Abstract
A heat-stable raw-starch-digesting amylase (RSDA) was generated through PCR-based site-directed mutagenesis. At 65°C, the half-life of this mutant RSDA, which, compared with the wild-type RSDA, lacks amino acids R178 and G179, was increased 20-fold. While the wild type was inactivated completely at pH 3.0, the mutant RSDA still retained 41% of its enzymatic activity. The enhancement of RSDA thermostability was demonstrated to be via a Ca2+-independent mechanism.
Raw starch is the most abundant source of glucose in the world. Industrially, starch granules are gelatinized in water to improve the conversion of starch molecules into maltodextrins and simple sugars by α-amylase and/or other amylolytic enzymes (13). This gelatinization of starch granules requires heating and a long reaction time. Therefore, finding enzymes capable of digesting raw starch directly would provide a possible replacement for such an expensive and energy-consuming process. Many organisms, including fungi, yeast, and bacteria, are known to produce raw-starch-digesting amylases (RSDAs) (1, 2, 3, 22). Some of these RSDAs have been produced and applied to the beverage, food, paper, and textile industries as well as to dishwashing and laundry detergent production (11). However, the relatively low reaction temperature optimal for RSDA on granular starch results in not only a low reaction rate but also a high risk of bacterial contamination. Therefore, increasing the thermostability of RSDAs for industrial utilization is an important goal of protein engineering.
Studies have shown that a loop area within domain B of α-amylase plays a crucial role in improving the thermostability of this protein (17). Deletion of an Arg-Gly dipeptide from both the liquefying α-amylase (LAMY) of Bacillus strain KSM-1378 and Bacillus amyloliquefaciens amylase (BAA) increased their thermostability (5, 18). In previous reports, an RSDA from a Cytophaga sp. has been shown to exhibit an excellent starch digestion activity on raw cornstarch (7, 8). By comparing the amino acid sequences of this RSDA and other amylases (5, 12, 19, 23), a loop structure within domain B in Cytophaga sp. RSDA similar to those in other amylases was also found. In this study, we demonstrated that removing Arg and Gly residues from the corresponding loop area in this RSDA improved the stability of the enzyme both at high temperatures and in stringent environments. To the best of our knowledge, this is the first thermostable RSDA that has been reported to date.
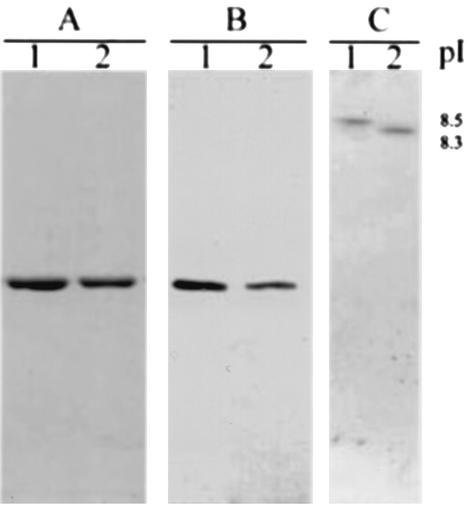
The mutant RSDA, with residues 178 and 179 deleted, was generated by site-directed mutagenesis by the method of splicing by overlap extension based on PCR (4). A recombinant plasmid (unpublished data), pHR, was constructed by ligation of pHY300PLK (TaKaRa, Shing, Japan) and a DNA fragment containing the rsda gene. To generate the deletion mutant, the nucleotide sequence of the rsda gene was PCR amplified on pHR by using primer 1 (5′-CGGACCGTACGATTTGTACGATCTAGG- 3′, where the BsiWI recognition sequence is underlined) and primer 2 (5′-CCCATGCTTTGCCGGTAAACTTAAAGATGCGGCTCAA-3′) (reaction 1). A separate PCR was done by using primer 3 (5′-GAGCCGCATCTTTAAGTTTACCGGCAAAGCAT-3′) and primer 4 (5′-TCCTTCTCGAGTCCATCCAATCACATCT -3′, where the XhoI recognition sequence is underlined) (reaction 2). Primers 2 and 3 are deletion mutation primers that lack the 6 nucleotides (5′-AGGGGA-3′ from the 532th to 537th nucleotide) that encode the Arg178 and Gly179 residues. In the second round of PCR, 80-ng samples of DNA fragments from each of reactions 1 and 2 were mixed and used as templates for further amplification with primers 1 and 4. The final product was isolated from the agarose gel, digested with BsiWI and XhoI, and reinserted into pHR linearized with the same enzymes. The newly constructed plasmid was designated pHRΔRG. All constructs were verified by DNA sequencing (14). The wild-type and deletion mutant RSDA proteins were purified from Bacillus subtilis(pHR) and B. subtilis(pHRΔRG) culture media through ammonium sulfate precipitation, DEAE-Sepharose CL-6B ion-exchanger chromatography, and Sephacryl S-200 gel filtration (7). The active fractions were then collected and pooled. The purified wild-type RSDA and mutant RSDA (RSDAΔRG) from B. subtilis appeared at the same location on a sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel as that of the RSDA from a Cytophaga sp., determined both by Coomassie brilliant blue staining (Fig. 1A) and by Western blot analysis (Fig. 1B). Isoelectrofocusing gel electrophoresis using the PhastSystem (Pharmacia, Uppsala, Sweden) showed that the pI value of the RSDAΔRG (8.3) was different from that of the wild-type RSDA (8.5) (Fig. 1C). These results indicate that removing the positive charge from the protein surface caused a shift of the pI value of the RSDAΔRG.
FIG. 1.
Identification of purified RSDA and RSDAΔRG produced by B. subtilis DB430 on sodium dodecyl sulfate-polyacrylamide gel eletrophoresis with Coomassie brilliant blue staining (A), Western blotting (B), and isoelectrofocusing electrophoresis (C) with Coomassie brilliant blue staining. Lanes 1 and 2, purified RSDA and RSDAΔRG, respectively.
The raw-starch-digesting activities of wild-type RSDA and RSDAΔRG were assayed by the method described by Itakor et al. (6), with slight modification, i.e., by using 4% (wt/vol) raw cornstarch suspended in 0.04 M imidazole buffer, pH 7.0, as a substrate and incubating at 45°C for 20 min. One unit of RSDA activity was defined as the amount of enzyme that produced an amount of reducing sugar equivalent to 1 mg of glucose per h (6). The specific activity of RSDAΔRG (3,415 U/mg) was 35% of that of wild-type RSDA (9,640 U/mg). This result implies that the deletion of residues 178 and 179 had an adverse effect on enzymatic activity. The enzymatic activities of RSDAs (7) were measured at concentrations of raw cornstarch from 0.5 to 15%, with kinetic parameters calculated by using the Lineweaver-Burk equation. The turnover numbers of RSDAΔRG and wild-type RSDA proteins were 3.5 and 4.5 s−1, respectively. In addition, the Km value of RSDAΔRG (27% [wt/vol]) was fivefold higher than that of wild-type RSDA (5.6% [wt/vol]). Therefore, these results suggest that the deletion of residues 178 and 179 leads to a lower catalytic activity and an increased Km value of the RSDA. It has been reported that the substrate-binding region in α-amylase does not include these two amino acid residues (9, 15). The deletion of residues Arg178 and Gly179 might change the three-dimensional structure of RSDA and result in a loss of its affinity to raw starch.
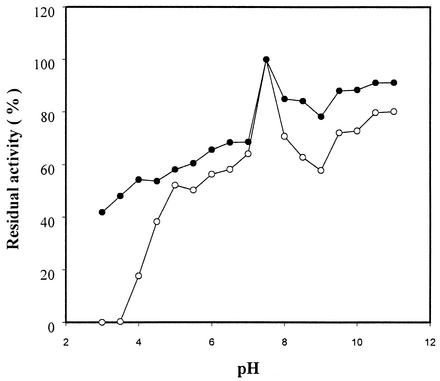
To determine the optimal pH range for RSDA, the pH of the incubation solution was adjusted with 100 mM universal buffer (10) and enzyme activity was measured from pH 3 to 11 with intervals of 0.5 pH unit. The optimal pH for both wild-type RSDA and RSDAΔRG was in the range of 5.5 to 7.5 (data not shown). After incubation at various pHs for 2 h at 4°C, the residual activity was measured at pH 7.0. RSDA incubated at pH 7.5 retained 100% of its activity and was used as the reference. At pH 3.0 and 11, RSDAΔRG exhibited 41 and 90%, respectively, of the reference RSDA activity. However, wild-type RSDA completely lost its amylase activity at pH 3.0, while at pH 11 it retained 80% of the activity of the reference RSDA (Fig. 2). RSDAΔRG appears to be more stable in stringent environments than wild-type RSDA. This characteristic allows the mutant enzyme to become more applicable to the food and detergent industries.
FIG. 2.
Effect of pH on the stability of RSDA (○) and RSDAΔRG (•). The enzymes were incubated at 4°C in buffers of various pHs for 2 h prior to the determination of residual activity.
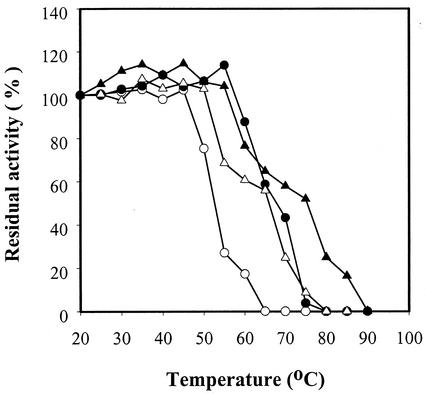
The optimal temperature of this enzyme was assayed between 20 and 90°C at 5°C intervals by using soluble starch. The optimal temperature curves of RSDAΔRG and wild-type RSDA were very similar, with temperature optima ranging from 35 to 55°C (data not shown). To determine the thermostability, RSDAs were incubated for 1 h at the temperatures indicated in Fig. 3 before being subjected to activity assays. Then the residual enzymatic activity acting on 4% raw starch was measured at 45°C in 0.04 M, pH 7.5, imidazole buffer for 20 min. As shown in Fig. 3, the residual activities of wild-type RSDA and RSDAΔRG at 55°C were 27 and 68% of that at 20°C, respectively, and wild-type RSDA was totally inactivated while mutant RSDA retained 56% of its activity during incubation at 65°C. To determine the thermostability of RSDAs, another set of tests was carried out in the presence of 2 mM CaCl2. At 65°C with 2 mM Ca2+, wild-type RSDA retained 60% of its enzymatic activity; however, in the absence of 2 mM Ca2+, its activity was lost completely (Fig. 3).
FIG. 3.
Thermostability of RSDA (○, •) and RSDAΔRG (▵, ▴). Both enzymes were incubated for 1 h without (open symbols) or with (fill symbols) 2 mM CaCl2 at different temperatures prior to the determination of the residual activity at 45°C.
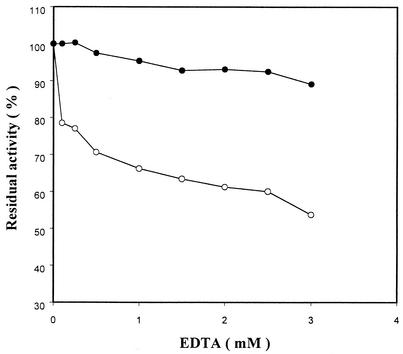
In a compensatory experiment, the enzymatic activity on raw cornstarch was measured with Ca2+ depleted from the enzyme solution by EDTA. As shown in Fig. 4, wild-type RSDA lost 50% of its activity in the presence of 3 mM EDTA, but the activity of RSDAΔRG was only slightly affected under the same reaction conditions. This result was different from what was observed with most α-amylases (5, 9, 16, 17). The enzymatic activity of LAMY was completely lost in the presence of 0.05 mM EDTA, and LAMYΔRG (mutant protein) under the same treatment lost 30 to 40% of its activity. In addition, it was reported that the thermostability of LAMYΔRG was improved by enhanced Ca2+ binding to the enzyme molecule (5).
FIG. 4.
Effect of EDTA on the activity of RSDA (○) and RSDAΔRG (•). The enzyme was incubated at 40°C for 30 min prior to the determination of residual enzyme activities with 4% raw cornstarch.
Suzuki et al. (18) constructed various chimeric genes from the structural genes of B. licheniformis α-amylase and BAA and analyzed the products. They showed that amino acid residues within two regions, i.e., region I (176th to 178th amino acids) and region II (266th to 269th amino acids), independently affected thermostability. Our data confirmed their observation that region I plays a major role in the thermostability of the α-amylase.
The deletion of two residues from RSDA will decrease the positive charge of the protein molecule, change the electrostatic interaction, and increase hydrophobicity. Our results indicate that RSDAΔRG possesses an enhanced half-life at higher temperatures. This phenomenon may be caused by delaying the reversible unfolding of the protein molecule or preventing irreversible inactivation, as mentioned by Tomazic and Klibanov (20, 21).
The mutant enzyme RSDAΔRG, made by site-directed mutagenesis of the Cytophaga sp. rsda gene, showed not only increased thermostability but also increased resistance to extreme pH conditions. These characteristics endow the mutant enzyme with broad applications in industry.
REFERENCES
- 1.Gawande, B. N., and A. Y. Patkar. 2001. Purification and properties of a novel raw starch degrading cyclodextrin glycosyltransferase from Klebsiella pneumoniae AS-22. Enzyme Microb. Technol. 22:735-743. [DOI] [PubMed] [Google Scholar]
- 2.Hamilton, L. M., C. T. Kelly, and W. M. Fogarty. 1999. Purification and properties of the raw starch degrading α-amylase of Bacillus sp. IMD 434. Biotechnol. Lett. 21:111-115. [Google Scholar]
- 3.Hamilton, L. M., C. T. Kelly, and W. M. Fogarty. 1999. Purification and properties of the raw starch degrading α-amylase of Bacillus sp. IMD 435. Proc. Biotechnol. 35:27-31. [Google Scholar]
- 4.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 5.Igarashi, K., Y. Hatada, K. Ikawa, H. Araki, T. Ozawa, T. Kobayashi, K. Ozaki, and S. Ito. 1998. Improved thermostability of a Bacillus α-amylase by deletion of an arginine-glycine residue is caused by enhanced calcium binding. Biochem. Biophys. Res. Commun. 248:372-377. [DOI] [PubMed] [Google Scholar]
- 6.Itakor, P., O. Shida, and N. Tsukadoshi. 1989. Screening for raw starch digesting bacteria. Agric. Biol. Chem. 53:53-60. [Google Scholar]
- 7.Jeang, C. L., Y. H. Lee, and L. W. Chang. 1995. Purification and characterization of a raw-starch digesting amylase from a soil bacterium—Cytophaga sp. Biochem. Mol. Biol. Int. 35:549-557. [PubMed] [Google Scholar]
- 8.Jeang, C.-L., L.-A. Chen, M.-Y. Chen, and R.-J. Shiau. 2002. Cloning of a gene encoding raw-starch-digesting amylase from a Cytophaga sp. and its expression in Escherichia coli. Appl. Environ. Microbiol. 68:3651-3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Machius, M., N. Declerck, R. Huber, and G. Wiegand. 1998. Activation of Bacillus licheniformis α-amylase through a disorder/order transition of the substrate-binding site mediated by a calcium-sodium-calcium metal triad. Structure 6:281-292. [DOI] [PubMed] [Google Scholar]
- 10.Mckenzie, H. A. 1969. pH and buffers, p. 485. In R. M. C. Dawson, D. C. Elliott, W. H. Elliott, and K. M. Jones (ed.), Data for biochemical research, 2nd ed. Oxford University Press, Bristol, United Kingdom.
- 11.Mikuni, K., M. Monma, and K. Kainuma. 1987. Alcohol fermentation of corn starch digesting by Chalara paradoxa amylase without cooking. Biotechnol. Bioeng. 29:729-732. [DOI] [PubMed] [Google Scholar]
- 12.Nakajima, R., T. Imanaka, and S. Aiba. 1985. Nucleotide sequence of the Bacillus stearothermophilus α-amylase gene. J. Bacteriol. 163:401-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nielsen, J. E., and T. V. Borchert. 2000. Protein engineering of bacterial α-amylases. Biochim. Biophys. Acta 1543:253-274. [DOI] [PubMed] [Google Scholar]
- 14.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanoja, R. R., J. Morlon-Guyot, J. Jore, J. Pintado, N. Juge, and J. P. Guyot. 2000. Comparative characterization of complete and truncated forms of Lactobacillus amylovorus α-amylase and role of the C-terminal direct repeats in raw-starch binding. Appl. Environ. Microbiol. 66:3350-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Savchenko, A., C. Vieille, S. Kang, and J. G. Zeikus. 2002. Pyrococcus furiosus alpha-amylase is stabilized by calcium and zinc. Biochemistry 41:6193-6201. [DOI] [PubMed] [Google Scholar]
- 17.Suvd, D., Z. Fujimoto, K. Takase, M. Matsumura, and H. Mizuno. 2001. Crystal structure of Bacillus stearothermophilus alpha-amylase: possible factors determining the thermostability. J. Biochem. (Tokyo) 129:461-468. [DOI] [PubMed] [Google Scholar]
- 18.Suzuki, Y., N. Ito, T. Yuuki, H. Yamagata, and S. Udaka. 1989. Amino acid residues stabilizing a Bacillus alpha-amylase against irreversible thermoinactivation. J. Biol. Chem. 264:18933-18938. [PubMed] [Google Scholar]
- 19.Takkinen, K., R. F. Pettersson, N. Kalkkinen, L. Palva, H. Soderlund, and I. Kaariainen. 1983. Amino acid sequence of alpha-amylase from Bacillus amyloliquefaciens deduced from the nucleotide sequence of the cloned gene. J. Biol. Chem. 258:1007-1013. [PubMed] [Google Scholar]
- 20.Tomazic, S. J., and A. M. Klibanov. 1988. Mechanisms of irreversible thermal inactivation of Bacillus α-amylase. J. Biol. Chem. 263:3086-3091. [PubMed] [Google Scholar]
- 21.Tomazic, S. J., and A. M. Klibanov. 1988. Why is one Bacillus α-amylase more resistant against irreversible thermoinactivation than another? J. Biol. Chem. 263:3092-3096. [PubMed] [Google Scholar]
- 22.Yamamoto, K., Z. Z. Zhang, and S. Kobayashi. 2000. Cycloamylose (cyclodextrin) glucanotransferase degrades intact granules of potato raw starch. J. Agric. Food Chem. 48:962-966. [DOI] [PubMed] [Google Scholar]
- 23.Yuuki, T., T. Nomura, H. Tezuka, A. Tsuboi, and H. Yamagata. 1985. Complete nucleotide sequence of a gene coding for heat- and pH-stable alpha-amylase of Bacillus licheniformis: comparison of the amino acid sequences of three bacterial liquefying alpha-amylases deduced from the DNA sequences. J. Biochem. (Tokyo) 98:1147-1156. [DOI] [PubMed] [Google Scholar]