel, a portion of the gel was stained with Coomassie blue R-250 to reveal the migration position of the 10-kDa protein. At the identical migration position on the unstained portion, a gel slice containing approximately 3.5 μg of the 10-kDa protein was excised, sealed in a dialysis membrane containing 1 ml of protein electrophoresis buffer lacking SDS, and submerged in an electrophoresis tank containing the same buffer for 4 h at 50 V. Following this treatment, elution of protein was judged to be complete by Coomassie blue staining of the gel slice. The buffer inside the dialysis membrane was removed, passed through a 0.2-μm-pore-size filter, and then concentrated to 30 μl with a Microcon-3 microconcentrator (Millipore). The retentate was brought up to 1 ml with dH2O and reconcentrated to 30 μl, and then 10 μl was tested for inhibition activity by plate bioassay.
Inhibitor function for the 10-kDa protein was also examined by applying at a flow rate of 0.5 ml per min a portion of fraction IV (which had been adjusted to 100 mM sodium phosphate, pH 6.8) to a Sephadex 200 size exclusion chromatography column (Amersham Biosciences), which had been preequilibrated with 100 mM sodium phosphate, pH 6.8. From analysis of fractions by their absorbance at 280 nm, a single protein peak was evident, and it was tested for the presence of the 10-kDa protein by SDS-PAGE and silver staining and tested for inhibitor function by plate bioassays.
Library construction and other molecular biological techniques.
S. ipomoeae strain 91-03 was grown in yeast extract-malt extract medium (17), which contained only 17% sucrose (to allow growth of S. ipomoeae in this medium), and genomic DNA was extracted as described previously (17). Purified genomic DNA was partially digested with Sau3AI so that most of the resulting fragments were in the range of 40 to 60 kb, as judged by pulsed-field gel electrophoresis, and the partially digested DNA was then dephosphorylated with calf intestinal alkaline phosphatase (CIAP). Cosmid vector pOJ446 (3) was digested with HpaI, dephosphorylated with CIAP, and further digested with BamHI to generate two vector arms. Then, per the manufacturer's instructions, cosmid arms and the partially digested genomic fragments were ligated and packaged into lambda phage heads by using a Gigapack III XL packaging extract (Stratagene), and the resulting particles were used to transduce E. coli strain XL1-Blue MR, with selection of transductants occurring on LB agar amended with 50 μg of apramycin sulfate/ml. Over 5,000 colonies were pooled, completely mixed in 20% glycerol, and stored at −80°C.
Based on the N-terminal amino acid sequence of the inhibitor protein along with the strong preference for G or C at the third codon position in Streptomyces open reading frames (ORFs) (2), the degenerate oligonucleotide 5′ GACGC(C/G)CC(C/G)GG(C/G)CACCC(C/G)GG(G/C)AAGCACTACCT(C/G)CAGGT(C/G)AACGT(C/G) was synthesized (by Operon Technologies, Inc.) and then radiolabeled by using [γ-32P]ATP and T4 polynucleotide kinase (25). For colony blot hybridizations, the stored cosmid library was plated on LB agar containing 50 μg of apramycin sulfate/ml, incubated for 20 h at 37°C, and transferred to nitrocellulose membranes. Following transfer, amplification of the library on LB agar containing chloramphenicol as well as subsequent lysis, denaturation, neutralization, and baking steps were done as described elsewhere (25). Baked membranes were wetted at 37°C and cleansed of bacterial debris (25), and hybridizations were then performed as previously described (4).
Nucleotide sequencing and analysis.
Automated nucleotide sequencing was performed as described previously (23). Database homology searches employing the BLASTP and BLASTX algorithms (1) with default settings were enabled by the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/). Analysis of nucleotide sequences for ORFs using the FRAME algorithm (2) was as provided by the Frameplot 2.3.2 website (http://watson.nih.go.jp/∼jun/cgi-bin/frameplot.pl).
Nucleotide sequence accession number.
The sequence determined in this study has been deposited in the GenBank database under accession no. AF545850.
RESULTS
Specificity and stability of S. ipomoeae group III inhibitor.
To begin characterization of interstrain inhibition in S. ipomoeae, we chose to isolate group III inhibitor since it was found to be released into the supernatant of group III broth cultures and since it was found to be stable for extended periods of storage; such factors would facilitate purification of functional inhibitor. The lack of correlation found previously (8) between inhibitor production and the presence of plasmids in S. ipomoeae suggested that interstrain inhibitors are chromosomally encoded in these bacteria. We therefore chose strain 91-03, a plasmidless group III strain (8), for purification of inhibitor and subsequent cloning of its structural gene.
S. ipomoeae group III inhibitor prevents the growth of group I and group II strains and therefore causes clear inhibition zones to form within lawns of susceptible S. ipomoeae cells growing on agar plates (8). To determine whether group III inhibitor has a similar effect on the growth of other bacteria, plate bioassays were performed in which spores of various other Streptomyces species or cells from a variety of other gram-positive or gram-negative bacteria were spread on plates and then overspotted with supernatant derived from an S. ipomoeae strain 91-03 culture. As expected, upon incubation, clear inhibition zones were evident within the lawns of both group I and II control strains; however, no inhibition of any other bacteria tested was apparent (Table 1). Several Streptomyces species were examined—in particular, three strains of the potato pathogen S. scabiei—as well other bacterial genera, including both soil-dwelling and non-soil-dwelling organisms.
TABLE 1.
Specificity of S. ipomoeae group III inhibitora
| Test organism | Sensitivityb |
|---|---|
| Streptomyces | |
| Streptomyces ipomoeae group I (78-61) | + |
| Streptomyces ipomoeae group II (88-35) | + |
| Streptomyces ipomoeae group III (88-03) | − |
| Streptomyces scabiei (ss-1) | − |
| Stretomyces scabiei (ss-2) | − |
| Streptomyces scabiei (ss-3) | − |
| Streptomyces coelicolor | − |
| “Streptomyces cyanogenus” | − |
| Streptomyces lividans | − |
| Streptomyces rochei | − |
| Other organisms | |
| Alcaligenes faecalis | − |
| Bacillus subtilis | − |
| Enterococcus faecalis | − |
| Mycobacterium smegmatis | − |
| Staphylococcus aureus | − |
| Streptococcus strain B | − |
| Enterobacter aerogenes | − |
| Escherichia coli | − |
| Proteus vulgaris | − |
| Pseudomonas aeruginosa | − |
| Serratia marcescens | − |
Sensitivity was determined by plate bioassays in which spores of the indicated Streptomyces species or cells of the other bacterial genera were spread on agar plates and overspotted with supernatant containing inhibitor produced by S. ipomoeae strain 91-03 (see Materials and Methods for details).
+, inhibition was observed; −, no inhibition was seen.
As mentioned above, group III inhibitor activity in isolated culture supernatants was found to be very stable, with no measurable loss of activity following storage either at 4°C for up to 1 month or at −20°C for over a year with repeated cycles of freezing and thawing. The stability of unpurified group III inhibitor was examined further by preincubating aliquots of strain 91-03 culture supernatant at different temperatures and pHs and then determining the effects of these treatments on inhibitor function directed against a group II strain by the plate bioassay method described above. While the majority of inhibitor function was retained at 25°C in the range of pH 4 to 10 (Table 2), group III inhibitor remained apparently completely stable when exposed to near neutral to alkaline conditions (i.e., pH 6 to 10) as long as the inhibitor was not also exposed to temperatures greater than approximately 40°C; with pretreatments of 50°C or higher, inhibition activity was greatly reduced or completely lost regardless of the accompanying pH (Table 2).
TABLE 2.
Stability of unpurified group III inhibitor present in culture supernatant following different temperature and pH treatmentsa
| Pretreatment | Inhibition zone (cm) | |
|---|---|---|
| Temp (°C) | pH | |
| 25 | 2.0 | 0.25 |
| 4.0 | 1.55 | |
| 6.0 | 2.00 | |
| 8.0 | 1.95 | |
| 10.0 | 2.05 | |
| 12.0 | 0.70 | |
| 40 | 2.0 | 0.20 |
| 4.0 | 0.45 | |
| 6.0 | 2.00 | |
| 8.0 | 1.90 | |
| 10.0 | 2.05 | |
| 12.0 | 0.20 | |
| 50 | 2.0 | 0.15 |
| 4.0 | 0.20 | |
| 6.0 | 0.20 | |
| 8.0 | 0.25 | |
| 10.0 | 0.25 | |
| 12.0 | 0 | |
| 60 | 2.0 | 0 |
| 4.0 | 0 | |
| 6.0 | 0 | |
| 8.0 | 0 | |
| 10.0 | 0 | |
| 12.0 | 0 | |
| 80 | 2.0 | 0 |
| 4.0 | 0 | |
| 6.0 | 0 | |
| 8.0 | 0 | |
| 10.0 | 0 | |
| 12.0 | 0 | |
Aliquots of supernatant from a S. ipomoeae strain 91-03 culture were adjusted to the indicated pH values and incubated at the indicated temperatures for 1 h except for the 80°C treatment, where the incubation time was only 20 min. The aliquots were then cooled, returned to the original pH value of the supernatant, and spotted onto plates seeded with group II strain 88-35 spores (see Materials and Methods for bioassay details). Following growth of the test strain, the diameter of the resulting inhibition zone was determined.
Bacteriolytic function of group III inhibitor.
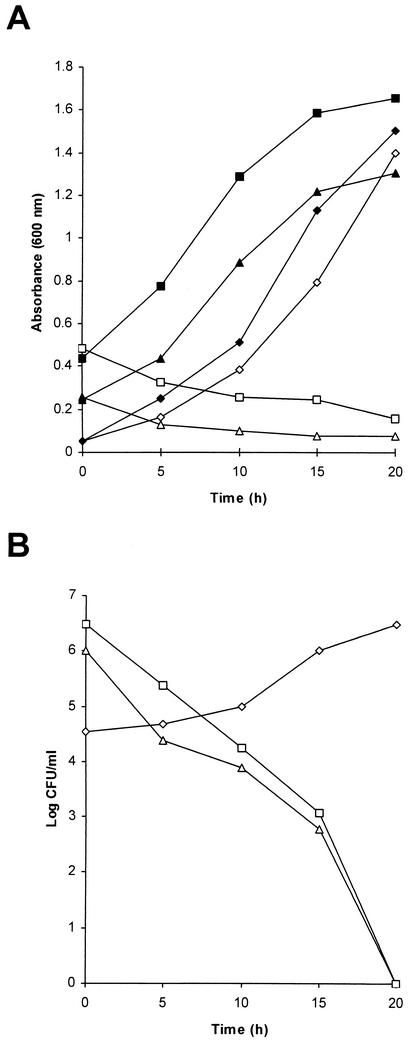
To investigate whether group III inhibitor causes cell death and possibly lysis of sensitive S. ipomoeae cells, supernatant from strain 91-03 was added to a final concentration of 20% to logarithmically growing cultures of representative group I, II, or III strains and samples were examined at subsequent times for absorbance at 600 nm (Fig. 1A) and for CFU by plating dilutions on agar media (Fig. 1B). Exposure to group III inhibitor caused a steady decline in absorbance for both S. ipomoeae strains 78-61 (group I) and 88-35 (group II), while similarly exposed cultures of strain 88-03 (group III) still showed a gradual increase in absorbance which approximated the increases seen for untreated control cultures of the three strains (Fig. 1A). The decrease in absorbance readings for group I and II strains suggested that lysis of sensitive cells occurred following exposure to group III inhibitor; consistent with this notion of bacteriolytic function for group III inhibitor were the viable counts obtained at the time points used for absorbance readings, which showed dramatic reductions in CFU for inhibitor-treated strains 78-61 and 88-35, while CFU for the similarly treated strain 88-03, as expected, showed a steady increase (Fig. 1B).
FIG. 1.
Bacteriolytic activity of group III inhibitor. Purified supernatant from an S. ipomoeae strain 91-03 culture was added to a final concentration of 20% (vol/vol) to logarithmically growing broth cultures of representative group I, II, or III strains, and at the indicated time points, samples of each culture were measured spectrophotometrically for absorbance at 600 nm (A) or were diluted serially and plated on agar media for subsequent determination of CFU (B). ▪, strain 88-35 (group II) without inhibitor; ▴, strain 78-61 (group I) without inhibitor; ⧫, strain 88-03 (group III) without inhibitor; □, strain 88-35 with inhibitor added; ▵, strain 78-61 with inhibitor added; ◊, strain 88-03 with inhibitor added.
Purification of group III inhibitor.
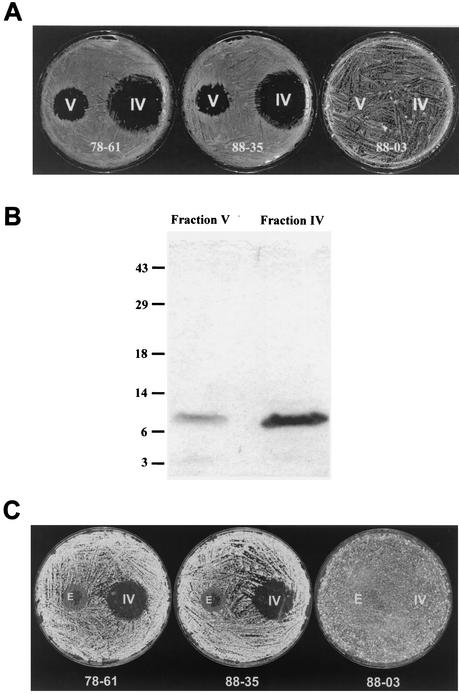
We found that group III inhibitory activity in culture supernatants was sensitive to treatment with proteinase K (data not shown), a result that suggested the group III inhibitor may belong to the class of proteinaceous antimicrobial substances produced by bacteria termed bacteriocins (16). Based on similar protocols used to purify bacteriocins from other bacteria, we purified the group III inhibitor of strain 91-03 approximately 2,000-fold (Table 3) using a sequential combination of ammonium sulfate precipitation (the precipitate from this step was referred to as fraction I), cation-exchange chromatography (the eluate containing inhibitory activity from this step was fraction II), and hydrophobic interaction chromatography. The eluate from the latter chromatography step, which contained inhibitor function (i.e., fraction III in Table 3), was then subjected to centrifugal ultrafiltration involving a filter with a molecular weight cutoff of 10,000, a process which yielded two fractions: one (fraction IV) retained proteins with molecular weights greater than 10,000 and contained approximately 90% of the inhibitor activity derived from fraction III (Table 3 and Fig. 2A), and the other (fraction V) represented the filtrate from the ultrafiltration and so contained proteins with molecular weights of less than 10,000 and also contained the remaining 10% of the purified group III inhibitor function (Table 3 and Fig. 2A). The appearance of inhibitor activity in both fractions following centrifugal ultrafiltration was consistent with the possibility that the molecular weight of the inhibitor might approximate the cutoff value of the filter; this notion was strengthened upon analysis of fractions IV and V by SDS-PAGE followed by Coomassie-blue staining of the gel, where it was found that a predominate protein band of approximately 10 kDa was present in both fractions and in relative concentrations that were consistent with the percent distribution of inhibitory activity in these two fractions (Fig. 2B).
TABLE 3.
Purification of the group III inhibitor from S. ipomoeae strain 91-03a
| Purification stage | Vol (ml) | Total protein (μg) | Total inhibition activity (IU) | Sp act (IU/μg) | Purification (fold) | Yield (%) |
|---|---|---|---|---|---|---|
| Culture supernatant | 1,000 | 1.5 × 106 | 8.2 × 107 | 5.4 × 101 | 1 | 100 |
| (NH2)2SO4 precipitate (fraction 1) | 250 | 1.2 × 105 | 2.5 × 107 | 2.0 × 102 | 4 | 30.5 |
| Cation-exchange chromatography (fraction II) | 24 | 5.2 × 103 | 1.6 × 107 | 3.0 × 104 | 565 | 19.5 |
| Hydrophobic interaction chromatography (fraction III) | 10 | 4.1 × 103 | 4.4 × 107 | 1.1 × 105 | 1,996 | 53.6 |
| Centrifugal ultrafiltrationb retentate (fraction IV) | 0.4 | 3.4 × 102 | 4.0 × 107 | 1.2 × 105 | 2,221 | 48.8 |
| Centrifugal ultrafiltration filtrate (fraction V) | 0.2 | 3.6 × 101 | 3.2 × 106 | 8.9 × 104 | 1,650 | 3.9 |
IU, inhibition unit. One unit was defined as the highest 10-fold dilution of supernatant or fraction that still resulted in a zone of inhibition at least 5 mm in diameter when 5 μl of that dilution was spotted onto a test lawn in plate bioassays.
The centrifugal ultrafiltration step involved a filter with a 10,000-molecular-weight cutoff.
FIG. 2.
Correlation between S. ipomoeae group III inhibitor activity and the 10-kDa protein species. (A) Plate bioassays involving fractions IV and V. Aliquots (4 μl) of fractions IV and V were spotted onto agar plates spread with spores of S. ipomoeae strain 78-61 (group I), 88-35 (group II), or 88-03 (group III), and the plates were then incubated for 3 days. (B) SDS-PAGE analysis followed by Coomassie blue R-250 staining of aliquots (10 μl) of fractions IV and V. The migration positions of molecular weight markers are indicated (molecular weights are in thousands). (C) Bioassays involving gel-eluted 10-kDa protein. A portion of fraction IV was electrophoresed under nondenaturing conditions (see Materials and Methods), a gel slice containing the 10-kDa protein was excised, and the protein was then electroeluted and tested along with fraction IV in plate bioassays involving strains 78-61 (group I), 88-35 (group II), and 88-03 (group III). E, eluted 10-kDa protein (10 μl of 30 μl total was spotted).
To relate inhibitor activity directly to the 10-kDa protein species, an aliquot of fraction IV was first electrophoresed on a nondenaturing gel and a portion of the gel was stained with Coomassie blue in order to reveal the migration position of the 10-kDa protein. The identical position on the unstained portion was then excised, the protein was electroeluted, and the resulting eluate was overspotted onto representative S. ipomoeae strains in plate bioassays. The eluted 10-kDa protein was found to inhibit the growth of representative group I and group II strains but not a group III isolate (Fig. 2C). Interestingly, while characteristic clearing of sensitive strains by the eluted 10-kDa protein was seen at the center of the overspot area, there was an additional concentric region surrounding this inner zone where growth was evident but aerial hyphal development was retarded (Fig. 2C). Since this particular inhibition pattern was never observed (e.g., even upon dilution of relevant fractions) prior to gel electrophoresis and electroelution of the 10-kDa protein, this result suggested that either these manipulations had altered the activity of this putative inhibitor protein or they had separated the 10-kDa factor from an additional component needed to give the characteristic full zone of clearing. To address the latter possibility, a portion of fraction IV was first subjected to size exclusion chromatography with a Sephadex 200 column at 4°C; only a single protein peak was observable by analysis of resulting fractions for their absorbance at 280 nm (data not shown). Peak fractions, which were found to give the characteristic full zone of clearing in bioassays, were subsequently examined by gel electrophoresis and silver staining and found to contain only the 10-kDa protein (data not shown). These results thus not only argue that the electrophoresis and/or elution manipulations described did in fact alter activity of the 10-kDa protein but also confirm the inhibitory function of this protein and allow its classification as a bacteriocin molecule, which we designate ipomicin.
Cloning and nucleotide sequencing of the ipomicin gene.
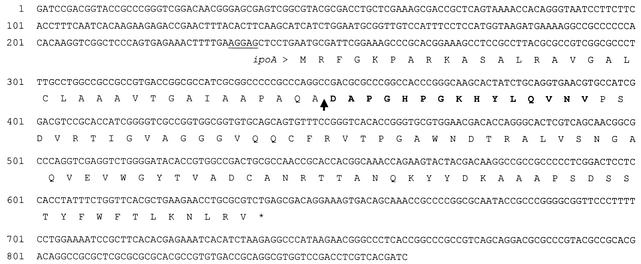
Following analysis of sufficient quantities of fraction IV by SDS-PAGE and subsequent transfer to an Immobilon-P membrane, microsequencing of the 10-kDa ipomicin protein present on this membrane was performed, and the resulting N-terminal 15-amino-acid sequence was used to design a degenerate DNA oligonucleotide specific for the corresponding portion of the gene (see Materials and Methods for details). The oligonucleotide was used as a radiolabeled probe in colony blot hybridization experiments to identify cloned versions of the gene that were present in a cosmid library, which consisted of 35-kb fragments of S. ipomoeae strain 91-03 genomic DNA inserted into the cosmid vector pOJ446 (3). Seven positive cosmid clones, which by restriction enzyme analysis appeared to be comprised of four distinct cosmid species, were identified by this method. Upon digestion of their DNAs with various restriction enzymes and subsequent Southern blotting using the same degenerate probe, the smallest hybridizing fragment for each cosmid was identified as a 0.85-kb Sau3AI fragment (a single 0.85-kb Sau3AI hybridizing fragment was also seen in Southern blots involving strain 91-03 chromosomal DNA and the same degenerate oligonucleotide probe [data not shown]). Subcloning of this small fragment from one of these cosmids (i.e., pSIP1) into the E. coli plasmid vector pSP72 was eventually achieved, and the complete 862-bp nucleotide sequence of the insert region of the resulting clone, pSIP8, was determined (Fig. 3).
FIG. 3.
Nucleotide sequence of the 862-bp Sau3AI fragment containing the ipoA gene and deduced amino acid sequence of the precursor form of ipomicin. Amino acids are indicated by their one-letter designations below the nucleotide sequence. A putative Shine-Dalgarno ribosome binding sequence is underlined. The N-terminal 15 amino acids of purified ipomicin protein, which were confirmed by protein sequencing, are in bold. The site of putative cleavage of a 35-amino-acid signal sequence, which would create the mature inhibitor protein, is indicated by an arrow.
Within the sequenced region, a 393-bp ORF was identified (and designated ipoA), which would therefore encode a full-length protein of 131 amino acids. A sequence (AGGAG) that nearly matches the consensus Shine-Dalgarno ribosome binding motif precedes ipoA by 7 bases. Beginning at the 36th codon of the ipoA ORF is the region encoding the exact 15-amino-acid sequence (Fig. 3) found at the N terminus of the purified ipomicin protein; thus, this mature form appears to be derived from the full-length precursor following processing in which a 35-amino-acid N-terminal signal sequence is removed. Following removal of the putative signal sequence, the resulting 96-amino-acid mature ipomicin polypeptide would have a calculated molecular weight of 10,434, which is in close agreement with the size estimate derived from SDS-PAGE, and a pI of 9.08, which matches the cationic nature of other gram-positive bacteriocins (16). The ipomicin protein was examined in database searches for relatedness to other known proteins using the BLASTP and BLASTX algorithms (1) along with their default settings; however, no significant similarities that might help reveal the function of this inhibitory molecule were evident.
The composite G+C content of the 862-bp insert of plasmid pSIP8 was found to be 63.7%. Streptomyces ORFs typically show a nonrandom distribution of G+C at the three codon positions, with the highest percent G+C occurring at the third position and intermediate and lowest percentages occurring at the first and second positions, respectively. While the FRAME algorithm (2), which measures such percent G+C distribution, predicted the existence of ipoA, whose third-position G+C content was calculated to be 86.4%, no other ORFs containing the typical nonrandom G+C distribution for Streptomyces genes were found in the sequenced region (data not shown).
DISCUSSION
Based on the colicin archetype from E. coli, bacteriocins initially included proteinaceous, plasmid-encoded molecules which are bactericidal only for closely related bacterial species and strains that possess specific membrane receptors. Colicin production was also found to be repressible, with induction often occurring under conditions lethal to producing cells (16, 27). As more bacteriocins have been discovered, including many from gram-positive sources, their classification has been expanded to include certain other extracellularly released, proteinaceous, ribosomally synthesized antimicrobial molecules that share some but not all of the properties of the original colicin prototype (16). The ipomicin protein of S. ipomoeae group III bacteria both is bacteriolytic and has an extremely narrow target spectrum (with the latter property being somewhat unusual for bacteriocins from gram-positive organisms) (16) but unlike colicins is chromosomally encoded. The latter assertion is based on previous work (8) which showed no correlation between the production of inhibitors and the presence of plasmids in S. ipomoeae; these results led to our isolation here of ipomicin from S. ipomoeae 91-03, a strain which was previously shown to lack any plasmid species, as determined by both pulsed-field agarose gel electrophoresis and alkaline lysis extraction techniques (8). While it remains undetermined whether ipomicin binds to specific cytoplasmic membrane receptors, its high target specificity suggests that this may be the case (16). Also, while inducible production of ipomicin was not investigated per se here, cells of S. ipomoeae 91-03 appeared to readily produce and release this inhibitor into the supernatant throughout growth in broth cultures with no obvious deleterious effects on the producing strain (data not shown).
The exact mechanism by which ipomicin causes lysis of sensitive S. ipomoeae group I and group II cells remains to be elucidated. Many low-molecular-weight bacteriocins from gram-positive sources appear to form pores in the cytoplasmic membranes of their target cells, a process that can ultimately lead to cell lysis (16). It is also possible that ipomicin instead represents some type of bacteriolytic enzyme, which is somehow specific for sensitive S. ipomoeae cells (16, 21).
Structural genes for bacteriocins are often found to be clustered with additional related genes, including those involved in translocation of and immunity to the bacteriocin (16). Although no additional ORFs were found within the 862-bp Sau3AI chromosomal sequence that contains the ipoA gene, additional related genes may still be located nearby. To address this possibility, we are determining the nucleotide sequence of a cloned 5.5-kb PvuII fragment derived from strain 91-03 which is known to be comprised in part by the 862-bp Sau3AI ipoA-containing sequence (our unpublished results).
Interestingly, the leucine residue at position 13 of the signal sequence of the ipoA gene product is encoded by TTA, an extremely rare codon in Streptomyces whose translation by the tRNA product of the Streptomyces coelicolor bldA gene under certain culture and genetic background conditions appeared to be more efficient later rather than earlier during growth of S. coelicolor liquid cultures (7). Whether similar regulation involving the rare TTA codon of ipoA confers some aspect of temporal control on ipomicin production during growth of S. ipomoeae is an interesting question which remains to be explored.
A proteinaceous substance with bacteriocin-like properties produced by Streptomyces virginiae has been reported (24). That inhibitor was found to be active against many streptomycete species but not against any other microorganisms tested. S. ipomoeae strains belonging to inhibition group II also produce a substance that is inhibitory to group I and group III strains (8). It will be interesting to determine whether this inhibitor is actually an additional bacteriocin as well as determining its relationship, if any, to ipomicin. Exposure to a combination of ipomicin and group II inhibitor would theoretically inhibit all 36 strains of S. ipomoeae that were previously analyzed for inhibitory reactions during cocultivation of strains (8); such treatment represents a potential biocontrol mechanism for protection of sweet potatoes from infection by pathogenic S. ipomoeae bacteria.
Acknowledgments
We thank Fred A. Rainey, Mark Batzer, and Gail Kilroy for assistance with automated sequencing, Huangen Ding for advice and assistance with size exclusion chromatography, and Rebecca Todd for providing numerous bacterial strains. Naomi Ward is gratefully acknowledged for performing the initial experiments that demonstrated sensitivity of group III inhibitor to proteinase K treatment. We are grateful to Matthew Ducote for assistance with preparation of certain figures, and we also thank him and Kevin Schully for helpful discussions and generous assistance during the course of these experiments.
This work was supported by the Louisiana Agricultural Experiment Station, Hatch Project LAB03475 (to G.S.P.).
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bibb, M. J., P. R. Findlay, and M. W. Johnson. 1984. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene 30:157-166. [DOI] [PubMed] [Google Scholar]
- 3.Bierman, M., R. Logan, K. O'Brien, E. T. Seno, R. N. Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43-49. [DOI] [PubMed] [Google Scholar]
- 4.Brasch, M. A., G. S. Pettis, S. C. Lee, and S. N. Cohen. 1993. Localization and nucleotide sequences of genes mediating site-specific recombination of the SLP1 element in Streptomyces lividans. J. Bacteriol. 175:3067-3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bukhalid, R. A., and R. Loria. 1997. Cloning and expression of a gene from Streptomyces scabies encoding a putative pathogenicity factor. J. Bacteriol. 179:7776-7783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Champness, W. C., and K. F. Chater. 1994. Regulation and integration of antibiotic production and morphological differentiation in Streptomyces spp., p. 61-93. In P. Piggot, J. C. P. Moran, and P. Youngman (ed.), Regulation of bacterial differentiation. American Society for Microbiology, Washington, D.C.
- 7.Chater, K. F. 1993. Genetics of differentiation in Streptomyces. Annu. Rev. Microbiol. 47:685-713. [DOI] [PubMed] [Google Scholar]
- 8.Clark, C. A., C. Chen, N. Ward-Rainey, and G. S. Pettis. 1998. Diversity within Streptomyces ipomoeae based on inhibitory interactions, rep-PCR, and plasmid profiles. Phytopathology 88:1179-1186. [DOI] [PubMed] [Google Scholar]
- 9.Clark, C. A., and D. R. LaBonte. 1992. Disease factors in breeding and biotechnology for sweetpotato, p. 484-494. In W. A. Hill, C. K. Bonsi, and P. A. Loretan (ed.), Sweetpotato technology for the 21st century. Tuskegee University, Tuskegee, Ala.
- 10.Clark, C. A., and A. Lawrence. 1981. Morphology of spore-bearing structures in Streptomyces ipomoea. Can. J. Microbiol. 27:575-579. [DOI] [PubMed] [Google Scholar]
- 11.Clark, C. A., and S. W. Matthews. 1987. Histopathology of sweet potato root infection by Streptomyces ipomoea. Phytopathology 77:1418-1423. [Google Scholar]
- 12.Clark, C. A., and J. W. Moyer. 1988. Compendium of sweet potato diseases. The American Phytopathological Society, St. Paul, Minn.
- 13.Clark, C. A., and B. Watson. 1983. Susceptibility of weed species of Convolvulaceae to root-infecting pathogens of sweet potato. Plant Dis. 67:907-909. [Google Scholar]
- 14.Healy, F. G., M. Wach, S. B. Krasnoff, D. M. Gibson, and R. Loria. 2000. The txtAB genes of the plant pathogen Streptomyces acidiscabies encode a peptide synthetase required for phytotoxin thaxtomin A production and pathogenicity. Mol. Microbiol. 38:794-804. [DOI] [PubMed] [Google Scholar]
- 15.Hirochika, H., and K. Sakaguchi. 1982. Analysis of linear plasmids isolated from Streptomyces: association of protein with the ends of the plasmid DNA. Plasmid 7:59-65. [DOI] [PubMed] [Google Scholar]
- 16.Jack, R. W., J. R. Tagg, and B. Ray. 1995. Bacteriocins of gram-positive bacteria. Microbiol. Rev. 59:171-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, England.
- 18.King, R. R., C. H. Lawrence, L. A. Calhoun, and J. B. Ristaino. 1994. Isolation and characterization of thaxtomin-type phytotoxins associated with Streptomyces ipomoeae. J. Agric. Food Chem. 42:1791-1794. [Google Scholar]
- 19.Lanoot, B., M. Vancanneyt, I. Cleenwerck, L. Wang, W. Li, Z. Liu, and J. Swings. 2002. The search for synonyms among streptomycetes by using SDS-PAGE of whole-cell proteins. Emendation of the species Streptomyces aurantiacus, Streptomyces cacaoi subsp. cacaoi, Streptomyces caeruleus, and Streptomyces violaceus. Int. J. Syst. Evol. Microbiol. 52:823-829. [DOI] [PubMed] [Google Scholar]
- 20.Loria, R., R. A. Bukhalid, B. A. Fry, and R. R. King. 1997. Plant pathogenicity in the genus Streptomyces. Plant Dis. 81:836-846. [DOI] [PubMed] [Google Scholar]
- 21.Navarre, W. W., and O. Schneewind. 1999. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 63:174-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perkins, D. D. 1962. Preservation of Neurospora stock cultures with anhydrous silica gel. Can. J. Microbiol. 8:591-594. [Google Scholar]
- 23.Pettis, G. S., and S. Prakash. 1999. Complementation of conjugation functions of Streptomyces lividans plasmid pIJ101 by the related Streptomyces plasmid pSB24.2. J. Bacteriol. 181:4680-4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roelants, P., and F. Naudts. 1964. Properties of a bacteriocin-like substance produced by Streptomyces virginiae. Antonie Leeuwenhoek 30:45-53. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Snapper, S. B., R. E. Melton, S. Mustafa, T. Kieser, and W. R. Jacobs, Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4:1911-1919. [DOI] [PubMed] [Google Scholar]
- 27.Tagg, J. R., A. S. Dajani, and L. W. Wannamaker. 1976. Bacteriocins of gram-positive bacteria. Bacteriol. Rev. 40:722-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]