Abstract
The hydrothermal vent clam Calyptogena magnifica (Bivalvia: Vesicomyidae) depends for its nutrition on sulfur-oxidizing symbiotic bacteria housed in its gill tissues. This symbiont is transmitted vertically between generations via the clam's eggs; however, it remains uncertain whether occasionally symbionts are horizontally transmitted or acquired from the environment. If symbionts are transmitted strictly vertically through the egg cytoplasm, inheritance of symbiont lineages should behave as if coupled to the host's maternally inherited mitochondrial DNA. This coupling would be obscured, however, with low rates of horizontal or environmental transfers, the equivalent of recombination between host lineages. Population genetic analyses of C. magnifica clams and associated symbionts from eastern Pacific hydrothermal vents clearly supported the hypothesis of strictly maternal cotransmission. Host mitochondrial and symbiont DNA sequences were coupled in a clam population that was polymorphic for both genetic markers. These markers were not similarly coupled with sequence variation at a nuclear gene locus, as expected for a randomly mating sexual population. Phylogenetic analysis of the two cytoplasmic genes also revealed no evidence for recombination. The tight association between vesicomyid clams and their vertically transmitted bacterial endosymbionts is phylogenetically very young (<50 million years) and may serve as a model for the origin and evolution of eukaryotic organelles.
Endosymbiotic associations have played major roles in evolution, from the ancient origins of the energy-producing organelles of eukaryotic cells (16, 28) to relatively young associations between invertebrate animals and chemoautotrophic bacteria in deep-sea hydrothermal vent communities (38). The degree of integration between host and symbiont varies greatly in these younger symbioses. For example, the mouthless and gutless adults of vestimentiferan tubeworms (Annelida: Siboglinidae) depend entirely on thiotrophic (sulfide-oxidizing) endosymbionts for their nutrition (6, 13), yet they acquire their symbionts anew each generation from the surrounding environment (9, 27). Although molecular studies and fossil evidence suggest that vestimentiferan tubeworms, and presumably their symbionts, have existed at hydrothermal vents for at least 100 million years (2, 19, 20), phylogenetic trees based on these environmentally acquired symbionts do not parallel those of their tubeworm hosts (12, 33). In contrast, vesicomyid clams appear to transmit their thiotrophic endosymbionts vertically between generations via their eggs (3, 10), yet the clams have retained a rudimentary gut (25). Molecular systematic studies suggest that vesicomyid clams radiated more recently than tubeworms, perhaps less than 50 million years ago (37), and evolutionary relationships among the symbionts closely parallel those of their clam hosts, i.e., they exhibit cospeciation (38).
Although phylogenetic patterns of cospeciation are expected with “strictly” vertical transmission of symbionts (30, 38), such patterns do not preclude some degree of horizontal transmission (i.e., symbionts are occasionally transferred between host individuals) or “leakage” among lineages within individual species. Moreover, such cospeciation patterns do not require vertical transmission, as they can also be found in some tightly integrated host-parasite systems maintained by horizontal or environmental transmission (18, 34, 35); thus, biologists should cautiously interpret cospeciation patterns as evidence for strictly vertical transmission. If symbiont transmission is strictly vertical through the cytoplasm of a host's eggs, the inheritance of symbiont lineages should parallel that of other maternally inherited cytoplasmic factors, like mitochondria (14, 38). Thus, in natural populations, variation in the symbiont genome will behave as if completely linked (i.e., coupled) with mitochondrial variants, and they should exhibit gametic phase disequilibrium. However, horizontal transfers between hosts, or acquisition of new symbionts from the environment, will decouple host and symbiont lineages and randomize their occurrence together. Such decoupling events are comparable to conventional genetic recombination in a Mendelian population; thus, a randomly mating population at equilibrium should exhibit no gametic phase disequilibrium, even if decoupling events are extremely rare. Unfortunately, tests of coupling due to vertical transmission of host mitochondrial and symbiont genomes are difficult. Often, host or symbiont lineages lack sufficient polymorphism to detect coupling. Even the application of rapidly evolving genetic markers might be misleading, because host and symbiont lineages might covary geographically as a consequence of shared historical population structures, as we shall demonstrate. To date, only one example exists that is consistent with intraspecific coupling of host mitochondrial and symbiont lineages due to strictly vertical transmission. The mitochondrial gene tree of the aphid Uroleucon ambrosiae is broadly congruent with gene trees based on its bacterial endosymbiont, Buchnera aphidicola, and two endosymbiont plasmids (14, 15).
Herein, we tested the hypothesis that host mitochondrial and endosymbiont genomes of the hydrothermal vent clam Calyptogena magnifica (Bivalvia: Vesicomyidae) are coupled in natural populations. This clam harbors a sulfur-oxidizing γ-subdivision proteobacterium in specialized cells contained within its gill tissues (5, 6, 13). Cytological and molecular evidence reveals that the symbionts are transmitted via the eggs of Calyptogena clams (3, 10). To date, these bacteria have not been found living freely in the marine environment, and attempts to culture the symbionts have failed (32); nonetheless, this negative evidence does not preclude the existence of a free-living form of the bacterium or rare horizontal transfers between hosts (38). To test our hypothesis, we examined covariation of host and symbiont DNA sequences from C. magnifica individuals sampled across the known range of this species. Additionally, to assess whether such covariation might result from shared historical population structures for host and symbiont lineages, we also examined nuclear gene markers that should exhibit no coupling within populations that have achieved gametic phase equilibrium.
MATERIALS AND METHODS
Biological samples.
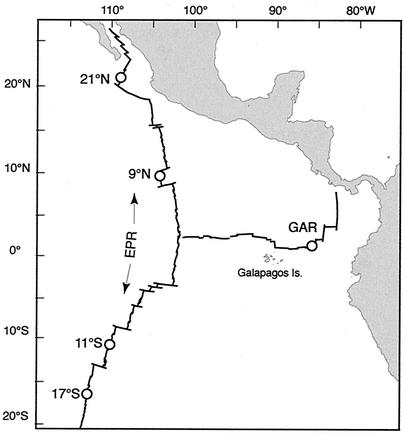
We examined DNA sequence variation in C. magnifica clams and associated symbionts sampled from the known range of the host species (Fig. 1). Specimens were collected using the submersible Alvin and stored at −80°C prior to genetic analyses. We examined 10 individuals from 21°N (Alvin dives 2230, 2231, 2232, and 2233), 13 from 9°N (Alvin dives 2353, 2358, 2370, 2503, and 2769), 9 from 11°S (Alvin dive 3323), and 38 from 17°S (Alvin dives 3294, 3328, and 3329) on the East Pacific Rise (EPR), and 10 from the Galapagos Rift (GAR) (Alvin dives 2223 and 2224).
FIG. 1.
Sampling localities in the EPR and GAR hydrothermal vents: 21°N (20°50′N, 109°06W); 9°N (9°50′N, 104°17′W); 11°S (11°18′S, 110°32′W); 17°S (17°25′S, 113°12′W); GAR (00°48′N, 86°10′W).
DNA extraction, PCR amplification, and sequencing.
The manufacturer's protocols were followed for the DNEasy kit (Qiagen, Inc., Chatsworth, Calif.) to extract total DNA from adductor muscle and gill tissues of each clam. Adductor DNA (endosymbiont-free tissue) was used to amplify a 570-bp region of the clam mitochondrial cytochrome oxidase c subunit I gene (mtCOI), a 410-bp region of the clam mitochondrial cytochrome b (cytb), and 1,150 bp of a noncoding anonymous single-copy nuclear DNA locus (Cmg24) of the clam. Primers and PCR conditions were described for mtCOI amplification by Peek et al. (37), for cytb by Dahlgren et al. (8), and for Cmg24 by Karl et al. (24). Gill DNA (endosymbiont-infected tissue) was used to amplify an 840-bp fragment of the symbiont internal transcribed spacer (ITS) region (symITS) of the ribosomal operon using the bacterium-specific primers and PCR conditions reported by Jensen et al. (23). We confirmed that these primers were symbiont specific by attempting amplification on adductor DNA (i.e., endosymbiont-free tissue used for the amplification of mitochondrial and nuclear genes) and failing to obtain any PCR product. A BLAST search (1) of the symITS sequences obtained in our study with sequences from GenBank showed that they were more similar to endosymbiont ITS sequences reported for other vesicomyid clams and, to a lesser degree, to the ITS sequence of uncultured free-living γ-subdivision proteobacterium, bacterial symbionts of vestimentiferan hydrothermal vent tubeworms, and other bacteria. We sequenced all PCR products in both directions, using ABI 377 or Licor 4000L sequencers. Sequences were proofread and aligned with Sequencher version 4.1 (Gene Codes Corp., Ann Arbor, Mich.). We found no genetic variation at cytb. Consequently this marker was not used for further analyses.
We cloned Cmg24 PCR amplification products from 10 individuals from 21°N, 10 from 9°N, and 10 from 17°S, using the PCR-Script Amp cloning kit (Stratagene, La Jolla, Calif.) and following the manufacturer's protocol. We sequenced one clone per individual, and the comparison among these sequences revealed the presence of a single polymorphic site. We then obtained PCR amplifications of the Cmg24 fragment for 75 clams. We sequenced directly all 75 PCR products on a Licor sequencer. These sequences did not reveal any additional polymorphic sites. The gel image produced by the Licor system allows one to determine whether an individual sample has more than one nucleotide at a particular position. We examined the gel image to establish whether an individual was homozygous (one band) or heterozygous (two bands) at the polymorphic site. We obtained the same scores for individual clams in repeated sequencing runs.
Population genetic analyses.
The program Arlequin 2000 (41) was used to estimate genetic differentiation between populations (FST), following the method of Hudson et al. (21), and to conduct exact tests of population genetic differentiation (40).
Independence tests.
If completely coupled, host mitochondrial and symbiont lineages should exhibit gametic phase disequilibrium. However, a small amount of horizontal or environmental transfers, which is equivalent to recombination, would eliminate this coupling on relatively short time scales. We used the likelihood ratio test (G test) as implemented with the JMP statistical program (version 4; SAS Institute, Cary, N.C.) to test for independence of mtCOI and symITS DNA sequences from individual clams. We also tested for nuclear-cytoplasmic disequilibrium between the nuclear gene Cmg24 and both cytoplasmic genes, symITS and mtCOI. In a randomly mating host population at gametic phase equilibrium, nuclear genes should vary independently of mitochondrial and symbiont genes.
Phylogenetic analyses.
We used PAUP*4.0b10 (42) to construct a gene tree of cytoplasmic haplotypes by combining mtCOI and symITS DNA sequences from individuals. To infer phylogenetic relationships among haplotypes, we used parsimony (equally weighted substitutions and characters), neighbor-joining based on Kimura-2-parameter distances (26), and maximum likelihood analyses, assuming empirical base frequencies, one substitution parameter, and equal rates among sites. We also conducted a partition homogeneity test (11) to examine whether combining sequence data from mtCOI and symITS was appropriate (i.e., each gene separately produces no significant conflicting tree topologies under the parsimony criteria). Parsimony analyses were performed with a branch and bound search, which guarantees finding the most parsimonious tree (43), and maximum likelihood analyses were performed with heuristic searches. An absence of homoplasy (i.e., recombination) in the combined mtCOI-symITS tree would be consistent with the hypothesis of strictly vertical transmission of symbiont and mitochondria. In contrast, detection of homoplasy would suggest the occurrence of recombination (i.e., horizontal or environmental transmission) of endosymbionts between clam lineages.
RESULTS
Geographical genetic variation.
We identified 6 mtCOI, 11 symITS, and 2 Cmg24 variants among the clams examined in this study (Table 1). Mitochondrial and symbiont cytoplasmic variants were observed in 16 different combinations (Table 2). We found low levels of genetic variation in the three markers. mtCOI variants differed by one to two nucleotide substitutions, symITS variants by one to five, and Cmg24 by one.
TABLE 1.
Frequencies of C. magnifica mitochondrial (mtCOI), nuclear (Cmg24), and symbiont (symITS) variants at EPR and GAR localities
| Gene and marker | GenBank accession no. | Frequency of marker at localitya:
|
||||
|---|---|---|---|---|---|---|
| 21°N | 9°N | 11°S | GAR | 17°S | ||
| mtCOI | ||||||
| 1 | AY191977 | 10 | 13 | 9 | 0 | 29 |
| 2 | AY191978 | 0 | 0 | 0 | 10 | 0 |
| 3 | AY191979 | 0 | 0 | 0 | 0 | 5 |
| 4 | AY191980 | 0 | 0 | 0 | 0 | 2 |
| 5 | AY191981 | 0 | 0 | 0 | 0 | 1 |
| 6 | AY191982 | 0 | 0 | 0 | 0 | 1 |
| symITS | ||||||
| A | AY191983 | 7 | 12 | 8 | 10 | 6 |
| B | AY191984 | 1 | 0 | 0 | 0 | 0 |
| C | AY191985 | 1 | 0 | 0 | 0 | 0 |
| D | AY191986 | 1 | 0 | 0 | 0 | 0 |
| E | AY191987 | 0 | 1 | 0 | 0 | 0 |
| F | AY191988 | 0 | 0 | 1 | 0 | 2 |
| G | AY191989 | 0 | 0 | 0 | 0 | 1 |
| H | AY191990 | 0 | 0 | 0 | 0 | 26 |
| I | AY191991 | 0 | 0 | 0 | 0 | 1 |
| J | AY191992 | 0 | 0 | 0 | 0 | 1 |
| K | AY191993 | 0 | 0 | 0 | 0 | 1 |
| Cmg24 | ||||||
| a | AY232659 | 20 | 22 | 18 | 20 | 8 |
| b | AY232660 | 0 | 0 | 0 | 0 | 62 |
We examined 80 clams for mtCOI and symITS and 75 for Cmg24 (i.e., 150 alleles).
TABLE 2.
Polymorphic nucleotide sites in mtCOI and symITS variantsa
| mtCOI variant | symITS variant |
|---|---|
| 11345 | 00000000000000000000011123356667 |
| 29062 | 78888888888999999999903685897896 |
| 98059 | 90123456789012345678902815754086 |
| 1 GTTGA | A CTTTTCATGATAACTTTTATAGGGCCACCTCT |
| 1 ..... | B ----------------------.......... |
| 1 ..... | C .......................A........ |
| 1 ..... | D ..............................T. |
| 1 ..... | E ..........................G..... |
| 1 ..... | F ......................A......... |
| 1 ..... | G ........................TT...... |
| 1 ..... | H ........................TT.....C |
| 1 ..... | I ........................TT.A...C |
| 1 ..... | J ........................TT..A..C |
| 1 ..... | K ........................TT...C.C |
| 2 ..C.. | A ................................ |
| 3 A.... | H ........................TT.....C |
| 5 ...A. | H ........................TT.....C |
| 4 .C... | H ........................TT.....C |
| 6 ....G | H ........................TT.....C |
The data indicate the 16 different combinations observed among the variants of these cytoplasmic genes. Numbers correspond to mtCOI variants, and letters correspond to symITS variants. Vertical numbers above nucleotides indicate the base position of the polymorphic nucleotides relative to the DNA fragments deposited in GenBank for mtCOI and symITS.
Population genetic analyses.
Evidence existed for genetic differentiation or population subdivision across the range of C. magnifica (Table 1). The sample from 17°S differed from all other EPR and GAR localities for novel alleles at all three marker loci. Exact tests of genetic differentiation between 17°S and the other localities were significant for symITS and Cmg24 (Table 3). In addition, the GAR sample differed significantly from all other localities for mtCOI.
TABLE 3.
FST values for populations of C. magnifica in the EPR and GAR localities
| Marker | Locality |
FST value between localitiesa
|
||||
|---|---|---|---|---|---|---|
| 21°N | 9°N | GAR | 11°S | 17°S | ||
| mtCOI | 21°N | 0 | 1* | 0 | 0.07 | |
| 9°N | 1* | 0 | 0.07 | |||
| GAR | 1* | 0.82* | ||||
| 11°S | 0.07 | |||||
| symITS | 21°N | 0 | 0 | 0 | 0.68* | |
| 9°N | 0 | 0 | 0.71* | |||
| GAR | 0 | 0.73* | ||||
| 11°S | 0.70* | |||||
| Cmg24 | 21°N | 0 | 0 | 0 | 0.81* | |
| 9°N | 0 | 0 | 0.81* | |||
| GAR | 0 | 0.81* | ||||
| 11°S | 0.81* | |||||
Asterisks mark population pairs that were significantly different according to exact tests of population differentiation (P < 0.05).
Tests of genic independence.
To test the hypothesis that symbiont lineages were coupled with the host's maternally inherited mitochondrial DNA, we needed to avoid the confounding effects of intergenic correlations due to population subdivision. We were able to test for coupling of cytoplasmic lineages among individual clams from only a single population, 17°S, where clams were simultaneously polymorphic for symbiont and mitochondrial gene markers. For the entire sample of 38 clams obtained from the 17°S region, sequence variants for each marker were lumped into two categories: (i) variants that were unique to this locality, versus (ii) variants that also occurred in the north. We conducted 2 × 2, likelihood ratio, contingency tests of linkage disequilibrium between all three pairwise combinations of genetic markers (Table 4) to test the null hypothesis of genic independence (i.e., gametic phase equilibrium). The null hypothesis was only rejected for the symITS-mtCOI combination of symbiont and mitochondrial markers (test i), which is expected under strictly vertical cytoplasmic cotransmission. The null hypothesis was not rejected for the genic combinations involving the nuclear marker, Cmg24-mtCOI and Cmg24-symITS (i.e., tests ii and iii), as expected for independent loci at nuclear-cytoplasmic equilibrium.
TABLE 4.
Likelihood ratio contingency test of linkage disequilibrium between host and symbiont genetic markers for C. magnifica clams collected at 17°Sa
| Test | Contrast | Frequency of gametic combination
|
G2b | Pc | |||
|---|---|---|---|---|---|---|---|
| N/N | N/S | S/N | S/S | ||||
| i | symITS-mtCOI | 8 (6.1) | 0 (1.9) | 21 (22.9) | 9 (7.1) | 4.951 | 0.026* |
| ii | Cmg24-mtCOI | 5 (5.9) | 3 (2.1) | 47 (46.1) | 15 (16.0) | 0.612 | 0.434 |
| iii | Cmg24-symITS | 2 (1.6) | 6 (6.4) | 12 (12.4) | 50 (49.6) | 0.134 | 0.714 |
Three tests were conducted: (i) symbiont versus host mitochondria; (ii) host nuclear versus host mitochondria; and (iii) host nuclear versus symbiont nuclear. Expected numbers appear in parentheses. Each gametic combination was labeled S, representing 17°S endemic variants, or N, representing alternative variants, as defined in the text and the legend for Fig. 2.
One degree of freedom.
∗, statistically significant at P < 0.05.
Phylogenetic coevolution of symbiont and mitochondrial lineages.
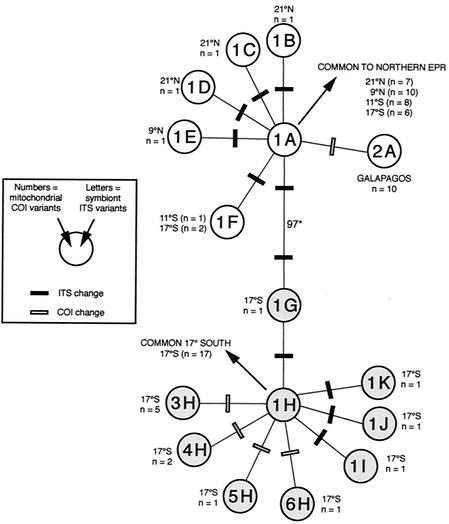
We used the 16 different combinations of the 6 mtCOI and 11 symITS variants found among the 78 clams examined (Table 2) to construct a gene tree of cytoplasmic haplotypes (Fig. 2). The same tree topology was obtained regardless of the phylogenetic method used (parsimony, distance, or maximum likelihood). Combining sequence data from mtCOI and symITS was appropriate for this phylogenetic analysis because a partitioning homogeneity test (11) failed to detect significant heterogeneity in tree topologies between the two genes.
FIG. 2.
Gene tree of cytoplasmic haplotypes combining host mitochondrial COI (mtCOI) and symbiont ITS (symITS) DNA sequences. Each circle represents a unique mtCOI-symITS haplotype combination (16 different combinations were observed among the 80 clams examined). Numbers inside the circles represent mtCOI haplotypes, and letters represent symITS variants (see Table 2). Gray circles indicate cytoplasmic haplotype combinations observed exclusively at 17°S. Combinations 1B, 1C, 1D, and 1E were observed only in clams from EPR localities other than 17°S. Combinations 1A and 1F were observed among clams from 17°S and other EPR localities. Combination 2A was the only combination observed among the clams examined from GAR. The bootstrap value (*) based on a branch and bound search indicates that 97% of 100 replicates supported the separation of the two groups defined by this node.
The coevolutionary path that reflects joint changes in symbiont and mitochondrial sequences in C. magnifica was clearly revealed in the combined phylogenetic tree for symITS and mtCOI sequences. As expected for coupled genetic systems, the phylogenetic distribution of symITS variants was not independent of clam mtCOI variants. The tree identified two groups that correspond with 17°S endemic combinations versus combinations found at populations to the north (11°S, 9°N, 21°N, and GAR). For EPR localities to the north of 17°S, all within-population variation was due to changes in symITS. The common haplotype (1-A) was found in 83.3% of the EPR individuals to the north of 17°S. The GAR sample exhibited a unique haplotype (2-A) that differed by a single substitution in mtCOI. In contrast, the southern population at 17°S exhibited variation in both its mitochondrial and bacterial sequences. The most common northern combination (1-A) also occurred at 17°S but was less frequent (15.8%). Two individuals (5.3%) had the combination (1-F) also found in the north, but all other 17°S individuals exhibited combinations found only at this locality. The most common southern haplotype (1-H) occurred in 44.7% of individuals at 17°S. The four mtCOI variants unique to 17°S (variants 3 to 6) only occurred with a symITS variant (type H) also unique to 17°S, representing 22.7% of the clams at this locality. Apparently, mtCOI variants 3 to 6 arose in a clam lineage that already had symITS type H. Likewise, the predominantly northern symITS variants (types A to F) arose in a clam lineage that already had the northern mtCOI type 1.
DISCUSSION
All our findings are consistent with coupling of mitochondrial and endosymbiont lineages due to strictly maternal cotransmission. As expected for tightly coupled genes, the pattern of geographical variation in symITS paralleled that of the clam mtCOI. Both genes exhibited novel groups of alleles found only at 17°S in the EPR. However, such geographical covariation is not necessarily a consequence of coupling due to strictly cytoplasmic cotransmission. For example, covariation between gene loci can result from a common history of population subdivision and genetic drift (31), a likely hypothesis since parallel gene frequency shifts also occurred in the nuclear marker Cmg24 (Table 1). Because nuclear genes assort independently of cytoplasmic factors in a sexually reproducing population, such geographical covariance may have resulted from historical processes that simultaneously affected nuclear and cytoplasmic genomes. However, in our analysis, we avoided the problem of shared historical population structure by testing the independence of gene associations (i.e., gametic phase equilibrium) within a single population.
Significant associations between the two cytoplasmic genomes were observed at 17°S, which is expected with coupling of mitochondrial and endosymbiont lineages due to strictly maternal cotransmission. Additionally, we demonstrated that variation at a nuclear gene was independent of the two cytoplasmic markers, as expected for a randomly mating population at gametic phase equilibrium. The null hypothesis of independence was not rejected for genic combinations involving the nuclear marker (Cmg24-mtCOI and Cmg24-symITS), as expected for independent loci at nuclear-cytoplasmic equilibrium. This latter result also provided evidence that cryptic subdivision (i.e., the Wahlund effect [45]) does not exist in the clams at 17°S and that this population comprises a randomly mating sexual population. A previous study of allozymes and nuclear DNA markers in C. magnifica also revealed no evidence for inbreeding or cryptic subdivision within populations of this species (24). Thus, the genic disequilibrium between symITS and mtCOI provides substantive evidence for strict coupling.
Phylogenetic analyses also reinforced the view that maternal cytoplasmic inheritance is the only mechanism for symbiont transmission in this clam species. We found no phylogenetic evidence for recombination between symbiont and mitochondrial markers when the two sequences were combined into cytoplasmic haplotypes. Lack of homoplasy in the combined tree topology is consistent with no horizontal or environmental transmission of symbionts (14). If occasional leakage of symbionts occurs between host lineages, mixed symbiotic and mitochondrial sequences and homoplasy are expected (14). If we had observed just one clam (out of 38 from 17°S) bearing symITS types A, F, or G with mtCOI types 3, 4, 5, or 6, the resulting homoplasy would suggest some endosymbiont horizontal or environmental transmission.
We considered additional factors that might have affected our present results. First, the inheritance of mitochondria is not strictly maternal in some marine bivalves. Some of them exhibit doubly uniparental inheritance, i.e., female mitochondrial lineages are transmitted through eggs and male mitochondrial lineages are transmitted through sperm (36, 46). If paternal transmission of mitochondrial DNA occurs in vesicomyid clams, it would mix mitochondria and symbionts from different lineages and eliminate the tight coupling observed in this study. We found no evidence for doubly uniparental inheritance in these clams, e.g., heteroplasmy of mtCOI sequences. Another potentially confounding factor is that the ribosomal operon of some bacteria may occur in multiple copies (7). Divergent symITS sequences might reflect paralogous rather than homologous variation and confound the association with mitochondrial variation. Yet, we found no evidence for paralogous variation in individual clams, despite multiple PCR amplifications of individuals from polymorphic populations. DNA sequencing unambiguously revealed only one symITS per individual. If multiple ribosomal operons exist in these bacteria, their ITS sequences might have converged on a single copy (concerted evolution). Alternatively, PCR primer bias might have favored amplification of a single copy, an unlikely prospect given the highly conserved ribosomal regions targeted by the ITS primers used in this study. Nevertheless, rather than produce artificial associations, incorporation of multiple copies of either mtCOI or symITS in our analyses might have obscured mitochondrial-symbiont associations.
We observed extremely low levels of genetic variation in the clam mtCOI marker. We used this marker because it shows a high degree of polymorphism within and among populations of several other hydrothermal vent species (22). However, our results indicate that, compared to other hydrothermal vent species, variation at this gene is very low in C. magnifica. Similarly, we did not observe any polymorphism at another mitochondrial marker (a fragment of cytb) among several clams, even though this marker is very variable in other hydrothermal vent species (22). These observations are consistent with a previous study that showed extremely low levels of genetic polymorphism for this clam species at other markers, including Cmg24 (24). These authors suggest that high rates of extinction of local populations and subsequent recolonization may have eroded genetic diversity in this clam species. C. magnifica is a late colonizer of hydrothermal vents, and thus vents may disappear before or shortly after this species settles. It is possible that continuous bottlenecks have dramatically reduced mitochondrial and nuclear genetic variability in this species. However, our data indicate that symITS evolves more rapidly than host mtCOI, a possible consequence of the less-constrained, noncoding nature of the ITS. Additionally, genes of vertically transmitted symbionts appear to exhibit accelerated nucleotide substitution rates compared to host mitochondrial genes (29) and compared to free-living bacterial counterparts (39). It is remarkable that despite the low genetic variation at mtCOI and the relatively limited sample from 17°S, we detected significant cytoplasmic associations at this locality.
In conclusion, the present results provide compelling evidence for genetic coupling due to strictly maternal cotransmission of host mitochondrial and symbiont genomes in a sexually reproducing species. The present evidence for strict maternal cotransmission suggests that the endosymbiotic bacterium may have lost its ability to live freely in the marine environment and reinfect C. magnifica clams. This is surprising because closely related, γ-subdivision proteobacterial endosymbionts of lucinid clams and vestimentiferan tubeworms appear to be environmentally acquired (4, 12, 17, 27). The evolution of vertical cotransmission by vesicomyid endosymbionts is associated with an accelerated rate of nucleotide substitution (39), a phenomenon also observed in vertically transmitted endosymbionts of aphids (29). If the clam endosymbiont has become completely dependent on its hosts for transmission, we expect it might also exhibit evidence for genome reduction, e.g., loss of genes essential for free-living bacteria, as has been observed in the Buchnera endosymbionts of aphids (44). Yet the association of these γ-subdivision proteobacteria with vesicomyid clams is relatively young, probably less than 50 million years old (38). The evolution of complete interdependence between the participants in this symbiosis may parallel ancient evolutionary processes by which eukaryotic cells acquired mitochondria, chloroplasts, and possibly other cytoplasmic organelles (28). Therefore, the vesicomyid-endosymbiont system may serve as a useful model for exploring early stages in the evolution of new cytoplasmic organelles.
Acknowledgments
This study was supported by NSF grants (OCE9633131 and OCE9910799) and generous funding from the Monterey Bay Aquarium Research Institute (the David and Lucille Packard Foundation).
We thank Ed DeLong, Shana Goffredi, Heide Schulz, Peter Smouse, Robbie Young, and two anonymous reviewers for helpful comments on the manuscript; Irvin Pan, Steven Hallam, and Yong Jin Won for technical assistance; and Tim Shank for providing additional clam specimens from the vicinity of 17°S. We are especially grateful to the crew and pilots of the R/V Atlantis and DSV Alvin for their efforts to obtain the specimens used in this study.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Black, M. B., K. Halanych, P. Maas, W. R. Hoeh, J. Hashimoto, D. Desbruyères, R. Lutz, and R. C. Vrijenhoek. 1997. Molecular systematics of deep-sea tube worms (Vestimentifera). Mar. Biol. 130:141-149. [Google Scholar]
- 3.Cary, S. C., and S. J. Giovannoni. 1993. Transovarial inheritance of endosymbiotic bacteria in clams inhabiting deep-sea hydrothermal vents and cold seeps. Proc. Natl. Acad. Sci. USA 90:5695-5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cary, S. C., W. Warren, E. Anderson, and S. J. Giovannoni. 1993. Identification and localization of bacterial endosymbionts in hydrothermal vent taxa with symbiont-specific polymerase chain reaction amplification and in situ hybridization techniques. Mol. Mar. Biol. Biotechnol. 2:51-62. [PubMed] [Google Scholar]
- 5.Cavanaugh, C. M. 1985. Symbiosis of chemolithotrophic bacteria and marine invertebrates from hydrothermal vents and reducing sediments. Biol. Soc. Wash. Bull. 6:373-388. [Google Scholar]
- 6.Childress, J. J., H. Felbeck, and G. N. Somero. 1987. Symbiosis in the deep sea. Sci. Am. 255:114-120. [Google Scholar]
- 7.Daffonchio, D., A. Cherif, and S. Borin. 2000. Homoduplex and heteroduplex polymorphisms of the amplified ribosomal 16S-23S internal transcribed spacers describe genetic relationships in the “Bacillus cereus group.” Appl. Environ. Microbiol. 66:5460-5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dahlgren, T. G., J. R. Weinberg, and K. M. Halanych. 2000. Phylogeography of the ocean quahog (Arctica islandica): influences of paleoclimate on genetic diversity and species range. Mar. Biol. 137:487-495. [Google Scholar]
- 9.DiMeo, C. A., A. E. Wilbur, W. E. Holben, R. A. Feldman, R. C. Vrijenhoek, and S. C. Cary. 2000. Genetic variation among endosymbionts of widely distributed vestimentiferan tubeworms. Appl. Environ. Microbiol. 66:651-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Endow, K., and S. Ohta. 1990. Occurrence of bacteria in the primary oocytes of vesicomyid clam Calyptogena soyoae. Mar. Ecol. Prog. Ser. 64:309-311. [Google Scholar]
- 11.Farris, J. S., M. Kallersjo, A. G. Kluge, and C. Bult. 1994. Testing significance of incongruence. Cladistics 10:315-319. [Google Scholar]
- 12.Feldman, R. A., M. B. Black, C. S. Cary, R. A. Lutz, and R. C. Vrijenhoek. 1997. Molecular phylogenetics of bacterial endosymbionts and their vestimentiferan hosts. Mol. Mar. Biol. Biotechnol. 6:268-277. [PubMed] [Google Scholar]
- 13.Fiala-Medioni, A., and M. LePennec. 1988. Structural adaptations in the gill of the Japanese subduction zone bivalves (Vesicomyidae) Calyptogena phaseoliformes and Calyptogena laubiere. Oceanol. Act. 11:185-192. [Google Scholar]
- 14.Funk, D. J., L. Helbling, J. J. Wernegreen, and N. A. Moran. 2000. Intraspecific phylogenetic congruence among multiple symbiont genomes. Proc. R. Soc. Lond. B 267:2517-2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Funk, D. J., J. J. Wernegreen, and N. A. Moran. 2001. Intraspecific variation in symbiont genomes: bottlenecks and the Aphid-Buchnera association. Genetics 157:477-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gray, M. W., G. Burger, and B. F. Lang. 1999. Mitochondiral evolution. Science 283:1476-1481. [DOI] [PubMed] [Google Scholar]
- 17.Gros, O., A. Darrasse, P. Durand, L. Frenkiel, and M. Moueza. 1996. Environmental transmission of sulfur-oxidizing bacterial gill endosymbiont in the tropical lucinid bivalve Codakia orbicularis. Appl. Environ. Microbiol. 62:2324-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hafner, M. S., and R. D. M. Page. 1995. Molecular phylogenies and host-parasite cospeciation: gophers and lice as a model system. Phil. Trans. R. Soc. Lond. B 349:77-83. [DOI] [PubMed] [Google Scholar]
- 19.Halanych, K. M., R. A. Lutz, and R. C. Vrijenhoek. 1998. Evolutionary origins and age of vesitimentiferan tube worms. Cah. Biol. Mar. 39:355-358. [Google Scholar]
- 20.Haymon, R. M., and R. A. Koski. 1985. Evidence of an ancient hydrothermal vent community: fossil worm tubes in cretaceous sulfide deposits of the Samail ophiolite, Oman. Biol. Soc. Wash. Bull. 6:57-65. [Google Scholar]
- 21.Hudson, R. R., D. D. Boos, and N. L. Kaplan. 1992. A statistical test for detecting geographic subdivision. Mol. Biol. Evol. 9:138-151. [DOI] [PubMed] [Google Scholar]
- 22.Hurtado, L. A. 2002. Evolution and biogeography of hydrothermal vent organisms in the Eastern Pacific Ocean. Ph.D. thesis. Rutgers University, New Brunswick, N.J.
- 23.Jensen, M. A., J. A. Webster, and N. Straus. 1993. Rapid identification of bacteria on the basis of polymerase chain reaction-amplified ribosomal DNA spacer polymorphisms. Appl. Environ. Microbiol. 59:945-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karl, S. A., S. J. Schutz, D. Desbruyères, R. A. Lutz, and R. C. Vrijenhoek. 1996. Molecular analysis of gene flow in the hydrothermal-vent clam Calyptogena magnifica. Mol. Mar. Biol. Biotechnol. 5:193-202. [Google Scholar]
- 25.Kennish, M. J., and R. A. Lutz. 1992. The hydrothermal vent clam, Calyptogena magnifica (Boss and Turner 1980): a review of existing literature. Rev. Aquat. Sci. 6:29-66. [Google Scholar]
- 26.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitution through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 27.Laue, B. E., and D. C. Nelson. 1997. Sulfur-oxidizing symbionts have not co-evolved with their hydrothermal vent tube worm hosts: an RFLP analysis. Mol. Mar. Biol. Biotechnol. 6:180-188. [PubMed] [Google Scholar]
- 28.Margulis, L. 1984. Early life. Jones and Bartlett, Boston, Mass.
- 29.Moran, N. A. 1996. Accelerated evolution and Muller's ratchet in endosymbiotic bacteria. Proc. Natl. Acad. Sci. USA 93:2873-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moran, N. A., M. A. Munson, P. Baumann, and H. Ishikawa. 1993. A molecular clock in endosymbiotic bacteria is calibrated using the insect hosts. Proc. R. Soc. Lond. B. 253:167-171. [Google Scholar]
- 31.Nei, M., and W. H. Li. 1973. Linkage disequilibrium in subdivided populations. Genetics 75:213-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nelson, D. C., and C. R. Fisher. 1995. Chemoautotrophic and methanotrophic endosymbiotic bacteria at deep-sea vents and seeps, p. 125-167. In D. M. Karl (ed.), Microbiology of deep-sea hydrothermal vent habitats. CRC Press, Boca Raton, Fla.
- 33.Nelson, K., and C. Fisher. 2000. Absence of cospeciation in deep-sea vestimentiferan tube worms and their bacterial endosymbionts. Symbiosis 28:1-15. [Google Scholar]
- 34.Nishiguchi, M. K., E. G. Ruby, and M. J. McFall-Ngai. 1998. Competitive dominance among strains of luminous bacteria provides an unusual evidence for parallel evolution in sepiolid squid-vibrio symbioses. Appl. Environ. Microbiol. 64:3209-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishiguchi, M. K. 2002. Host recognition is responsible for symbiont composition in environmentally transmitted symbiosis. Microb. Ecol. 44:10-18. [DOI] [PubMed] [Google Scholar]
- 36.Passamonti, M., and V. Scali. 2001. Gender-associated mitochondrial DNA heteroplasmy in the venerid clam Tapes philippinarum (Mollusca Bivalvia). Curr. Genet. 39:117-124. [DOI] [PubMed] [Google Scholar]
- 37.Peek, A., R. Gustafson, R. Lutz, and R. Vrijenhoek. 1997. Evolutionary relationships of deep-sea hydrothermal vent and cold-water seep clams (Bivalvia: Vesicomyidae): results from the mitochondrial cytochrome oxidase subunit I. Mar. Biol. 130:151-161. [Google Scholar]
- 38.Peek, A. S., R. A. Feldman, R. A. Lutz, and R. C. Vrijenhoek. 1998. Cospeciation of chemoautotrophic bacteria and deep sea clams. Proc. Natl. Acad. Sci. USA 95:9962-9966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peek, A. S., R. C. Vrijenhoek, and B. S. Gaut. 1998. Accelerated evolutionary rate in sulfur-oxidizing endosymbiotic bacteria associated with the mode of symbiont transmission. Mol. Biol. Evol. 15:1514-1523. [DOI] [PubMed] [Google Scholar]
- 40.Raymond, M., and F. Rousset. 1995. An exact test for population differentiation. Evolution 49:1280-1283. [DOI] [PubMed] [Google Scholar]
- 41.Schneider, S., D. Roessli, and L. Excoffier. 2000. Arlequin, a software package for population genetics data analysis, 2.000 ed. Genetics and Biometry Laboratory, Department of Anthropology, University of Geneva, Geneva, Switzerland.
- 42.Swofford, D. L. 1998. PAUP*: phylogenetic analysis using parsimony (*and other methods), 4th ed. Sinauer, Sunderland, Mass.
- 43.Swofford, D. L., G. J. Olsen, P. J. Waddell, and D. M. Hillis. 1996. Phylogenetic inference, p. 407-514. In D. M. Hillis, C. Moritz, and P. K. Mable (ed.), Molecular systematics, 2nd ed. Sinauer Associates, Sunderland, Mass.
- 44.Tamas, I., L. Klasson, B. Canback, A. K. Naslund, A. S. Eriksson, J. J. Wernegreen, J. P. Sandstrom, N. A. Moran, and S. G. Andersson. 2002. 50 million years of genomic stasis in endosymbiotic bacteria. Science 296:2376-2379. [DOI] [PubMed] [Google Scholar]
- 45.Wahlund, S. 1928. Zusammensetzung von Populationen und Korrelationerscheinungen vom Standpunkt der Vererbungslehre aus Betrachtet. Hereditas 11:65-106. [Google Scholar]
- 46.Zouros, E., A. O. Ball, C. Saavedra, and K. R. Freeman. 1994. An unusual mitochondrial DNA inheritance in the blue mussel Mytilus. Proc. Natl. Acad. Sci. USA 91:7463-7467. [DOI] [PMC free article] [PubMed] [Google Scholar]