Abstract
One strategy to obtain better yields of secreted proteins has been overexpression of single endoplasmic reticulum-resident foldases or chaperones. We report here that manipulation of the unfolded-protein response (UPR) pathway regulator, HAC1, affects production of both native and foreign proteins in the yeast Saccharomyces cerevisiae. The effects of HAC1 deletion and overexpression on the production of a native protein, invertase, and two foreign proteins, Bacillus amyloliquefaciens α-amylase and Trichoderma reesei endoglucanase EGI, were studied. Disruption of HAC1 caused decreases in the secretion of both α-amylase (70 to 75% reduction) and EGI (40 to 50% reduction) compared to the secretion by the parental strain. Constitutive overexpression of HAC1 caused a 70% increase in α-amylase secretion but had no effect on EGI secretion. The invertase levels were twofold higher in the strain overexpressing HAC1. Also, the effect of the active form of T. reesei hac1 was tested in S. cerevisiae. hac1 expression caused a 2.4-fold increase in the secretion of α-amylase in S. cerevisiae and also slight increases in invertase and total protein production. Overexpression of both S. cerevisiae HAC1 and T. reesei hac1 caused an increase in the expression of the known UPR target gene KAR2 at early time points during cultivation.
Saccharomyces cerevisiae has been used in production of numerous heterologous proteins, not least because it is considered a safe host for biotechnical processes. A feature that makes S. cerevisiae a good host for production of proteins, especially foreign proteins, is the fact that S. cerevisiae cells are able to perform most posttranslational modifications that occur in higher eukaryotes, including folding, subunit assembly, glycosylation, and proteolytic processing of secreted polypeptides. A major limitation in its use as a production host has been its limited capacity to secrete proteins.
Many genetic methods have been used to manipulate S. cerevisiae in order to obtain better productivity of foreign proteins. Approaches like the use of appropriate promoters and their modified versions (26) and the use of fusions of signal peptides to the heterologous proteins for efficient targeting to the secretory pathway (1) have been used successfully for production of some proteins (reviewed in reference 25). Also, manipulation of glycosylation of the foreign protein has been shown to increase secretion (29). There are also some cases in which overexpression of genes of the secretory pathway has been shown to increase protein secretion. One such example is the S. cerevisiae Sso proteins, which act in the fusion of secretory vesicles to the plasma membrane (28). These proteins have been shown to increase the secretion of Bacillus amyloliquefaciens α-amylase and the native protein invertase when they are overexpressed.
Misfolded heterologous proteins accumulate in the endoplasmic reticulum (ER) (15), and it seems that the efficiency of protein folding and assembly in the ER is the step that determines the efficiency of foreign protein production. In order to reduce heterologous protein accumulation and to facilitate secretion, the effects of overexpression of ER-resident chaperones and foldases have been tested in S. cerevisiae. Overproduction of Kar2p has been shown to stimulate the secretion of bovine prochymosin 26-fold but to have no effect on the secretion of plant thaumatin (9). Coexpression of Kar2 and PDI acted synergistically to increase secretion of single-chain antibody fragments (34). Also, PDI overexpression can increase secretion of heterologous proteins; secretion of human growth factor B was enhanced 10-fold, and production of Schizosaccharomyces pombe acid phosphatase was enhanced fourfold (24). Increased amounts of Bip/Kar2 or PDI are not always beneficial for foreign protein production, since overexpression of PDI has also been found to decrease the production of recombinant proteins and to cause accumulation of the proteins in mammalian cells (6). The results discussed above indicate that overexpression of chaperones and foldases does not always overcome the secretion block and that the physical characteristics of the secreted protein and/or adjusting the expression level of the folding factors may be critical in designing a system to improve foreign protein production.
The expression of genes for several ER-resident chaperones and foldases is controlled by the unfolded-protein response (UPR) pathway. Conditions that overload the ER with unfolded and misfolded proteins are known to activate the UPR (16). Also, environmental stresses that perturb ER functions, such as glucose starvation, changes in the ER redox status, perturbation of cellular calcium balance, inhibition of protein glycosylation, or viral infection, have been shown to activate the UPR. The UPR pathway is triggered by a reduction in the level of free Kar2/Bip in the ER, possibly resulting from the binding of Kar2/Bip to misfolded polypeptides (2). Several genes that function in the UPR pathway have been characterized in S. cerevisiae. HAC1 encodes a bZIP-type transcription factor with amino acid similarity to the mammalian ATF/CREB proteins (21). An unconventional splicing event, resulting in cleavage of a 250-base intron from the mRNA, activates HAC1. The spliced HAC1 is translated into a 238-amino-acid Hac1p protein, which is the active form that functions in the UPR pathway (4, 17, 20, 32). In S. cerevisiae the expression of several ER-resident chaperones and foldases has been shown to be activated by the UPR (reviewed in reference 12). There are indications that the UPR regulates expression of genes other than just those that function in the ER. It has been shown recently that genes encoding functions that occur throughout the secretory pathway are induced by Hac1p upon UPR induction (35).
We have previously cloned functional homologues of S. cerevisiae HAC1 from the filamentous fungi Trichoderma reesei and Aspergillus nidulans (hac1 and hacA, respectively) and have shown that the induction of the genes involves two steps. These steps are splicing of an intron that is 20 bases long and truncation of the mRNA at the 5′ flanking region (30a). Truncation removes an upstream open reading frame from the mRNA, possibly increasing translation initiation at the right start codon.
The aim of this study was to examine the role of the UPR pathway in foreign and native protein production in S. cerevisiae. We deleted the HAC1 gene and thus inactivated this pathway. On the other hand, we activated the UPR constitutively by expressing an induced form of S. cerevisiae HAC1 or its functional homologue hac1 from the filamentous fungus T. reesei. Because one central factor in the UPR pathway was manipulated, all functions under Hac1p control throughout the secretory pathway should be affected. Our results show that this is a novel way to increase the production of both native and foreign proteins in S. cerevisiae.
MATERIALS AND METHODS
Strains and media.
The S. cerevisiae strains used in this study were DBY746 (Matα his3Δ1 leu2-3,112 trp1-289 ura3-52 Cyhr) of D. Botstein and BMA64-1A (Mata ura3-1 trp1Δ2 leu2-3,112 his3-11 ade2-1 can1-100). The effect of constitutive UPR induction on α-amylase was studied in a derivative of DBY746 designated H1256 (S. Keränen, VTT Biotechnology). In this strain expression of the B. amyloliquefaciens α-amylase is driven by the S. cerevisiae ADH1 promoter in a construct integrated into the TRP1 locus.
The cultivation medium used for α-amylase production was S. cerevisiae synthetic complete (SC) medium with appropriate amino acids (33), which was buffered to pH 6.0 with 2% succinic acid and supplemented with 2% glucose as the carbon source and 10 mM CaCl2. Cultivation for invertase production was done in SC medium containing 2% sucrose as the carbon source. To measure the production of T. reesei EGI and total secreted proteins, the strains were grown in SC medium (33) with 2% glucose. All shake flask cultures were inoculated to an initial optical density at 600 nm (OD600) of 0.2, incubated at 30°C, and shaken at 250 rpm.
Construction of strains and plasmids.
The S. cerevisiae HAC1 gene was disrupted by replacing it in the genome with a DNA fragment containing the G418 antibiotic resistance cassette flanked by 48-bp sequences from the 5′ and 3′ ends of the HAC1 open reading frame. The G418 resistance cassette consists of the Escherichia coli kanamycin resistance gene cloned between the promoter and terminator of the Ashbya gossypii TEF gene encoding translation elongation factor 1. The DNA fragment used in disruption of the S. cerevisiae HAC1 gene was produced by PCR from the kanMX2 module (37) with the oligonucleotide primers 5′-CCA CCT ACG ACA ACA ACC GCC ACT ATG GAA ATG ACT GAT TTT GAA CTA CTT GCC TCG TCC CCG CCG GGT CAC (forward primer) and 5′-AAT TAT ACC CTC TTG CGA TTG TCT TCA TGA AGT GAT GAA GAA ATC ATT GAC ACT GGA TGG CGG CGT TAG TAT CGA (reverse primer). The fragment was transformed into S. cerevisiae strain BMA64-1A. The transformants were first selected for resistance to kanamycin as described by Wach et al. (37). The resistant clones were then screened for inositol dependence on mineral medium (36) without inositol. The HAC1 disruption in inositol-dependent clones was verified by Southern hybridization performed as described by Sambrook et al. (31). In the HAC1 disruptant strain, the α-amylase was expressed from the ADH1 promoter in multicopy plasmid YEpαa6 with the LEU2 marker gene (26), in which the α-amylase gene was cloned between the S. cerevisiae ADH1 promoter and terminator. T. reesei EGI was expressed from plasmid pMP311 (22), in which the endoglucanase cDNA was cloned between the S. cerevisiae PGK1 promoter and terminator in a multicopy vector with the LEU2 marker gene.
To make a construct for constitutive UPR induction, the HAC1 gene fragment was amplified by PCR from S. cerevisiae chromosomal DNA. This fragment starts 24 bp before the translation started codon of the HAC1 gene and ends with a translation stop codon inserted after the proline codon at amino acid position 220 of the deduced protein. The oligonucleotide primers used were 5′-ATC GCA GGA TTC CCA CCT ACG ACA ACA ACC GCC ACT (forward primer) and 5′-TAC AGC GGA TCC CTA TGG ATT ACG CCA ATT GTC AAG (reverse primer). The intronless, active form of HAC1 was cloned between the promoter and terminator of the S. cerevisiae PGK1 gene into the S. cerevisiae single-copy vector pKK1 carrying the LEU2 selectable marker gene. The final expression plasmid was designated pMS109.
The active form of the T. reesei hac1 (30a) cDNA containing the protein-coding region without the 20-bp intron was produced by performing PCR with the forward primer 5′-CCG CAA CAC GAC ACG GCA GGC AAC and the reverse primer 5′-CTA GGT AGA CGT TGT ATT TTG from a plasmid carrying this cDNA form. The PCR fragment was cloned into the multicopy expression vector pAJ401 (30) between the S. cerevisiae PGK1 promoter and terminator. The resulting plasmid was designated pMS132.
RNA isolation was done with a Trizol kit according to the instructions of the manufacturer. Northern hybridizations were done as described by Sambrook et al. (31). S. cerevisiae transformations were done by the method of Gietz et al. (8).
Activity assays.
The α-amylase activity in S. cerevisiae culture supernatants was measured with the Phadebas amylase test (Pharmacia) performed according to the instructions of the manufacturer. For the invertase assay, equal amounts of cells, as determined by OD600 measurement, were harvested from different cultures, washed with 5 ml of 10 mM NaN3, and resuspended in 0.2 M sodium acetate buffer (pH 5.0) with 10 mM NaN3. The invertase activity of the cells was measured by incubating them with 0.166 M sucrose in 0.2 M sodium acetate buffer (pH 5.0) for 6 min. The reaction was stopped by adding 1 volume of 0.5 M KPO4 (pH 7.0) and by separating the cells rapidly from the reaction mixture by filtration. The glucose formed in the reaction mixture was measured with a GOD Perid kit (Boehringer Mannheim) used according to the manufacturer's protocols.
The endoglucanase activity of cultures was measured with the substrate 4-methylumbelliferyl-d-lactoside (Sigma). Supernatant samples were incubated at 50°C for 3 h in reaction mixtures containing 0.25 mg of the substrate per ml and 0.1 M glucose in 50 mM sodium acetate (pH 5.0). Each reaction was stopped by adding 2 volumes of 1 M Na2CO3, and the absorbance of the mixture was measured at a wavelength of 370 nm.
To measure the total amount of proteins secreted into the culture medium, three parallel cultures were grown in big shake flasks for 4 days, and 45-ml samples were withdrawn from the cultures daily. The proteins were precipitated with 10% (vol/vol) trichloroacetic acid overnight at 4°C. After centrifugation for 15 min at 4°C and 4,800 × g, the pellets were dissolved in 2% Na2CO3 in 0.1 N NaOH buffer, and the amounts of protein were measured by the method of Lowry et al. (14).
RESULTS
Effect of S. cerevisiae HAC1 disruption.
First, we wanted to test the effect of HAC1 disruption on protein production. Our hypothesis was that induction of the UPR would be beneficial for the secretion of proteins and that a lack of the UPR would cause a reduction in protein production. The HAC1 gene was disrupted by replacing it in the S. cerevisiae genome by a kanamycin resistance cassette. S. cerevisiae HAC1 mutants are known to require inositol for growth (20), and thus, the possible disruptants were tested for inositol dependence. Several inositol-dependent clones were found, and HAC1 disruption was verified by Southern hybridization (data not shown).
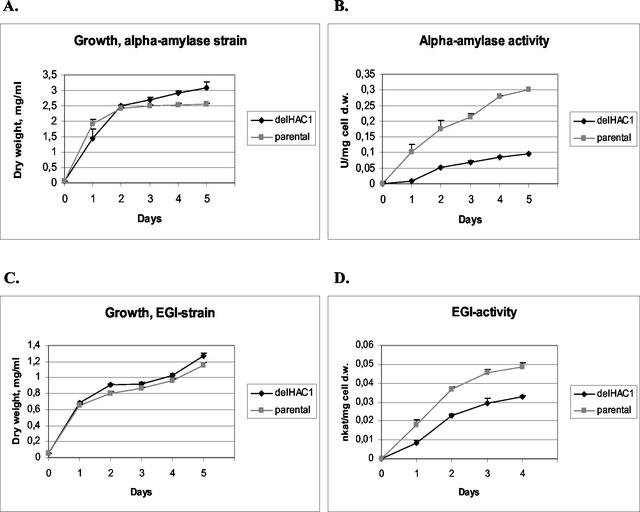
The effects of HAC1 disruption on the production of two heterologous proteins, the B. amyloliquefaciens α-amylase (27) and the T. reesei endoglucanase EGI (22), as well as a native protein, invertase, were tested. To test the effect of HAC1 deletion on the secretion of α-amylase, the α-amylase expression vector (YEpαa6) was transformed into the HAC1 disruptant strain and its parental strain. Three α-amylase-producing transformants from both strains were grown in shake flasks in SC medium. Samples were taken daily, and the OD600 and α-amylase activities were measured. The HAC1 disruptant strain produced 25 to 30% of the amount of α-amylase produced by the wild-type control strain (Fig. 1B). To test the effect of HAC1 disruption on production of T. reesei endoglucanase EGI, the EGI expression vector (pMP311) was transformed into the HAC1 disruptant strain, and three transformants from this strain and three transformants from the parental strain transformed with the pMP311 vector were grown in shake flasks. The level of endoglucanase EGI produced by the HAC1 disruptant was about 50 to 60% of the level produced by the parental strain (Fig. 1D). There were slight differences in the growth of the HAC1 disruptants carrying EGI and α-amylase expression cassettes and the parental strains. In the parental strain cultures the cell densities were slightly lower than those in the disruptant cultures, and the difference was clearer for the α-amylase-producing strain (Fig. 1A and C). To test the effect of HAC1 disruption on the production of invertase, the HAC1 disruptant and the parental strain were cultivated in SC medium with 2% sucrose as the carbon source. The amounts of invertase in the periplasmic space of the strains were measured daily for 5 days. There was no significant difference between the strains in terms of production of invertase (data not shown).
FIG. 1.
Effects of HAC1 deletion (delHAC1) on the growth of strains producing B. amyloliquefaciens α-amylase (A) and T. reesei endoglucanase EGI (C) and on the secretion of B. amyloliquefaciens α-amylase (B) and T. reesei endoglucanase EGI (D). One katal was equal to 1 mol of substrate consumed per s. The results were obtained from parallel cultures of the HAC1 deletant strains and the host (BMA64-1A). The error bars indicate standard deviations. d.w., dry weight.
Effect of constitutive S. cerevisiae HAC1 overexpression.
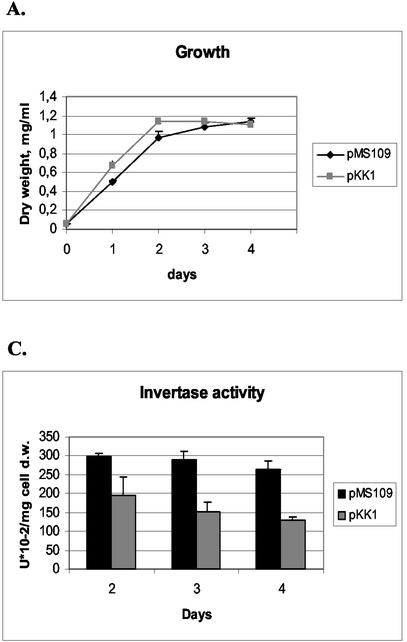
In order to constitutively induce the UPR in S. cerevisiae, a truncated version of the S. cerevisiae HAC1 gene was expressed from a single-copy plasmid from the PGK1 promoter. The truncated version did not include the 250-bp intron of HAC1 that in normal conditions prevents translation of the mRNA. Therefore, the mRNA expressed from the PGK1 promoter in the plasmid was translated to form the active Hac1 protein and caused constitutive induction of the UPR. The HAC1 expression plasmid pMS109 and the control plasmid pKK1 without the HAC1 sequences were transformed into an S. cerevisiae strain producing B. amyloliquefaciens α-amylase from an integrated construct. Four pMS109 transformants and four strains transformed with the vector pKK1 were cultivated for 5 days in shake flasks, and samples were taken daily to monitor S. cerevisiae growth and α-amylase production. The growth of the pMS109 transformants appeared to be somewhat retarded compared with the growth of the control (Fig. 2A). This is in accordance with the previously reported finding that constitutive expression of Hac1p causes slower growth of S. cerevisiae (4, 13).
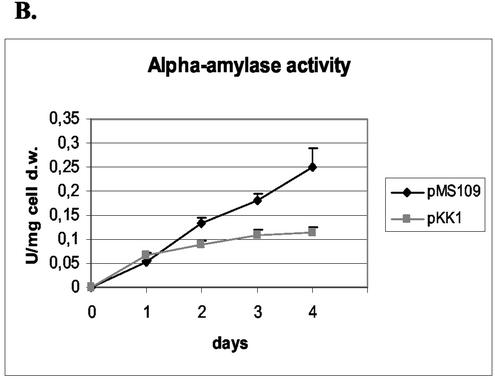
FIG. 2.
Effects of S. cerevisiae HAC1 overexpression on growth (A), secretion of B. amyloliquefaciens α-amylase (B), and production of invertase (C) were assayed from the HAC1-overexpressing strain (pMS109). The parental strain (H1256 transformed with the pKK1 vector) was used as a control. Data obtained with parallel cultures of the HAC1 transformants and the host strain are shown. The error bars indicate standard deviations. d.w., dry weight.
The amounts of the Bacillus α-amylase secreted by each of the pMS109 transformants were larger than the amounts produced by any of the control pKK1 transformants. The average level of production of the pMS109 transformants was twofold higher at the end of the cultivation than the average level of production of control clones (Fig. 2B). We also tested whether expression of the induced form of HAC1 could enhance production of the other foreign protein, endoglucanase EGI of T. reesei. No beneficial effect, however, was detected (data not shown).
To analyze the effect of constitutive UPR induction on S. cerevisiae invertase production, four clones transformed with pMS109 and four clones transformed with the pKK1 vector of the α-amylase-producing strain were cultivated with 2% sucrose as the carbon source for 5 days. The amounts of the invertase in the periplasmic space of the strains were measured, and they were about 1.6- to 2-fold greater in the pMS109 strains than in the pKK1 strains at all time points that were tested (Fig. 2C). The effect on total protein secretion was tested by using three clones transformed with the pMS109 vector and three clones transformed with the pKK1 vector of the α-amylase-producing strain. The strains were grown in 2% glucose for 4 days, samples from the culture supernatants were taken daily, and the total proteins produced were analyzed by using trichloroacetic acid-precipitated samples. The results revealed no clear differences among the strains (data not shown).
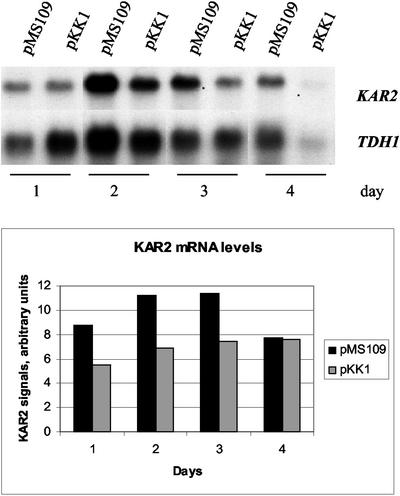
It has been shown that the promoter of the S. cerevisiae KAR2 gene encoding an ER chaperone contains an unfolded protein response element (UPRE) and is induced by the UPR (3). The levels of expression of the KAR2 gene in the pMS109 and control strains at different times were studied by Northern hybridization. The levels of KAR2 expression were about 1.5- to 2-fold higher on days 1, 2, and 3 of cultivation in the strain expressing the constitutive HAC1 gene than in the control. During day 4 the levels of expression were similar in the two strains (Fig. 3).
FIG. 3.
Effect of HAC1 overexpression on expression of the KAR2 gene. The KAR2 signals were quantified and normalized to the TDH1 signal intensities.
Effect of expression of the T. reesei hac1 gene on protein production in S. cerevisiae.
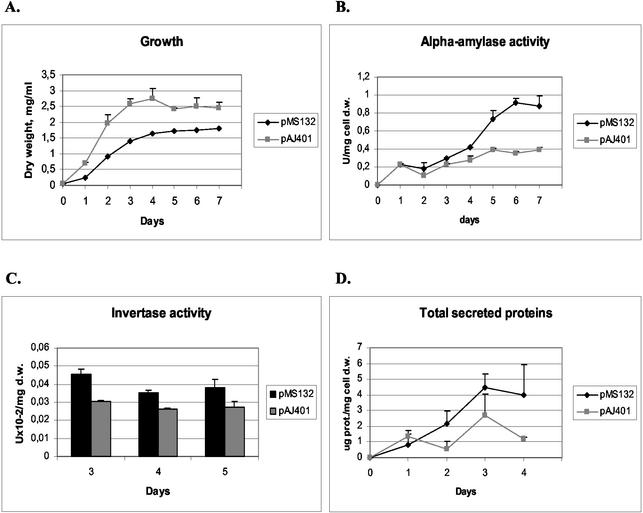
The T. reesei hac1 cDNA was expressed without its 5′ flanking region and the 20-base intron from plasmid pMS132. This plasmid and the empty vector pAJ401 were transformed into the S. cerevisiae strain producing Bacillus α-amylase. Three strains carrying pMS132 and three strains carrying the control vector were grown, and growth and α-amylase production were assayed. The results showed that per biomass the amount of α-amylase produced by the pMS132 transformant was larger than the amount produced by the pAJ401 transformant from day 3 until the end of the experiment. The difference between the two strains was highest on day 6, about 2.4-fold (Fig. 4B). Growth of the pMS132 transformants was slower, and the final biomass was lower than that of the control transformants (Fig. 4A).
FIG. 4.
Effect of expression of the active form of T. reesei hac1. (A) Growth of the strain expressing the hac1 construct (pMS132) compared to growth of the control. (B) Secretion of B. amyloliquefaciens α-amylase in the pMS132 and parental strains. (C) Invertase production by the pMS132 and parental strains. (D) Amounts of total secreted proteins in the pMS132 and parental strains. The parental strain (H1256 transformed with the pAJ401 vector) was used as a control. Data obtained with parallel cultures of the T. reesei hac1 transformants and the host strain are shown. The error bars indicate standard deviations. d.w., dry weight; prot., protein.
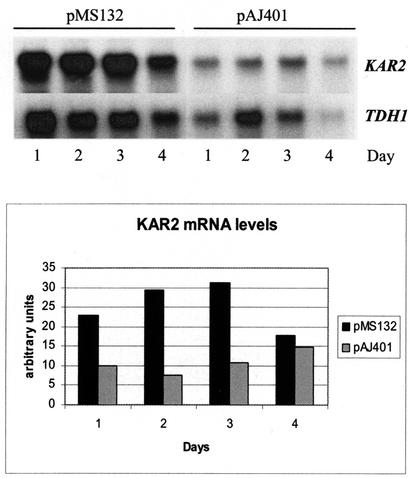
Northern hybridizations with the KAR2 gene as a probe showed that the level of KAR2 expression was higher in the pMS132 transformants at the beginning of cultivation. However, the difference between the pMS132 and control transformants was reversed on day 4 of cultivation, and no difference was observed later in the experiment (Fig. 5). This may have been a result of activation of the UPR by α-amylase production in the control strain at a late culture stage.
FIG. 5.
Effect of T. reesei hac1 expression on expression of KAR2. The KAR2 signals were quantified and normalized to TDH1 signal intensities.
As described above, the effect of T. reesei hac1 overexpression on production of a native protein, invertase, was tested. Four pMS132 transformants and four pAJ401 transformants were grown for 5 days on 2% sucrose to induce the invertase gene, and cell samples for invertase assays were taken daily. The results showed that more invertase was produced by the pMS132 transformants than by the control transformants (Fig. 4C). These results indicate that overexpression of the heterologous T. reesei hac1 gene can have an effect on protein production in S. cerevisiae similar to the effect produced by overexpression of the homologous HAC1 gene. Also, an experiment was performed as described previously to study the effect of T. reesei hac1 on the levels of total secreted proteins. Three pMS132 transformants and three pAJ401 transformants were grown for 4 days, and samples of the culture supernatants were taken daily. The results showed that there was a slight increase in the amount of total secreted proteins in the strains overexpressing hac1 (Fig. 4D).
DISCUSSION
It is generally believed that processes occurring in the ER may be the rate-limiting steps in secretion of proteins and in particular secretion of proteins of heterologous origin. This may be due to saturation of the machinery involved in proper protein folding and assembly, which leads to accumulation of misfolded proteins or protein aggregates in the ER. For this reason the levels of ER proteins that assist in the proper folding of newly synthesized proteins have been considered important for efficient secretion.
In our first experiment the role of the UPR in production of foreign proteins by S. cerevisiae was addressed by deletion of the HAC1 gene encoding the UPR regulator that upregulates the expression of folding factors in the ER. In a HAC1 disruptant strain, in which UPR induction did not occur (4, 20), both the production of Bacillus α-amylase and the production of T. reesei endoglucanase EGI were clearly reduced (Fig. 1B and D). These results suggest that at least some of the secretion pathway components that are regulated by the UPR pathway are involved in secretion of these proteins and that UPR pathway induction may be beneficial for the production of these proteins and foreign proteins in general. Interestingly HAC1 disruption did not retard the growth of these strains producing the foreign proteins. Assuming that at least parts of the foreign proteins are misfolded, this indicates that S. cerevisiae has mechanisms besides the UPR that can deal with the slowly folding foreign proteins in the ER. One such mechanism is the ER-associated degradation (ERAD) pathway. Thus, it may be that the foreign proteins produced in a strain with HAC1 deleted are degraded to some extent by the ERAD pathway. It has been shown that when the UPR function is deleted, the ERAD pathway is sufficient to eliminate the misfolded proteins from the ER, provided that the cell does not encounter unusual stress (19, 35). One possible explanation is that the extent of protein misfolding in these strains is relatively low, and thus the growth is not impaired. This explanation is supported by the observation that expression of the single-chain antibody 4-4-20 (scFv) in S. cerevisiae decreases the growth rate of the yeast cells. The growth rate is recovered with long expression times, but it has been shown that the recovery requires a functional UPR pathway (11).
Another possible explanation for the observed reduction in EGI and α-amylase levels is the general slowdown of the secretory functions caused by the HAC1 deletion. Since HAC1 has been shown to be nonessential for growth under normal conditions (21), the material (e.g., cell wall components and membrane proteins) needed for growth have to be efficiently passaged through the secretory pathway even when HAC1 is deleted. This could indicate that the production of foreign proteins is selectively decreased in the strains with HAC1 deleted. Also, since there are different secretory vesicle populations (5, 10, 23), the foreign proteins could be transported in a different vesicle population than the population that transports the proteins needed for growth. Thus, transport of the vesicles containing the foreign proteins would be regulated by the UPR. The fact that no reduction in invertase production was observed also favors this explanation.
Instead of overexpressing just one or two chaperone or foldase genes or other individual factors of the secretory pathway, in our approach we upregulated the whole ER folding machinery by constitutive UPR induction and examined the effect on protein secretion. Apparently, with this approach multiple functions involved in the secretory process can be enhanced since DNA microarray experiments with S. cerevisiae have shown that the UPR pathway regulates the transcription of about 380 genes through the action of Hac1p (35). A total of 103 of the genes with known functions are involved in secretion or biogenesis of secretory organelles. These include, quite expectedly, genes for the ER-resident chaperones and foldases. More surprising was the finding that genes encoding diverse functions, such as translocation, processing of the polypeptides, protein degradation, vesicle trafficking, lipid biosynthesis, and vacuolar protein sorting, were also induced by induction of the UPR. This supports a model in which the UPR improves secretion by enhancing the folding of misfolded proteins and also reduces the level of misfolded proteins by activating ERAD (35). Another group has shown by using DNA microarray technology that under ER stress conditions the expression of genes involved in cell wall biogenesis, protein secretion, and processing in the ER is induced (7).
By overexpressing the constitutively active form of S. cerevisiae HAC1 or T reesei hac1, we were able to increase secretion of both Bacillus α-amylase and invertase in S. cerevisiae (Fig. 2 and 4). The enhancement of α-amylase production with the T. reesei hac1 gene was about 2.4-fold, and the enhancement with the S. cerevisiae HAC1 gene was about 1.7-fold. On the other hand, invertase production was increased more by the S. cerevisiae HAC1 construct than by the T. reesei hac1 expression plasmid. This suggests that the optimal extent of UPR induction is different for different proteins. Furthermore, we could not detect improved production of the endoglucanase EGI (data not shown). This result could indicate that expression of this protein is not very dependent on the level of the UPR pathway components. This is in accordance with the observation that HAC1 deletion affected production of this protein less than it affected production of α-amylase. The results for total protein secretion show that the production of total proteins was higher in strains overexpressing the T. reesei hac1 gene, whereas no clear effect was seen in the strains overexpressing S. cerevisiae HAC1. The observed difference could have been caused by the higher level of induction of the UPR by the T. reesei hac1 construct.
It has been shown that constitutive UPR induction in S. cerevisiae reduces the growth rate of the cells (4, 13). The growth rate of the strain overexpressing the active form of the S. cerevisiae HAC1 gene was similar to that of the parental strain, but the final cell densities remained slightly lower until day 4 (Fig. 2A). The effect on growth was more pronounced with the T. reesei hac1 construct; in this case the final cell density of the UPR-induced cells was clearly lower than that of the parental strain, although the maximal growth rates of the strains were similar. This is consistent with the presumably higher level of expression of the T. reesei hac1 gene obtained with the multicopy vector, which may have induced the UPR more strongly than the single-copy S. cerevisiae HAC1 construct. Furthermore, the S. cerevisiae HAC1 expression construct does not contain the suggested C-terminal 18-amino-acid activation domain region (18). In order to use this approach optimally to increase protein production in S. cerevisiae, a balance has to be found between the increase in the protein-folding capacity in the ER and the growth inhibition.
Previous work on enhancing protein production by overexpression of individual foldase or chaperone genes has concentrated on foreign proteins. We showed that production of S. cerevisiae invertase could also be enhanced. However, overexpression of the UPR regulator Hac1p, like the foldase-chaperone overexpression approach, is not effective for every protein, as shown here by the lack of enhancement of T. reesei endoglucanase EGI production. This may be at least partially because different proteins may have problems with secretion at different levels of the secretory pathway. Both foreign proteins analyzed in this study are reasonably well produced, and EGI can even be secreted at levels of 100 to 200 mg/liter by brewer's yeast in bioreactor cultures (38). The fungal protein EGI is hypermannosylated by S. cerevisiae compared to the mannosylation in the native host T. reesei (22), and the bacterial α-amylase also gains glycosylation, although at a more moderate level (27). In the case of EGI, the enzyme is secreted mainly during the stationary phase of cultures and is retained in the membranous intracellular fraction at earlier stages of culture (22). In the case of Bacillus α-amylase, 70% of the protein produced is secreted, whereas only 30% is associated with the cells (27) and it seems that secretion into the culture medium is not the major limiting step.
We still do not completely understand the features of proteins that affect their secretion and what specific problems different proteins may encounter in heterologous hosts. In this situation, manipulation of the folding and secretory processes by the UPR regulator Hac1p, which controls a variety of functions in the protein production pathway, may turn out to be a very useful general approach. Our work with Aspergillus niger var. awamori has shown that this strategy is also beneficial with this industrially important filamentous fungus (M. Valkonen, M. Ward, H. Wang, M. Penttilä, and M. Saloheimo, submitted for publication).
Acknowledgments
We thank Riitta Nurmi for excellent technical assistance and Michael Ward and Huaming Wang for fruitful discussions.
This work was supported by Genencor International Inc. and the Finnish Technology Agency (Tekes).
REFERENCES
- 1.Baldari, C., J. A. Murray, P. Ghiara, G. Cesareni, and C. L. Galeotti. 1987. A novel leader peptide which allows efficient secretion of a fragment of human interleukin 1 beta in Saccharomyces cerevisiae. EMBO J. 6:229-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bertolotti, A., Y. Zhang, L. M. Hendershot, H. P. Harding, and D. Ron. 2000. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat. Cell Biol. 2:326-332. [DOI] [PubMed] [Google Scholar]
- 3.Cox, J. S., C. E. Shamu, and P. Walter. 1993. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell 73:1197-1206. [DOI] [PubMed] [Google Scholar]
- 4.Cox, J. S., and P. Walter. 1996. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell 87:391-404. [DOI] [PubMed] [Google Scholar]
- 5.David, D., S. Sundarababu, and J. E. Gerst. 1998. Involvement of long chain fatty acid elongation in the trafficking of secretory vesicles in yeast. J. Cell Biol. 143:1167-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis, R., K. Schooley, B. Rasmussen, J. Thomas, and P. Reddy. 2000. Effect of PDI overexpression on recombinant protein secretion in CHO cells. Biotechnol. Prog. 16:736-743. [DOI] [PubMed] [Google Scholar]
- 7.Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen, G. Storz, D. Botstein, and P. O. Brown. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11:4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gietz, D., A. St Jean, R. A. Woods, and R. H. Schiestl. 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20:1425.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harmsen, M. M., M. I. Bruyne, H. A. Raue, and J. Maat. 1996. Overexpression of binding protein and disruption of the PMR1 gene synergistically stimulate secretion of bovine prochymosin but not plant thaumatin in yeast. Appl. Microbiol. Biotechnol. 46:365-370. [DOI] [PubMed] [Google Scholar]
- 10.Harsay, E., and A. Bretscher. 1995. Parallel secretory pathways to the cell surface in yeast. J. Cell Biol. 131:297-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kauffman, K. J., E. M. Pridgen, F. J. Doyle III, P. S. Dhurjati, and A. S. Robinson. 2002. Decreased Protein Expression and Intermittent Recoveries in BiP Levels Result from Cellular Stress during Heterologous Protein Expression in Saccharomyces cerevisiae. Biotechnol. Prog. 18:942-950. [DOI] [PubMed]
- 12.Kaufman, R. J. 1999. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 13:1211-1233. [DOI] [PubMed] [Google Scholar]
- 13.Kawahara, T., H. Yanagi, T. Yura, and K. Mori. 1997. Endoplasmic reticulum stress-induced mRNA splicing permits synthesis of transcription factor Hac1p/Ern4p that activates the unfolded protein response. Mol. Biol. Cell 8:1845-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 15.Marquardt, T., and A. Helenius. 1992. Misfolding and aggregation of newly synthesized proteins in the endoplasmic reticulum. J. Cell Biol. 117:505-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Menzel, R., F. Vogel, E. Kargel, and W. H. Schunck. 1997. Inducible membranes in yeast: relation to the unfolded-protein-response pathway. Yeast 13:1211-1229. [DOI] [PubMed] [Google Scholar]
- 17.Mori, K., T. Kawahara, H. Yoshida, H. Yanagi, and T. Yura. 1996. Signalling from endoplasmic reticulum to nucleus: transcription factor with a basic-leucine zipper motif is required for the unfolded protein-response pathway. Genes Cells 1:803-817. [DOI] [PubMed] [Google Scholar]
- 18.Mori, K., N. Ogawa, T. Kawahara, H. Yanagi, and T. Yura. 2000. mRNA splicing-mediated C-terminal replacement of transcription factor Hac1p is required for efficient activation of the unfolded protein response. Proc. Natl. Acad. Sci. USA 97:4660-4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ng, D. T., E. D. Spear, and P. Walter. 2000. The unfolded protein response regulates multiple aspects of secretory and membrane protein biogenesis and endoplasmic reticulum quality control. J. Cell Biol. 150:77-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nikawa, J., M. Akiyoshi, S. Hirata, and T. Fukuda. 1996. Saccharomyces cerevisiae IRE2/HAC1 is involved in IRE1-mediated KAR2 expression. Nucleic Acids Res. 24:4222-4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nojima, H., S. H. Leem, H. Araki, A. Sakai, N. Nakashima, Y. Kanaoka, and Y. Ono. 1994. Hac1: a novel yeast bZIP protein binding to the CRE motif is a multicopy suppressor for cdc10 mutant of Schizosaccharomyces pombe. Nucleic Acids Res. 22:5279-5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Penttilä, M. E., L. Andre, M. Saloheimo, P. Lehtovaara, and J. K. Knowles. 1987. Expression of two Trichoderma reesei endoglucanases in the yeast Saccharomyces cerevisiae. Yeast 3:175-185. [DOI] [PubMed] [Google Scholar]
- 23.Roberg, K. J., N. Rowley, and C. A. Kaiser. 1997. Physiological regulation of membrane protein sorting late in the secretory pathway of Saccharomyces cerevisiae. J. Cell Biol. 137:1469-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson, A. S., V. Hines, and K. D. Wittrup. 1994. Protein disulfide isomerase overexpression increases secretion of foreign proteins in Saccharomyces cerevisiae. Bio/Technology 12:381-384. [DOI] [PubMed] [Google Scholar]
- 25.Romanos, M. A., C. A. Scorer, and J. J. Clare. 1992. Foreign gene expression in yeast: a review. Yeast 8:423-488. [DOI] [PubMed] [Google Scholar]
- 26.Ruohonen, L., M. K. Aalto, and S. Keränen. 1995. Modifications to the ADH1 promoter of Saccharomyces cerevisiae for efficient production of heterologous proteins. J. Biotechnol. 39:193-203. [DOI] [PubMed] [Google Scholar]
- 27.Ruohonen, L., P. Hackman, P. Lehtovaara, J. K. Knowles, and S. Keränen. 1987. Efficient secretion of Bacillus amyloliquefaciens alpha-amylase by its own signal peptide from Saccharomyces cerevisiae host cells. Gene 59:161-170. [DOI] [PubMed] [Google Scholar]
- 28.Ruohonen, L., J. Toikkanen, V. Tieaho, M. Outola, H. Söderlund, and S. Keränen. 1997. Enhancement of protein secretion in Saccharomyces cerevisiae by overproduction of Sso protein, a late-acting component of the secretory machinery. Yeast 13:337-351. [DOI] [PubMed] [Google Scholar]
- 29.Sagt, C. M., B. Kleizen, R. Verwaal, M. D. de Jong, W. H. Muller, A. Smits, C. Visser, J. Boonstra, A. J. Verkleij, and C. T. Verrips. 2000. Introduction of an N-glycosylation site increases secretion of heterologous proteins in yeasts. Appl. Environ. Microbiol. 66:4940-4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saloheimo, A., B. Henrissat, A. M. Hoffren, O. Teleman, and M. Penttilä. 1994. A novel, small endoglucanase gene, egl5, from Trichoderma reesei isolated by expression in yeast. Mol. Microbiol. 13:219-228. [DOI] [PubMed] [Google Scholar]
- 30a.Saloheimo, M., M. Valkonen, and M. Penttilä. 2003. Activation mechanisms of the HACI-mediated unfolded protein response in filamentous fungi. Mol. Microbiol. 47:1149-1161. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Shamu, C. E. 1997. Splicing together the unfolded-protein response. Curr. Biol. 7:R67-R70. [DOI] [PubMed] [Google Scholar]
- 33.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:3-21. [DOI] [PubMed] [Google Scholar]
- 34.Shusta, E. V., R. T. Raines, A. Pluckthun, and K. D. Wittrup. 1998. Increasing the secretory capacity of Saccharomyces cerevisiae for production of single-chain antibody fragments. Nat. Biotechnol. 16:773-777. [DOI] [PubMed] [Google Scholar]
- 35.Travers, K. J., C. K. Patil, L. Wodicka, D. J. Lockhart, J. S. Weissman, and P. Walter. 2000. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101:249-258. [DOI] [PubMed] [Google Scholar]
- 36.Verduyn, C., E. Postma, W. A. Scheffers, and J. P. Van Dijken. 1992. Effect of benzoic acid on metabolic fluxes in yeasts: a continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast 8:501-517. [DOI] [PubMed] [Google Scholar]
- 37.Wach, A., A. Brachat, R. Pohlmann, and P. Philippsen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793-1808. [DOI] [PubMed] [Google Scholar]
- 38.Zurbriggen, B. D., M. E. Penttilä, L. Viikari, and M. J. Bailey. 1991. Pilot scale production of a Trichoderma reesei endo-beta-glucanase by brewer's yeast. J. Biotechnol. 17:133-146. [DOI] [PubMed] [Google Scholar]