Abstract
The microbial production of l-(+)-lactic acid is rapidly expanding to allow increased production of polylactic acid (PLA), a renewable, biodegradable plastic. The physical properties of PLA can be tailored for specific applications by controlling the ratio of l-(+) and d-(−) isomers. For most uses of PLA, the l-(+) isomer is more abundant. As an approach to reduce costs associated with biocatalysis (complex nutrients, antibiotics, aeration, product purification, and waste disposal), a recombinant derivative of Escherichia coli W3110 was developed that contains five chromosomal deletions (focA-pflB frdBC adhE ackA ldhA). This strain was constructed from a d-(−)-lactic acid-producing strain, SZ63 (focA-pflB frdBC adhE ackA), by replacing part of the chromosomal ldhA coding region with Pediococcus acidilactici ldhL encoding an l-lactate dehydrogenase. Although the initial strain (SZ79) grew and fermented poorly, a mutant (SZ85) was readily isolated by selecting for improved growth. SZ85 exhibited a 30-fold increase in l-lactate dehydrogenase activity in comparison to SZ79, functionally replacing the native d-lactate dehydrogenase activity. Sequencing revealed mutations in the upstream, coding, and terminator regions of ldhL in SZ85, which are presumed to be responsible for increased l-lactate dehydrogenase activity. SZ85 produced l-lactic acid in M9 mineral salts medium containing glucose or xylose with a yield of 93 to 95%, a purity of 98% (based on total fermentation products), and an optical purity greater than 99%. Unlike other recombinant biocatalysts for l-lactic acid, SZ85 remained prototrophic and is devoid of plasmids and antibiotic resistance genes.
Polylactic acid (PLA) is being developed as a renewable alternative for conventional petroleum-based plastics (2, 8, 28). A new plant began operation in 2002 that is expected to increase the world production of l-(+)-lactic acid by 2.5-fold (5, 16, 18). PLA is a versatile plastic that can be tailored to specific applications by altering the ratio of the l-(+)- and d-(−)-lactic acid isomers (12, 23, 37). Optically pure isomers can be produced as separate products by microbial fermentation of carbohydrates (11, 17) using chiral-specific l-(+)- or d-(−)-lactate dehydrogenase (LDH) enzymes. The success of renewable commodity chemicals such as lactic acid is critically tied to production costs. Although glucose is the primary commercial feedstock, additional sugars such as pentoses (xylose and arabinose) and cellobiose from renewable lignocellulose are potentially available and may prove less expensive.
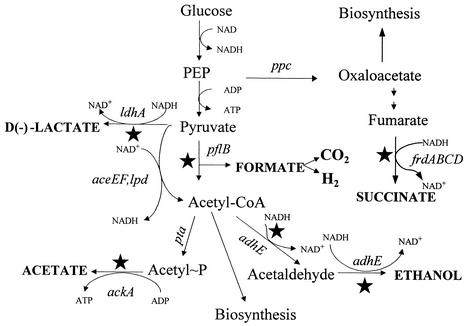
A variety of microorganisms can be used to produce optically pure lactic acid isomers (17). These typically require the addition of complex nutrients due to native biosynthetic limitations, adding costs for nutrients, product purification, and waste disposal. Alternative biocatalysts for lactic acid production are being engineered in yeasts (1) and Escherichia coli (7, 14, 40), the two most widely used microbial platforms for biotechnology (8). Native E. coli produces a mixture of acidic and neutral fermentation products (9) (Fig. 1). Two groups have reported the metabolic engineering of this organism for the production of d-lactic acid using different genetic approaches. E. coli strains developed by Chang et al. (7) contained mutations in phosphotransacetylase (pta) and phosphoenolpyruvate carboxylase (ppc) and produced high yields of lactic acid but required dicarboxylic acids or complex nutrients for growth. Although omitting the ppc mutation eliminated the requirement for dicarboxylic acids, yield was significantly diminished by the accumulation of succinate. An alternative approach by Zhou et al. (40) combined mutations in four genes: pyruvate formatelyase (pflB), acetate kinase (ackA), alcohol dehydrogenase (adhE), and fumarate reductase (frdBC). The resulting strain (SZ63) produced high yields of d-lactic acid from sugars during growth in mineral salts medium without additional nutrients and contained no antibiotic resistance genes or plasmids.
FIG. 1.
Fermentation pathways in native and recombinant E. coli. Solid stars denote enzymatic steps which have been eliminated in strains SZ79 and SZ85 by internal deletions of corresponding genes. In these strains, the P. acidilactici ldhL coding region and transcriptional terminator were integrated into ldhA downstream from the ldhA promoter.
Two different genetic approaches have been used to engineer E. coli for the production of l-lactic acid. In both, recombinant genes encoding l-LDH enzymes were used to replace native fermentation pathways. Early studies by Clark and colleagues (9, 26) demonstrated that strains of E. coli containing mutations in pyruvate formatelyase (pflB) and LDH (ldhA) are unable to metabolize glucose anaerobically in mineral salts media unless supplied with an alternative pathway for NADH oxidation. Subsequently, these double mutants have been used to isolate plasmids expressing LDH genes from a variety of different microorganisms (3, 13, 15, 30). Although resulting strains produced single isomers as a dominant fermentation product, none were evaluated as potential biocatalysts for lactic acid production. Recently, Dien et al. (14) constructed analogous E. coli pfl ldhA double mutants containing a plasmid expressing the Streptococcus bovis ldh gene (39) and demonstrated the production of over 70 g of l-lactic acid per liter at high yields (0.8 g/g of glucose) using complex media. An alternative approach was reported by Chang et al. (7) in which a double mutant (ldhA pta) and a triple mutant (ldhA pta ppc) were constructed as hosts for a plasmid expressing the Lactobacillus casei ldh gene (20). Although l-lactic acid was produced during fermentation in complex medium, product titers and yields were lower than those reported by Dien et al. (14). None of these recombinants was capable of fermenting sugars to lactic acid in mineral salts medium without the inclusion of nutrient supplements and antibiotics (7, 14).
In this study, we describe a new E. coli biocatalyst containing five chromosomal deletions (pflB, ackA, adhE, ldhA, and frdBC) and a chromosomally integrated l-(+)-LDH gene (ldhL) from Pediococcus acidilactici (15). The resulting strain (SZ85) contains no plasmids or antibiotic resistance genes and produces high yields of optically pure l-(+)-lactic acid from glucose and xylose in a mineral salts medium.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture media.
The E. coli strains and plasmids used in this study are listed in Table 1. DH5α, TOP10F′, and S17-1 were used as hosts for plasmid constructions. During strain construction, cultures were grown at either 30, 37, or 42°C in Luria-Bertani broth (27) (per liter: 10 g of Difco tryptone, 5 g of Difco yeast extract, and 5 g of sodium chloride) containing 2% glucose or on this medium solidified with agar (1.5%). Antibiotics were used as needed at the following concentrations: kanamycin (50 μg ml−1), tetracycline (12.5 or 6.25 μg ml−1), and ampicillin (50 μg ml−1).
TABLE 1.
Plasmids and strains used in this study
| Plasmid or strain | Relevant characteristics | Source or reference |
|---|---|---|
| Plasmids | ||
| pCR2.1-TOPO | bla kan; TOPO TA cloning vector | Invitrogen |
| pLOI2302 | AscI linkers inserted into pUC19 at NdeI and SapI sites | 40 |
| pLOI2224 | kan; R6K conditional integration vector | 25 |
| pLOI2065 | bla; FRT-tet-FRT cassette | 40 |
| pGID150 | ery, source of ldhL gene from P. acidilactici | 15 |
| pLOI2392 | kan bla ldhA; ldhA (PCR) from E. coli W3110 cloned into pCR2.1-TOPO | This study |
| pLOI2393 | kan bla ldhL; ldhL (PCR) from pGID150 cloned into pCR2.1-TOPO vector | This study |
| pLOI2394 | bla; pLOI2302 after deletion of multiple cloning site (KpnI-HindIII) | This study |
| pLOI2395 | bla; EcoRI fragment (ldhA) from pLOI2392 cloned into pLOI2394 at EcoRI | This study |
| pLOI2396 | kan bla; ldhL-FRT-tet-FRT KpnI-SmaI fragment (FRT-tet-FRT) from pLOI2065 cloned into KpnI-BamHI (blunted) sites of pLOI2393 | This study |
| pLOI2397 | kan bla; ldhA′-ldhL-FRT-tet-FRT-′ldhA ApaI (blunted)-KpnI fragment (ldhL-FRT-tet-FRT) from pLOI2396 cloned into ldhA at HincII-KpnI sites of pLOI2395 | This study |
| pLOI2398 | kan; ldhA′-ldhL-FRT-tet-FRT-′ldhA AscI fragment (ldhA′-ldhL-FRT-tet-FRT-′ldhA) from pLOI2397 cloned into AscI site of pLOI2224 | This study |
| Strains | ||
| DH5α | ΔlacZM15 recA | Bethesda Research Laboratory |
| TOP10F′ | lacIq (episome) | Invitrogen |
| S17-1 | thi pro recA hsdR RP4-2 tet::Mu aphA::Tn7 λpir | 35 |
| SZ63 | W3110 ΔfocA-pflB::FRT ΔfrdBC ΔadhE::FRT ackA::FRT | 40 |
| SZ75, SZ76, SZ77, and SZ78 | tet W3110 ΔfocA-pflB::FRT ΔfrdBC ΔadhE::FRT ackA::FRT ΔldhA::(ldhL-frt-tet-frt) | This study |
| SZ79, SZ80, SZ81, and SZ82 | W3110 ΔfocA-pflB::FRT ΔfrdBC ΔadhE::FRT ackA::FRT ΔldhA::(ldhL-frt) strains SZ75 to SZ79, respectively, after deletion of tet | This study |
| SZ83, SZ84, SZ85, SZ86, SZ87, SZ88, SZ89, SZ90, SZ91, and SZ92 | Mutants of SZ79 with increased expression of ldhL | This study |
Strains engineered for lactic acid production were grown in M9 medium (27) (per liter: 6 g of Na2HPO4, 3 g of KH2PO4, 1 g of NH4Cl, 0.5 g of NaCl, 246.5 mg of MgSO4 · 7H2O, 14.7 mg of CaCl2, 1 mg of thiamine) containing 1% glucose. Stock cultures were maintained on M9 plates containing 1.5% agar and 2% glucose. M9 medium containing 5% sugar (glucose or xylose) was used for pH-controlled fermentations. Seed cultures were grown in M9 medium containing 1% xylose or 1% glucose as appropriate.
Genetic methods.
Plasmid pGID150 was used as a source of the ldhL gene from P. acidilactici (15), an isolate from corn silage. Standard methods were used for PCR amplification, plasmid construction, analyses of DNA fragments, and DNA sequencing (34). Electroporation conditions for chromosomal integration have been previously described (10, 25, 40). Plasmid DNA was isolated using a Qiaprep Spin miniprep kit. A QIAquick gel extraction kit was used to isolate DNA fragments from agarose gels. DNA sequencing was provided by the University of Florida Interdisciplinary Center for Biotechnology Research (with Applied Biosystems autosequencers) and by the Microbiology and Cell Science Sequencing Core (with a Licor long-read autosequencer), using dye-termination methods.
Integration of P. acidilactici ldhL into SZ63.
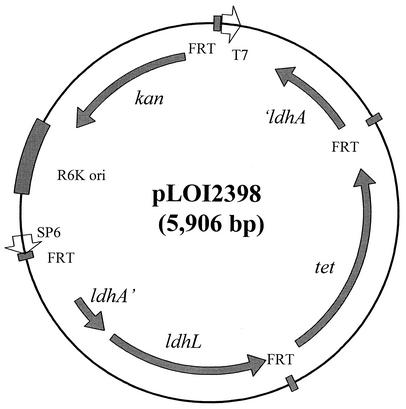
Plasmid pLOI2398 was constructed with a conditional R6K replicon to facilitate the integration of ldhL into the chromosomal ldhA gene of SZ63. The ldhL gene coding region and transcriptional terminator region (1 kbp) were amplified by PCR from pGID150 and cloned into a pCR2.1-TOPO vector to produce pLOI2393 (4.9 kbp). The forward primer (5′-3′ AAGGAGAAAGTCTTATGTCTAATATTCAAAATCA) included part of the E. coli ldhA ribosomal binding region (bold) and the amino terminus of P. acidilactici ldhL (underlined). The reverse primer (5′ to 3′, GTTTGGGGAAGGGACATAAAAATAGGTACAAAA) was downstream from the putative transcriptional terminator region. A 1.7-kbp fragment (SmaI-KpnI) containing a tet gene flanked by two FRT sites for the Saccharomyces cerevisiae FLP recombinase (25, 40) was isolated from pLOI2065 and directionally cloned into pLOI2393 between unique BamHI (Klenow-treated) and KpnI sites. In the resulting plasmid pLOI2396 (6.6 kbp), transcription of both ldhL and tet is oriented in the same direction.
To facilitate transfer of LDH genes into the λ-pir conditional R6K vector, a derivative of pUC19 was previously constructed in which AscI linkers were inserted into the SapI and NdeI sites which border the polylinker region (40). This plasmid was further modified by deleting most of the polylinker region (KpnI-HindIII region; blunt-end ligation) to produce pLOI2394 (2.7 kbp) containing a unique EcoRI site flanked by two AscI sites.
The coding region for the E. coli ldhA gene (1 kbp) was amplified by PCR using chromosomal DNA from strain W3110 as the template and Sigma Genosys ORFmer primers. After cloning into pCR2.1-TOPO to produce pLOI2392 (4.9 kbp), the EcoRI fragment containing ldhA was isolated and cloned into the EcoRI site of pLOI2394 to produce pLOI2395 (3.7 kbp). The ApaI (treated with T4 DNA polymerase to produce blunt end)-KpnI fragment (2.7 kbp) containing ldhL and tet was isolated from pLOI2396 and directionally cloned into pLOI2395 (HincII to KpnI sites) to produce pLOI2397 (6.4 kbp). In this plasmid, ldhA, ldhL, and tet genes are transcribed in the same direction. (Note that this construct included a partial deletion of the ldhA gene.) The AscI fragment (4 kbp) containing these three genes was isolated from pLOI2397 and cloned into the R6K integration vector pLOI2224 (25) to produce pLOI2398 (5.9 kbp). Plasmid pLOI2398 (Fig. 2) contains resistance genes for both kanamycin and tetracycline. This plasmid pLOI2398 was transferred into E. coli SZ63 by electroporation followed by selection for resistance to tetracycline. Integrants were confirmed by the absence of plasmid, sensitivity to kanamycin, and PCR analysis.
FIG. 2.
Integration vector used for chromosomal insertion of P. acidilactici ldhL. Four recombinase sites are present on this plasmid (shaded and labeled FRT).
Fermentation.
E. coli W3110 (wild type) and engineered derivatives were initially tested for lactic acid production using 18-ml screw-cap tubes. Single colonies from fresh plates were resuspended in M9 medium (1 ml) containing 1% glucose. Fermentation tubes were inoculated with approximately 0.05 ml of this suspension, filled to the brim, sealed, and incubated at 37°C for 48 h. Broth samples were analyzed by high-performance liquid chromatography (HPLC).
Strains constructed for lactic acid production were maintained on M9 plates containing 2% glucose and tested in a New Brunswick Bioflow 3000 10-liter fermentor (8 liter of M9 medium containing 5% sugar). Seed cultures were prepared by inoculating colonies from fresh M9 plates into 2-liter flasks containing 600 ml of M9 medium with 1% glucose (37°C; 20 h; 200 rpm). Cells were harvested by centrifugation and used to inoculate fermentation vessels (initial cell density of 33 mg [dry cell weight] per liter). Vessels were incubated at 37°C (pH 7.0) with agitation (200 rpm, single upflow marine impeller). Broth pH was controlled by automatic addition of 45% (wt/wt) KOH.
LDH assay.
LDH activity was assayed by measuring NADH oxidation (A340) using a Beckman DU 640 spectrophotometer (6, 26). Cells were harvested from 18-ml anaerobic tubes by centrifugation, washed once in Tris-maleate buffer (100 mM Tris-maleate, 1 mM dithiothreitol [pH 6.5]), and resuspended in this buffer at a density of approximately 0.33 mg (dry weight) per ml. Cells were permeablized by treating 0.1 ml of this suspension with 2 drops of chloroform and vigorously mixing for 15 s with a vortex mixer. After allowing the chloroform to settle, the upper layer containing permeabilized cells was used to assay LDH activity. The assay mixture contained 30 μl of sodium pyruvate (1 M; pH 7.5), 30 μl of NADH (6.4 mM), 400 μl of morpholinepropanesulfonic acid buffer (50 mM; pH 7.0), 530 μl of distilled H2O, and 10 μl of crude enzyme. Rates were measured at room temperature for 5 min. LDH activity (initial rate) is reported as micromole of NAD+ produced per minute per milligram of cell protein.
Analyses.
Cell mass was estimated by measuring the optical density at 550 nm (OD550) as previously described (24). At an OD550 of 1.0, each liter contained approximately 330 mg of cells (dry weight). Total protein was calculated as 55% of cell dry weight. Organic acids and sugars were measured by HPLC (38). A chiral column was used to analyze the isomeric purity of lactic acid (29, 40).
Nucleotide sequence accession numbers.
The sequences for the DNA regions containing the integrated ldhL gene in SZ79 and SZ85 have been deposited in GenBank and assigned accession numbers AY205157 and AY205156, respectively.
RESULTS
Construction of biocatalyst for the production of l-(+)-lactic acid.
E. coli strain SZ63 (focA-pflB frdBC adhE ackA) was previously developed to demonstrate the production of optically pure d-lactic acid in mineral salts medium (40). Using SZ63 as the parent, a new biocatalyst for the production of l-lactic acid was engineered by replacing part of the native ldhA coding region with the coding region and transcriptional terminator from P. acidilactici ldhL. Since the ldhL gene contains a weak ribosomal-binding region (Shine-Dalgarno sequence, AAGGG) (15), this region was replaced with the corresponding sequence for ldhA. In the integration vector, pLOI2398 (R6K replicon), the promoterless ldhL was oriented in the same direction as ldhA to allow expression from the native ldhA promoter.
A total of 238 tetracycline-resistant colonies were recovered after electroporation of pLOI2398, representing both single and double crossover events. Of these, 12 clones were sensitive to kanamycin (potential double crossover integrants). These 12 clones were further examined by PCR using primer sets for ldhA, ldhL, the ldhA ORFmer forward and ldhL reverse primers, and the ldhL forward and ldhA ORFmer reverse primer pair. Four clones containing double crossover events were retained and designated E. coli SZ75, SZ76, SZ77, and SZ78, respectively. Each was transformed with plasmid pFT-A (temperature conditional) containing an inducible FLP recombinase (33). Chromosomal tet genes were eliminated by induction of the recombinase. After curing the temperature-sensitive pFT-A plasmid by growth at 42°C, resulting strains were designated E. coli SZ79, SZ80, SZ81, and SZ82, respectively. These strains were devoid of antibiotic resistance markers, plasmids, and auxotrophic requirements.
Increasing the expression of ldhL after integration into E. coli.
The anaerobic growth of integrants containing the P. acidilactici ldhL gene was poor in M9 medium in comparison to that of SZ63 (parent), as illustrated by the results with SZ79 (Table 2). Cell mass and LDH activity for SZ79 were both approximately 5% of SZ63 levels under these growth conditions (Table 2). The poor performance of SZ79 and other integrants was hypothesized to result from weak expression of the P. acidilactici ldhL gene.
TABLE 2.
Comparison of LDH activity (M9 medium containing 1% glucose)
| Strain | PCR result for LDH gene
|
Anaerobic growth (OD550) | LDH activity (μmol/mg of protein per min) | |
|---|---|---|---|---|
| ldhA | ldhL | |||
| SZ63 | + | − | 0.100 | 0.814 |
| SZ79 | − | + | 0.005 | 0.042 |
| SZ83 | − | + | 0.135 | 1.348 |
| SZ84 | − | + | 0.170 | 1.163 |
| SZ85 | − | + | 0.185 | 1.347 |
| SZ86 | − | + | 0.115 | 1.198 |
| SZ87 | − | + | 0.170 | 1.282 |
| SZ88 | − | + | 0.160 | 1.242 |
| SZ89 | − | + | 0.110 | 1.316 |
| SZ90 | − | + | 0.190 | 1.339 |
| SZ91 | − | + | 0.180 | 1.243 |
| SZ92 | − | + | 0.190 | 1.179 |
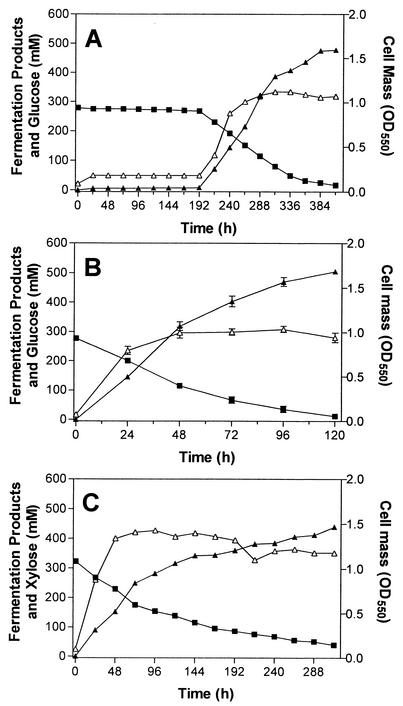
Mutants of SZ79 were selected for improved growth in M9 medium (8 liters; 5% glucose) using a 10-liter fermentation vessel. The fermentor (37°C; 200 rpm; pH 7.0; controlled by addition of 45% NaOH) was inoculated with E. coli SZ79 at an initial cell density of 33 mg/liter (OD550, 0.10). Cell mass doubled during the first 24 h, accompanied by production of small amounts of acids from glucose (Fig. 3A). Both cell mass and sugar concentration remained essentially unchanged for the following 7 days. Growth and acid production resumed after 8 days. Glucose metabolism was nearly complete after 17 days with a yield of 0.91 g of lactic acid/g of sugar metabolized (Table 3). Samples of fermentation broth were removed during the second growth phase and spread on M9 (2% glucose) plates to test for contamination. No contamination was evident, and these plates were used to isolate potential mutants. Ten colonies (SZ83-SZ92) were selected from the 12-day broth sample and tested for the presence of ldhL and absence of ldhA by PCR, for anaerobic growth in M9 (1% glucose), and for LDH activity (Table 2). Analysis of PCR products indicated that all products retained ldhL and lacked ldhA. Anaerobic growth (OD550) and LDH activity in the new clones averaged over 30-fold higher than results for the parent (SZ79). Based on these results, all new isolates were considered siblings. One, designated SZ85, was selected for further investigation.
FIG. 3.
Batch fermentation. (A) Fermentation of 5% glucose by SZ79 during selection of mutants for improved growth. (B) Fermentation of 5% glucose by SZ85. (C) Fermentation of 5% xylose by SZ85. Small amounts of succinate, formate, acetate, and ethanol were also detected (see Table 4). Symbols: ▪, sugar (glucose or xylose); ▴, lactate; ▵, OD550.
TABLE 3.
Effect of medium pH on ldhA and ldhL expression during anaerobic growtha
| Strain (gene) | Buffer | pH
|
LDH activity (μmol/mg of protein per min) | Activity ratiob | |
|---|---|---|---|---|---|
| Initial | Final | ||||
| W3110 (ldhA) | None | 6.8 | 5.3 | 0.519 | 2.2 |
| W3110 (ldhA) | HEPES | 8.0 | 7.5 | 0.232 | |
| SZ63 (ldhA) | None | 6.8 | 5.6 | 1.127 | 1.7 |
| SZ63 (ldhA) | HEPES | 8.0 | 7.9 | 0.680 | |
| SZ79 (ldhL) | None | 6.8 | 5.9 | 0.029 | 1.7 |
| SZ79 (ldhL) | HEPES | 8.0 | 7.9 | 0.017 | |
| SZ85 (ldhL) | None | 6.8 | 5.6 | 1.428 | 1.3 |
| SZ85 (ldhL) | HEPES | 8.0 | 7.9 | 1.099 | |
Colonies were inoculated into 18-ml screw-cap tubes filled to the brim with Luria-Bertani containing 1% glucose, sealed, and incubated anaerobically for 6 h. High levels of inocula and longer incubation times (12 h) were required for SZ79 due to poor growth. Where indicated, HEPES (0.2 M) was included to prevent acidification of medium.
Activity ratios were calculated by dividing activity present during growth without buffer by that from buffered cultures (26).
Regulation of P. acidilactici ldhL expression in recombinant E. coli.
The promoterless ldhL was integrated into ldhA, downstream from the ldhA promoter. Since transcription of the ldhA gene is known to be activated by growth at low pH (6, 19, 26), the effect of pH on the expression of ldhL was investigated (Table 3). Expression of ldhL (SZ79 and SZ85) and native ldhA (W3110 and SZ63) was higher when cells were grown in unbuffered medium that was allowed to acidify (pH 5.3 to 5.6) than in cells grown in buffered medium (pH 7.5 to 7.9), consistent with expression from the ldhA promoter. The effect of pH was larger for the wild-type organism, W3110, than for any of the genetically modified strains. LDH activities for SZ63, the strain engineered to produce d-lactic acid using the native LDH, were over twice that of the unmodified parental strain (W3110). The activity ratio for LDH in SZ63 (ldhA) was identical to that for the integrated derivative SZ79 expressing P. acidilactici ldhL. The smallest difference in activity ratio was observed for SZ85, a mutant of SZ79. Strain SZ85 also expressed the highest levels of LDH activity.
Comparison of DNA sequences in the ldhL region of SZ79 (parent) and SZ85 (mutant).
To investigate the mutational basis for the 30-fold increase in l-LDH activity (Table 3), DNA regions (2,825 bp) containing the integrated ldhL gene in SZ79 and SZ85 were sequenced and compared. The sequenced DNA included 906 bp upstream of the ldhA start codon (587 bp upstream from the native ldhA start codon, a 245-bp N-terminal segment of ldhA, 59 bp of vector DNA, and 15 bp of synthetic DNA containing a duplicate of the ldhA Shine-Dalgarno region), the ldhL coding region (972 bp), and 947 bp downstream of the ldhA transcriptional terminator (77 bp containing the putative ldhL transcriptional terminator, 138 bp of vector DNA, 535 bp containing the C terminus of ldhA, and an additional 197 bp downstream from ldhA). To facilitate sequencing, primers were designed that allowed each region to be subcloned into pCR2.1-TOPO as three overlapping fragments of approximately 1,000 bp. All clones were fully sequenced in both directions using the Licor autosequencer, 70% of which were independently confirmed by shorter reads using Applied Biosystems autosequencers. For comparison, the original P. acidilactici ldhL gene and downstream region (1,110 bp total) were also sequenced directly from pGID150, using M13 forward and reverse primers. All sequence data were in complete agreement.
The DNA sequence (972 bp) for the ldhL region from pGID150 was identical to the DNA sequence of the subcloned PCR product in SZ79. Both differed from the published sequence in six base positions (published to new observation): (i) codon 28, GAA (glu) to CAA (gln); (ii) codon 29 GAA (glu) to CAA (gln); (iii) codon 43, GTA (val) to GAT (asp); (iv) codon 107, CTT (leu) to GTT (val); (v) codon 221 (double mutation), AAA (lys) to GCA (ala). Alignments of translated sequences for genes encoding similar l-(+)-LDH (lack of allosteric regulation by fructose-1,6-bisphosphate) from two related organisms (Lactobacillus plantarum and Lactobacillus pentosus) were in complete agreement with the new ldhL sequence at four of these positions (15). These differences appear to be corrections. The two-base change in codon 221 (lys to ala) was surprising and may represent mutations acquired during passage of plasmid pGID150. Interestingly, either lysine or alanine are found at this position in l-(+)-LDHs from other bacteria (15).
The promoter region of ldhA and the region within the N-terminal fragment of the ldhA coding region can be presumed to contain a promoter(s) for the integrated P. acidilactici ldhL. Although the promoter for ldhA has not been rigorously defined, a putative sigma 70 promoter region is present approximately 100 bp upstream. No mutations were observed in SZ85 within the putative ldhA promoter region. One mutation was observed further upstream (A to G; 522 bp upstream from the ldhA start codon) and a second mutation was observed in the N-terminal fragment of ldhA at amino acid 55 (from SZ79 GAC to GGC in SZ85). It is possible that the mutation in the N terminus of ldhA allowed this region to serve as an additional promoter for ldhL. Although there is no clear match with consensus regions for sigma factors, an AT-rich region is appropriately spaced downstream from this mutation which could serve as a −10 site for sigma 38. No mutations were observed in the vector DNA or synthetic DNA. Note that this region contains two transcriptional terminators to prevent synthesis of a fusion protein.
Five mutations were found in the ldhL coding region of SZ85: (i) codon 15, GAC (asp) to GGC (gly); (ii) codon 63, AAC (asn) to AGC (ser); (iii) codon 221 (double mutation), GCA (ala) to AAA (lys); and (iv) codon 228, AAG (lys) to GAG (glu). None of these (old or new) codons are rare in E. coli, and none of the changes would be predicted to alter the efficiency of transcription. The change in codon 221 corrected an apparent mutation of ldhL in plasmid pGID150, restoring the original sequence (15). Two G-to-A mutations were found in the terminator region of ldhL. No mutations were observed in the truncated C-terminal region of ldhA (535 bp) or downstream region (197 bp). The observed mutations in the ldhL coding region and terminator region of SZ85 could contribute to the increase in l-(+)-LDH activity by increasing message stability, improving protein folding or stability, and increasing enzyme activity.
Production of l-lactic acid during pH-controlled batch fermentation.
Fermentations of glucose and xylose to lactic acid were compared in M9 medium containing 5% sugar, using E. coli SZ85 expressing P. acidilactici ldhL as the biocatalyst (Fig. 3B and C). With xylose, the growth phase continued for approximately 1 day longer than with glucose, resulting in a higher cell yield (Table 4). Despite higher cell mass with xylose, volumetric and specific productivities with glucose were twofold higher than with xylose. Lactic acid was the dominant fermentation product from both sugars, with low levels of other acids (Table 4). During the first 72 h, lactic acid production from xylose was approximately 60% of that from glucose. Xylose fermentation rates declined progressively thereafter, and 15% of the xylose remained unmetabolized when fermentation was terminated (312 h). Lactic acid yields based on metabolized xylose were 0.93 g/g of xylose. Glucose was exhausted after 120 h with yields averaging 0.95 g/g of glucose metabolized.
TABLE 4.
Comparison of sugar fermentation by engineered strains of E. coli W3110a
| Strain (sugar) | Cell mass (mg/liter) | Sugar utilized (mM) | Lactate (mM) | Yieldb (% total; % metabolized) | Max. vol. prod.c
|
Max. spec. prod.d
|
Coproduct concn (mM)
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Productivity (mmol liter−1 per h) | Time (h) | Productivity (mmol per g of cell dry weight h−1) | Time (h) | Succinate | Formate | Acetate | Ethanol | |||||
| SZ79 ldhL (glucose) | 370 | 262 | 478 | 86; 91 | 4.03 | 288 | 23.7 | 216 | 4.3 | <0.5 | 5.1 | <1.0 |
| SZ85 ldhL (glucose) | 347 ± 31 | 266 ± 9 | 505 ± 12 | 94; 95 | 7.18 ± 0.69 | 24 | 20.8 ± 1.4 | 24 | 1.88 ± 0.19 | <0.5 | 1.8 ± 0.5 | <1.0 |
| SZ85 ldhL (xylose) | 472 | 285 | 439 | 82; 93 | 3.54 | 48 | 9.8 | 24 | 6.75 | <0.5 | 3.4 | <1.0 |
Data represent an average of two or more fermentations. Standard deviations were calculated for data with three or more replicates.
Percentage of maximum theoretical yield for l-lactic acid (1 g of lactic acid per g of sugar). These values were calculated as a percentage of total sugar added to the fermentation vessels and as a percentage of sugar metabolized during fermentation.
Max. vol. prod., maximum volumetric productivity for l-lactic acid.
Max. spec. prod., maximum specific productivity for l-lactic acid.
Optical purity of lactic acid.
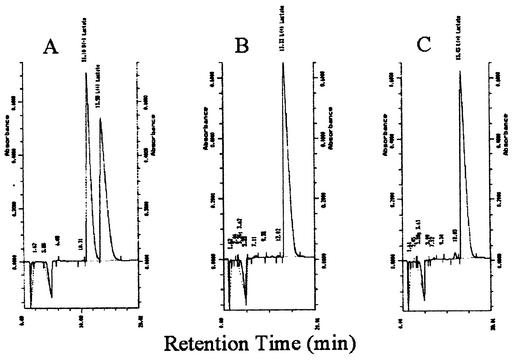
The optical purity of lactic acid produced by SZ85 was determined by HPLC using a chiral column to separate the d-(−) and l-(+) isomers (29). Based on these analyses, SZ85 was found to produce l-(+)-lactic acid with a chiral purity of at least 99.5% (Fig. 4).
FIG. 4.
Analysis of optical purity of l-lactate in fermentation broth using a chiral column. Absorbance is plotted relative to retention times (in minutes). Retention times are also noted above peaks. (A) Standards: a mixture of 10 mM l-(+)-lactic acid and 10 mM d-(−)-lactic acid. (B) Broth from SZ85. This broth contained 500 mM l-(+)-lactic acid and was diluted 40-fold prior to analysis on the chiral column. d-(−)-Lactic acid was estimated from the peak area at 12.02 min. (C) Diluted broth from SZ85 to which 0.05 mM d-(−)-lactic acid was added as an internal standard. Purity of the l-(+)-lactic acid from SZ85 was estimated to exceed 99.5%.
DISCUSSION
Lactic acid bacteria (17), fungi (17, 36), genetically engineered yeasts (1, 4, 31, 32), and genetically engineered bacteria (7, 14, 21, 22) have been investigated as potential biocatalysts for the production of optically pure l-(+)-lactic acid. Although several of these organisms are presumed to be in commercial use, none have been reported to grow and produce lactic acid efficiently in mineral salts media containing sugar. All have additional nutrient requirements which are typically met with complex supplements. Lactic acid yields from the most efficient of these biocatalysts ranged from 85 to 92% of the maximum theoretical yield (1 g per g of sugar metabolized). In most studies, unmetabolized glucose remained in the broth at the end of fermentation (5 to 20 g liter−1).
Our laboratory previously described E. coli SZ63 (focA-pflB frdBC adhE ackA) for the production of optically pure d-lactic acid (>95% of theoretical yield based on total sugar added) in mineral salts medium (40). A derivative of this strain was constructed for l-lactic acid production by functionally replacing the native ldhA gene encoding d-LDH in SZ63 with the ldhL gene encoding l-LDH from P. acidilactici. The resulting organism, strain SZ85, produced l-lactic acid from glucose in mineral salts medium with a yield of 95% (94% of total sugar), a purity of 99% with regard to total fermentation products, and an optical purity for the l-(+) isomer exceeding 99%. Although lower than glucose, lactic acid yields for xylose were also high (93% of metabolized sugar and 82% of total sugar) and were similar in optical purity.
Strain E. coli SZ85 has proven to be exceptionally stable during storage and transfers. Colonies were quite uniform upon transfer. Performance in seed cultures and in fermentations has been highly reproducible over a 12-month period of study. To eliminate the opportunity for reversion, chromosomal deletions were introduced into key genes for all alternative fermentation pathways rather than point mutations or gene disruptions. This strain has an absolute requirement for the functional expression of P. acidilactici ldhL for NADH oxidation during anaerobic growth. Antibiotic resistance genes and plasmids used during construction were eliminated to maximize the potential utility of SZ85. l-Lactic acid titers of up to 5% were achieved in simple batch fermentations with complete utilization of glucose; higher levels may be possible with fed-batch strategies but have not been investigated. In contrast to SZ85, previous strains of E. coli examined for l-lactic acid production contained plasmids and antibiotic resistance genes and required complex nutrients (7, 14). Although fermentation rates for SZ85 in mineral salts medium were lower than previously reported for complex media using other recombinant E. coli (7, 14), product yield and purity were highest using SZ85 as the biocatalyst.
Opportunities for further improvement of E. coli SZ85 include increases in volumetric productivity and acid tolerance. In recent studies (4, 31, 32), an engineered strain of Kluyveromyces lactis expressing the bovine LDH gene from a plasmid was reported to produce up to 6% lactic acid at pH 4.5, reducing the amount of base required for maintenance of pH by approximately 20% and reducing the potential for contamination (4, 31). Conditions for growth and lactic acid production by the engineered K. lactis strain included requirements for continuous aeration and supplements (adenine, uracil, and antibiotics). The lactic acid yield for the engineered K. lactis strain was high (85% based on metabolized glucose after 474 h). However, substantial levels of glucose remained unmetabolized at the end of this fermentation. Lower levels of glucose were fermented completely by the engineered K. lactis strain without pH control or base addition (31). In contrast, SZ85 completely metabolized glucose with a higher yield of l-lactic acid (95%) in a simple stirred-tank fermentor containing mineral salts medium and control of only pH and temperature. The high yield and purity of l-lactic acid produced by SZ85, simplicity of fermentation conditions, lack of aeration, and lack of complex nutritional requirements offer potential advantages for l-lactic acid production.
Acknowledgments
We thank J. Delcour (Université Catholique de Louvain, Louvain-la-Neuve, Belgium) for sharing plasmid pGID150 containing the P. acidilactici ldhL. Analyses of lactic acid optical purity were kindly provided by G. J. Luli and B. E. Wood (B.C. International, Dedham, Mass.).
This research was supported by grants from the U.S. Department of Agriculture (00-52104-9704 and 01-35504-10669) and the U.S. Department of Energy (FG-96ER20222).
Footnotes
Florida Agricultural Experiment Station Journal Series no. R-09069.
REFERENCES
- 1.Adachi, E., M. Torigoe, M. Sugiyama, J. Nikawa, and K. Shimizu. 1998. Modification of metabolic pathways of Saccharomyces cerevisiae by the expression of lactate dehydrogenase and deletion of pyruvate decarboxylase genes for the lactic acid fermentation at low pH value. J. Ferment. Bioeng. 86:284-289. [Google Scholar]
- 2.Arntzen, C. E., and B. E. Dale. 1999. Biobased industrial products, priorities for research and commercialization. National Academy Press, Washington, D.C. [PubMed]
- 3.Bernard, N., T. Ferain, D. Garmyn, P. Hols, and J. Delcour. 1991. Cloning of the d-lactate dehydrogenase gene from Lactobacillus delbrueckii subsp. bulgaricus by complementation in Escherichia coli. FEBS Lett. 290:61-64. [DOI] [PubMed] [Google Scholar]
- 4.Bianchi, M. M., L. Brambilla, F. Protani, C. Liu, J. Lievense, and D. Porro. 2001. Efficient homolactic fermentation by Kluyveromyces lactis strains defective in pyruvate utilization and transformed with heterologous LDH gene. Appl. Environ. Microbiol. 67:5621-5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boswell, C. 2001. Bioplastics aren't the stretch they once seemed. Chem. Market Rep. 260:15-18. [Google Scholar]
- 6.Bunch, P. K., F. Mat-Jan, N. Lee, and D. P. Clark. 1997. The ldhA gene encoding the fermentative lactate dehydrogenase of Escherichia coli. Microbiology 143:187-195. [DOI] [PubMed] [Google Scholar]
- 7.Chang, D. E., S. Shin, J. Rhee, and J. Pan. 1999. Homofermentative production of d- or l-lactate in metabolically engineered Escherichia coli RR1. Appl. Environ. Microbiol. 65:1384-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chotani, G., T. Dodge, A. Hsu, M. Kumar, R. LaDuca, D. Trimbur, W. Weyler, and K. Sanford. 2000. The commercial production of chemicals using pathway engineering. Biochim. Biophys. Acta 1543:434-455. [DOI] [PubMed] [Google Scholar]
- 9.Clark, D. P. 1989. The fermentation pathways of Escherichia coli. FEMS Microbiol. Rev. 63:223-234. [DOI] [PubMed] [Google Scholar]
- 10.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Datta, R., S. P. Tsai, P. Bonsignore, S. H. Moon, and J. R. Frank. 1995. Technological and economic potential of poly(lactic acid) and lactic acid derivatives. FEMS Microbiol. Rev. 16:221-231. [Google Scholar]
- 12.de Jong, S. J., B. van Eerdenbrugh, C.F. van Nostrum, J. J. Kettenes-van den Bosch, and W. E. Hennink. 2001. Physically crosslinked dextran hydrogels by stereocomplex formation of lactic acid oligomers: degradation and protein release behavior. J. Control. Release 71:261-275. [DOI] [PubMed] [Google Scholar]
- 13.Delcour, J., N. Bernard, D. Garmyn, T. Ferain, and P. Hols. 1993. Molecular genetics of lactate dehydrogenases from lactic acid bacteria. LAIT 73:127-131. [Google Scholar]
- 14.Dien, B. S., N. N. Nichols, and R. J. Bothast. 2001. Recombinant Escherichia coli engineered for the production of l-lactic acid from hexose and pentose sugars. J. Indust. Microbiol. Biotechnol. 27:259-264. [DOI] [PubMed] [Google Scholar]
- 15.Garmyn, D., T. Ferain, N. Bernard, P. Hols, and J. Delcour. 1994. Cloning, nucleotide sequence, and transcriptional analysis of the Pediococcus acidilactici l-(+)-lactate dehydrogenase gene. Appl. Environ. Microbiol. 61:266-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gross, R. A., and B. Kalra. 2002. Biodegradable polymers for the environment. Science 297:803-807. [DOI] [PubMed] [Google Scholar]
- 17.Hofvendahl, K., and B. Hahn-Hagerdal. 2000. Factors affecting the fermentative lactic acid production from renewable resources. Enzyme Microb. Technol. 26:87-107. [DOI] [PubMed] [Google Scholar]
- 18.Jarvis, L. 2001. Lactic acid outlook up as polylactide nears market. Chem. Market Rep. 259:5, 14.
- 19.Jiang, G. R., S. Nikolova, and D. P. Clark. 2001. Regulation of the ldhA gene, encoding the fermentative lactate dehydrogenase of Escherichia coli. Microbiology 147:2437-2446. [DOI] [PubMed] [Google Scholar]
- 20.Kim, S. F., S. J. Kim, and M. Y. Pack. 1991. Cloning and nucleotide sequence of the Lactobacillus casei lactate dehydrogenase gene. Appl. Environ. Microbiol. 57:2413-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kyla-Nikkila, K., M. Hujanen, M. Leisola, and A. Palva. 2000. Metabolic engineering of Lactobacillus helventicus CNRZ32 for production of pure l-(+)-lactic acid. Appl. Environ. Microbiol. 66:3835-3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lapierre, L., J. E. Germond, A. Ott, M. Delley, and B. Mollet. 1999. d-Lactate dehydrogenase gene (ldhD) inactivation and resulting metabolic effects in the Lactobacillus johnsonii strains La1 and N312. Appl. Environ. Microbiol. 65:4002-4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lunt, J. 1997. Large-scale production, properties and commercial applications of polylactic acid polymers. Polymer Degrad. Stabil. 59:145-152. [Google Scholar]
- 24.Martinez, A., S. W. York, L. P. Yomano, V. L. Pineda, F. C. Davis, J. C. Shelton, and L. O. Ingram. 1999. Biosynthetic burden and plasmid burden limit expression of chromosomally integrated heterologous genes (pdc, adhB) in Escherichia coli. Biotechnol. Prog. 15:891-897. [DOI] [PubMed] [Google Scholar]
- 25.Martinez-Morales, F., A. G. Borges, A. Martinez, K. T. Shanmugam, and L. O. Ingram. 1999. Chromosomal integration of heterologous DNA in Escherichia coli with precise removal of markers and replicons during construction. J. Bacteriol. 181:7143-7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mat-Jan, F., K. Y. Alam, and D. P. Clark. 1989. Mutants of Escherichia coli deficient in the fermentative lactate dehydrogenase. J. Bacteriol. 171:342-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 28.Ohara, H., H. Okuyama, S. Sawa, Y. Fujii, and K. Hiyama. 2001. Development of industrial production of high molecular weight poly-l-lactate from renewable resources. Nippon Kagaku Kaishi 6:323-331. [Google Scholar]
- 29.Omole, O. O., D. R. Brocks, G. Nappert, J. M. Naylor, and G. A. Zello. 1999. High-performance liquid chromatographic assay of (±)-lactic acid and its enantiomers in calf serum. J. Chromatogr. B 727:23-29. [DOI] [PubMed] [Google Scholar]
- 30.Ostendorp, R., W. Liebl, H. Schurig, and R. Jaenicke. 1993. The l-lactate dehydrogenase gene of the hyperthermophilic bacterium Thermotoga maritima cloned by complementation in Escherichia coli. Eur. J. Biochem. 216:709-715. [DOI] [PubMed] [Google Scholar]
- 31.Porro, D., M. Bianchi, B. M. Ranzi, L. Frontali, M. Vai, A. A. Winkler, and L. Alberghina. August2002. Yeast strains for the production of lactic acid transformed with a gene encoding lactic dehydrogenase. U.S. patent 6,429,006.
- 32.Porro, D., M. B. Michele, B. Luca, M. Rossella, B. Davide, C. Vittorio, L. Jefferson, C. L. Liu, M. R. Bianca, F. Laura, and A. Lilia. 1999. Replacement of a metabolic pathway for large-scale production of lactic acid from engineered yeasts. Appl. Environ. Microbiol. 65:4211-4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Posfai, G., M. D. Koob, H. A. Kirkpatrick, and F. C. Blattner. 1997. Versatile insertion plasmids for targeted genome manipulations in bacteria: isolation, deletion, and rescue of the pathogenicity island LEE of the Escherichia coli O157:H7 genome. J. Bacteriol. 179:4426-4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 35.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 36.Skory, C. D. 2000. Isolation and expression of lactate dehydrogenase genes from Rhizopus oryzae. Appl. Environ. Microbiol. 66:2343-2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsuji, F. 2002. Autocatalytic hydrolysis of amorphous-made polylactides: effects of l-lactide content, tacticity, and enantiomeric polymer blending. Polymer 43:1789-1796. [Google Scholar]
- 38.Underwood, S. A., M. L. Buszko, K. T. Shanmugam, and L. O. Ingram. 2002. Flux through citrate synthase limits the growth of ethanologenic Escherichia coli KO11 during xylose fermentation. Appl. Environ. Microbiol. 68:1071-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wyckoff, H. A., J. Chow, T. R. Whitehead, and M. A. Cotta. 1997. Cloning, sequence, and expression of the l(+) lactate dehydrogenase of Streptococcus bovis. Curr. Microbiol. 34:367-373. [DOI] [PubMed] [Google Scholar]
- 40.Zhou, S., T. B. Causey, A. Hasona, K. T., Shanmugam, and L. O. Ingram. 2002. Production of optically pure d-lactic acid in mineral salts medium by metabolically engineered Escherichia coli W3110. Appl. Environ. Microbiol. 69:399-407. [DOI] [PMC free article] [PubMed] [Google Scholar]