Abstract
The rate of microbial respiration can be described by a rate law that gives the respiration rate as the product of a rate constant, biomass concentration, and three terms: one describing the kinetics of the electron-donating reaction, one for the kinetics of the electron-accepting reaction, and a thermodynamic term accounting for the energy available in the microbe's environment. The rate law, derived on the basis of chemiosmotic theory and nonlinear thermodynamics, is unique in that it accounts for both forward and reverse fluxes through the electron transport chain. Our analysis demonstrates how a microbe's respiration rate depends on the thermodynamic driving force, i.e., the net difference between the energy available from the environment and energy conserved as ATP. The rate laws commonly applied in microbiology, such as the Monod equation, are specific simplifications of the general law presented. The new rate law is significant because it affords the possibility of extrapolating in a rigorous manner from laboratory experiment to a broad range of natural conditions, including microbial growth where only limited energy is available. The rate law also provides a new explanation of threshold phenomena, which may reflect a thermodynamic equilibrium where the energy released by electron transfer balances that conserved by ADP phosphorylation.
Understanding the rate at which microbes respire in biological and geochemical systems is central to developing quantitative descriptions of a broad range of problems in microbiology, from the propagation of disease to the attenuation of contaminants in drinking-water supplies. Microbiologists have in recent years expended considerable effort in investigating microbial respiration rates under specific conditions (22, 33, 36, 41).
In virtually all cases, the results of such studies have been cast in terms of a semi-empirical rate law such as the Monod equation (15). Application of such rate laws is limited in two senses. First, because of their semi-empirical nature, they are best suited to interpolating the results of a set of experiments within the range of chemical conditions tested (7). Second, none of the laws accounts for the thermodynamic effects of the energy available in the cell's environment. As such, there is no basis for applying a rate law derived for a laboratory experiment, where the available energy is typically maintained at a high level to provide for microbial growth at acceptable rates, to natural conditions, where much less energy may be available. As a result, the rate laws invariably predict positive activities even where there is no energy available to drive the cell's metabolism forward.
In a recent paper (13), we performed, on the basis of the chemiosmotic model of cellular respiration (19) and nonlinear nonequilibrium thermodynamics (6), a rigorous analysis of the problem of respiration in eukaryotic cells by mitochondria. In our analysis, we accounted for a simultaneous forward and backward flux of electrons through the respiratory chain. The resulting rate law is the product of a rate constant, the biomass concentration, and three terms: one describing the kinetic effect of the electron-donating process, one for the electron-accepting process, and a thermodynamic term that accounts for the amount of energy available in the environment. We showed that this rate law correctly predicts respiration rates in mitochondria over a broad range of chemical conditions and available energy.
Respiring bacteria are similar to mitochondria in that they employ an electron transport chain to transduce energy from their environment (27). As such, we might expect that a rate law describing mitochondrial respiration would also apply to respiration by prokaryotic cells. In this paper, we demonstrate that this is indeed the case. By casting the rate law in general terms, we provide a basis for extrapolating experimental studies over a range of chemical conditions. Especially, since the rate law honors the requirement of consistency between chemical kinetics and thermodynamics (6), it allows respiration rates to be evaluated where only limited energy is available.
RATE LAW DERIVATION
Cellular respiration.
According to chemiosmotic theory (19), respiration is driven by a redox reaction. In its simplest form, this reaction
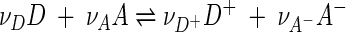 |
(1) |
represents the transfer of electrons from a donating species D to an accepting species A. Here D+ and A− are the oxidized and reduced forms of D and A, and νD, νA, etc., are reaction coefficients. We chose a simple form for reaction 1, but the theory presented here applies equally well to redox reactions involving other chemical species, such as H+, H2O, and so on, as described in the Appendix.
Reaction 1 is the combination of a half-cell reaction that donates electrons to the respiratory chain
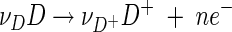 |
(2) |
and one that accepts electrons from the chain
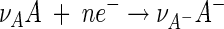 |
(3) |
(Fig. 1). Here n is the number of electrons transferred in reaction 1.
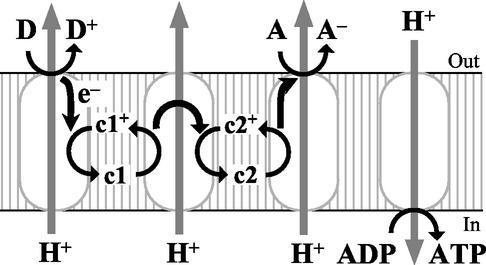
FIG. 1.
Generalized pathway of microbial respiration (13). Electrons derived from a donating species, D, are transferred through the respiratory chain containing three redox enzymes and two coenzymes, c1 and c2, to an accepting species, A. Energy liberated is conserved by translocating protons out of membrane, building up a proton motive force. The proton motive force is consumed by a proton-translocating ATP synthase to produce ATP from ADP. Reaction centers (ovals) are, from left to right, three redox enzymes and the ATP synthase.
The respiratory chain conserves a part of the energy, ΔG, released by reaction 1 by translocating protons out of the cell's membrane, generating a proton motive force. The proton motive force is then consumed in synthesizing ATP, the cell's energy currency, from ADP and phosphate, Pi,
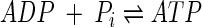 |
(4) |
Coupling reactions 1 and 4 gives the overall reaction
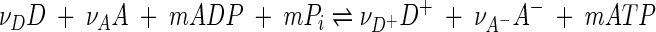 |
(5) |
representing microbial respiration. Here m is the number of ATP molecules synthesized per n electrons transferred.
Thermodynamic drive.
The net thermodynamic driving force, f, for microbial respiration is given as
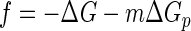 |
(6) |
from ΔG, the free energy change of the redox reaction (a negative change releases energy, which becomes available to microorganisms), and the phosphorylation potential, ΔGp (24), the free energy change of reaction 4. ΔGp is about 50 kJ mol−1 under typical physiological conditions (44).
The value of ΔG depends on ΔGo the standard Gibbs free energy change, as well as concentrations [D], [A], etc., of the chemical species involved, according to
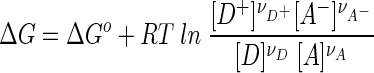 |
(7) |
Here R is the gas constant and T is the absolute temperature.
In general, reactions with negative free energy changes can proceed spontaneously. The negative signs in equation 6 arise because a negative free energy change of reaction reflects a positive thermodynamic drive. The term mΔGp in equation 6 represents the energy conserved into ATP by reaction 5. Since the value of ΔGp is positive, the energy utilized to phosphorylate ADP diminishes the driving force for the overall reaction (reaction 5).
Net respiration rate.
As is any complex chemical reaction, microbial respiration (reaction 5) is composed of a series of individual steps: substrate transport, electron transfer, proton translocation, ADP phosphorylation, and so on. Each step occurs in the reaction, sometimes more than once. The net respiration rate, ν (given as molar concentration per second), is the difference between the forward and reverse rates, ν+ and ν−, so that
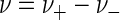 |
(8) |
The forward rate exceeds the reverse rate when the net driving force f is positive, and the overall reaction proceeds forward. When the net driving force is negative, conversely, the reverse rate dominates and the reaction proceeds backward.
The relationship between thermodynamic driving force and the overall reaction rate is explained by the theory of nonlinear nonequilibrium thermodynamics. A principal result of this field of study (6) shows that the ratio of the forward and reverse reaction rates varies with the driving force f according to
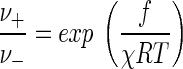 |
(9) |
In this equation, χ is a quantity known as the average stoichiometric number (34), which is defined as the ratio of the free energy change of the overall reaction (reaction 5) to the sum of the free energy changes for each elementary step. Depending on the reaction mechanism, an elementary step may occur more than once in the overall reaction. Where each of the steps occurs just once, the ratio and hence χ reduces to unity. For cellular respiration, however, the value of χ seldom takes a value of one. Instead, it can be taken to a good approximation as the number of times the rate-determining step occurs in the overall reaction.
Combining equations 8 and 9, the net respiration rate is
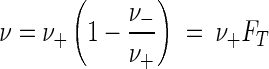 |
(10) |
where FT is the thermodynamic potential factor,
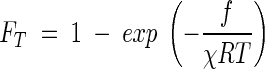 |
(11) |
This factor can range from 0 to 1 when there is a forward drive for respiration (f > 0) and from −∞ to 0 when the force drives respiration backward (f < 0).
The forward respiration rate, ν+, can vary up to the maximum rate, νmax (molar concentration per second), which occurs under optimum conditions, i.e., sufficient energy, high electron donor and acceptor concentrations, and low concentrations of the reaction products in the environment. This value is given by
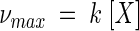 |
(12) |
where k is the intrinsic rate constant (moles per gram per second) and [X] is the biomass concentration (grams per liter). In the general case, however, the concentrations of species in solution affect the reaction kinetics, and the maximum rate is unlikely to be observed. In our previous study (13), we derived in a rigorous manner the relation between ν+ and νmax. We showed that this relation, given as
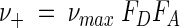 |
(13) |
depends on two unitless factors, FD and FA, which account for the kinetics of the electron-donating and -accepting half-reactions (reactions 2 and 3). The value of kinetic factor FD is given by
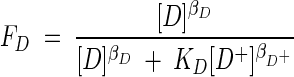 |
(14) |
and the value of FA is given by
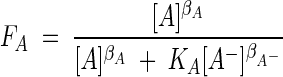 |
(15) |
Here, βD, βA, and so on are exponents whose values depend on details of the mechanism of electron transport and KD and KA are constants that reflect the standard free energy changes of the electron-donating and -accepting reactions (13).
The values of FD and FA can vary from near 0 to almost 1 depending on the concentrations of the reactant and product species appearing in the donating and accepting half-reactions. The values of the exponents (βD, βA, etc.) are not predicted by theory and so must be inferred from observation; as a first guess, however, they may be taken to be equal to unity, as we assume throughout the remainder of this paper.
Combining equations 10, 12, and 13, the overall rate of microbial respiration is described by the general rate law
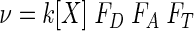 |
(16) |
The respiration rate, then, is the product of the maximum rate (since k[X] = νmax), the kinetic factors FD and FA, and the thermodynamic factor, FT. This equation clearly shows how kinetic and thermodynamic factors control microbial respiration.
Some of the parameters, such as m and χ, needed to evaluate the rate law can be determined or estimated from knowledge of a cell's physiology, as discussed in subsequent sections of this paper. Parameters such as k, KD, and KA, on the other hand, are most directly determined by fitting the rate law to experimental observations.
Biomass growth.
The energy conserved as ATP constitutes the cell's energy resource for maintaining and synthesizing biomass. Respiration, therefore, can lead to a change in biomass concentration [X], and this change may need to be accounted for in integrating the rate law (equation 16) over time.
Microbial growth may be limited by the rate at which energy is captured during respiration or by the availability of nutrients needed to synthesize biomass. Where energy conservation rather than nutrients is the factor limiting growth, the rate of biomass synthesis (grams per liter per second) is commonly taken to vary with the respiration rate, ν, according to the empirical equation (21)
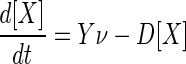 |
(17) |
where Y is the growth yield (grams of biomass produced per mole of reaction) and D is the decay constant (reciprocal seconds).
Energy of phosphorylation.
In the derivation above, we describe the energy conserved by respiration in terms of two variables: the free energy change of the phosphorylation reaction, ΔGp, and the number of ATP molecules synthesized in the overall reaction, m. We treat these values as constants, an appropriate assumption when respiration is at steady state. Indeed, many microorganisms, such as Escherichia coli and Wolinella succinogenes, are able to maintain a relatively constant proton motive force and phosphorylation potential, ΔGp, during respiration across large variations in the energy, ΔG, available from the environment (38, 39). It is important to keep in mind, nonetheless, that the value of ΔGp depends on the concentration ratio of ATP to ADP in the cytoplasm, as well as the concentration of free phosphate ion (24).
A microorganism in nature can regulate ATP production by its respiratory chain by changing the value of m to reflect physiological conditions (11). Indeed, the ability of microorganisms to adjust the amount of energy conserved during respiration to meet cellular energetic and physiological demands is well known (14, 26, 28). A microorganism can, for example, decrease the effective value of m by synthesizing alternative redox enzymes that translocate fewer protons than normal. By decreasing the value of m, an organism increases its respiration rate by increasing f and hence FT, as can be seen in equations 6 and 11.
Average stoichiometric number.
The value of χ in our model can be taken as the number of times the rate-determining step occurs per overall reaction. The rate-determining step in many common configurations of the respiratory chain is in our experience related to proton translocation across a redox enzyme. The value of χ therefore reflects the mechanism of electron transfer through this enzyme: it is the number of protons translocated by the enzyme in the overall reaction.
This value, in turn, depends on how the respiration reaction is written, i.e., the number of electrons transferred. It is therefore seldom appropriate to assume a priori that χ equals unity. The value of FT, on the other hand, does not depend on how the reaction is written, because both the thermodynamic driving force, f, in the numerator of equation 11 and χ in the denominator vary proportionately with the number of electrons transferred, n. In comparing different metabolisms, therefore, it is convenient to express χ as a value per electron transferred.
In the respiratory chain of Escherichia coli, most electrons passing from NADH to O2 are transmitted through NADH dehydrogenase II and quinol oxidase bo3 (40). Transfer across the oxidase is the rate-determining step. Two protons are translocated per electron transferred, giving the value of χ per electron transferred of 2. As a second example, electrons passing from NADH to O2 in Paracoccus denitrificans traverse NADH dehydrogenase I, the bc1 complex, and cytochrome c oxidase, translocating per electron transferred a total of five protons (43). Only proton translocation at cytochrome c oxidase, however, is rate determining (10). Two protons are translocated here per electron transferred, so the value of χ per electron transferred is, by chance, also 2.
There are at least two significant cases in which the value of χ does not reflect proton translocation. First, transport of an electron-donating or accepting species across the cell membrane may comprise the rate-determining step. Such transport, especially for large exobiotic molecules, can require the expenditure of considerable energy (25). Second, the passage of electrons to an extracellular electron acceptor, such as iron-bearing minerals, may become the rate-determining step due to the distance over which electrons must be transferred (20).
DISCUSSION
According to the rate law we developed (equation 16), kinetic as well as thermodynamic factors control the rate of microbial respiration. Where energy available in the environment is high compared to that conserved by ADP phosphorylation, the thermodynamic potential factor, FT, approximates unity and, for a given biomass concentration, the respiration rate is controlled solely by kinetic factors.
Kinetic control.
The oxidation of organic compounds by dioxygen (2, 30, 32) commonly provides sufficient energy that the thermodynamic term can be neglected. In such cases, the rate law becomes
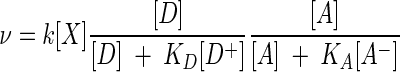 |
(18) |
where we have taken the various exponents β to be unity. This equation is closely related to rate laws in common use today. Where the species concentrations [D+] and [A−] can be taken to be constant, equation 18 reduces to the dual Monod equation (1):
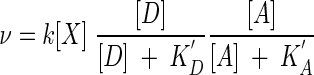 |
(19) |
where the constant terms KD[D+] and KA[A−] are replaced by the familiar half-saturation constants, denoted here as KD and KA. When [A] is similarly constant, the rate law takes the form
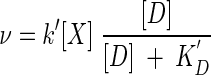 |
(20) |
which is the Monod equation. Here k′ is the product of k and the constant value of FA.
When [D+] and [A−] vary, on the other hand, we see from equation 18 that the accumulation of product species serves to decrease the reaction rate. As the concentrations of these species increase, so do the terms KD[D+] and KA [A−], decreasing the respiration rate when they control the denominator. Buildup of product species, it should be noted, can also inhibit respiration by decreasing the thermodynamic driving force (13). This effect is accounted for in equation 16 by the thermodynamic potential factor, FT, as discussed below (see “Arsenate reduction”).
Thermodynamic control.
In many natural environments, the depletion of the electron-donating or -accepting species or the buildup of reaction products has diminished the ability of the environment to supply energy to a cell. Notable examples of such environments are the interiors of microbial mats and the deep reaches of stratified water columns (18, 35).
Where the energy available in the environment is only slightly greater than that conserved by ADP phosphorylation (mΔGp), the net driving force, f, and hence FT approach zero. The thermodynamic factor becomes a dominant control on respiration rate and must be included in the rate law. When the respiration reaction (reaction 5) is at equilibrium, the energy available from the environment balances the energy conserved. In this case, f is zero and so is FT. Electrons proceed forward and backward through the transport chain at the same rate, giving a net respiration rate of zero. If the available energy drops below that needed to synthesize ATP (i.e., mΔGp), then f and hence FT become negative. The values of νmax, FD, and FA are invariably positive, so that the respiration rate, the product of these variables and FT, is negative.
In this case, ATP hydrolysis drives electrons backward through the respiratory chain, needlessly consuming the cell's energy stores, unless the microbe disables the chain. Only in special cases, such as producing reducing potential as NADH from NAD+, does a microbe allow electrons to run backward through its respiratory chain. To prevent respiration from reversing, microorganisms employ certain regulatory mechanisms, such as curtailing synthesis of critical redox enzymes, to disengage the respiratory chain as the overall reaction approaches equilibrium. Therefore, although it can theoretically decrease to −∞, microbial regulation ensures that under most circumstances, the value of FT ranges from 0 to 1.
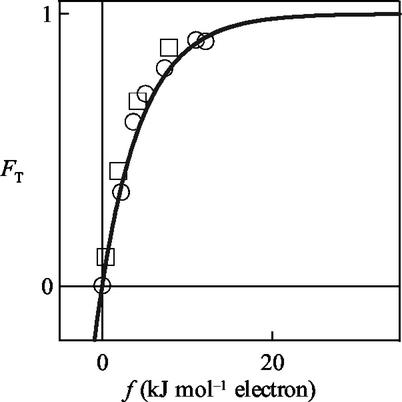
The relationship between the driving force, f, and the thermodynamic potential factor, FT, is well illustrated by the data in Fig. 2, reported by Rottenberg and Gutman (29, their Fig. 3). In their experiments on the mitochondrial respiratory chain, succinate donates electrons to the chain and NAD+ accepts electrons from the chain. About 4/3 moles of ATP is hydrolyzed per mole of succinate consumed. The amount of mitochondrial protein in the experiments and the concentrations of the chemical species except ADP and Pi were held constant, so that νmax, FD, and FA do not vary. The respiration rate, therefore, depends only on FT, which can be seen to approach unity when f is greater than about 20 kJ per mol of electrons. As predicted by our analysis, FT decreases toward zero as f becomes small.
FIG. 2.
Relationship between the thermodynamic driving force, f, and the thermodynamic potential factor, FT, for electron transfer through mitochondrial respiratory chain (13), as reported by Rottenberg and Gutman (29; their Fig. 3). Data points are FT, estimated as the ratio of the observed electron transfer rate, ν, relative to the observed or extrapolated highest rate (i.e., νmax × FD × FA). The thermodynamic driving force is estimated by equation 6, using reported concentrations of chemical species. The solid line is FT predicted by equation 11 at various thermodynamic driving forces, calculated using an average stoichiometric number, χ, per electron transferred of 2. Symbols: □ constant ΔG of −27.5 kJ per mol of electrons; ○, constant ΔG of −30.2 kJ per mol of electrons.
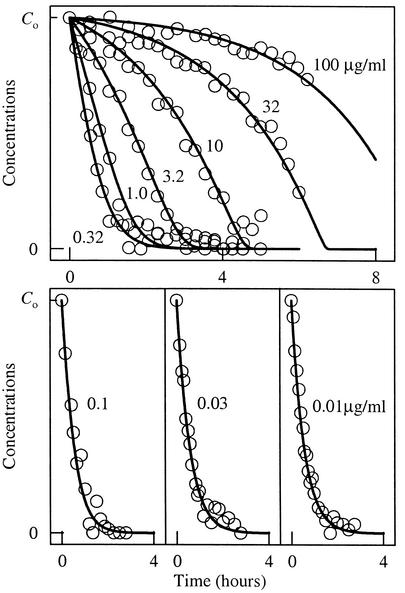
FIG. 3.
Comparison of benzoate degradation by Pseudomonas sp., as observed in experiments conducted by Simkins and Alexander (31), with predictions of the new rate law. The initial benzoate concentrations, C0, are 100, 32, 10, 3.2, 1.0, 0.32, 0.1, 0.03, and 0.01 μg ml−1, as labeled for each line. Benzoate concentration (lines) is predicted for each experiment by integrating equation 24, as described in the text.
Benzoate degradation.
To show how the new rate law can be applied where thermodynamic effects can be neglected, we consider the degradation of benzoate by a Pseudomonas strain, as reported in an experimental study by Simkins and Alexander (31). Benzoate (Ben−) oxidizes according to the reaction
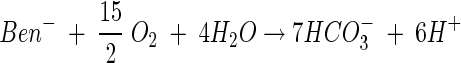 |
(21) |
To donate electrons to the respiratory chain, benzoate first degrades to intermediates of formate, acetaldehyde, and pyruvate, consuming two oxygen molecules per benzoate (9). Then, the 22 electrons derived from these intermediates are utilized for oxidative phosphorylation. About 5 protons are translocated per electron transferred (43), and 3 protons are consumed to synthesize each ATP (37), giving a total of about 37 ATP per benzoate.
We can show that in this case the thermodynamic effect is small. As discussed above, for aerobic respiration the value of χ per electron transferred is 2. In reaction 21, 22 electrons are transferred though the respiratory chain (n = 22), giving a χ of 44. Substituting these values into equation 11, FT is given by
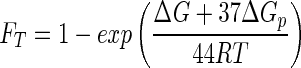 |
(22) |
The experiments were conducted in contact with the atmosphere, and the growth medium contained a pH buffer. We assume that pH in the experiments was buffered to 7 and that the O2 and HCO3− concentrations were 0.27 and 0.06 mM, respectively, corresponding to the partial pressures in the atmosphere of O2 and CO2. To calculate ΔG from equation 7, we note that the standard Gibbs free energy change of the reaction is −3.1 × 103 kJ mol−1. From equation 22, we can quickly calculate that the value of FT remains close to unity for any possible benzoate concentration (mathematically, until [Ben−] approaches 10−200 M).
The concentration of only benzoate varies in the experiments, so that FD can be written as
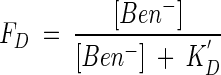 |
(23) |
and FA remains constant. Now, the rate law (equation 16) takes the form
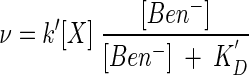 |
(24) |
of the Monod equation. This result (equation 24) predicts, using a single set of kinetic parameters, the results of the gamut of benzoate oxidation experiments undertaken by Simkins and Alexander (31), as shown in Fig. 3. We modeled these experiments by integrating the rate law (equation 24), using equation 17 to describe microbial growth. We set a growth yield, Y, of 60 g of biomass per mol of benzoate consumed, a decay constant D of zero, and the initial biomass concentrations reported in the study, which range from 1.8 × 10−3 to 3.0 × 10−3 g liter−1. We found best-fit values of 1.2 × 10−6 mol g−1 s−1 for k′ and 3.14 × 10−6 M for KD.
In the original study, the results of each experiment were fit separately to one of four different rate laws (the logistic, Monod, Michaelis-Menten, and first-order equations), depending on the pattern of the data, using a unique set of kinetic parameters, even though there is no reason to suspect that the mechanism by which benzoate oxidizes varied among the experiments. It is perhaps not surprising that the results of the entire set of experiments can be predicted using the Monod equation applied with a single set of parameters, although this has not been noted previously.
The result is significant, nonetheless, in that instead of empirically selecting an equation to fit the data, as is common practice, the law was reached in a rigorous manner by simplifying the general rate law to a specific form applicable to the experimental conditions of interest. In applying the experimental results to a field situation or differing conditions in the laboratory, we would have no guidance in selecting an empirical form for the rate law. However, the new rate law, because of its generality, affords the possibility of finding the correct rate law directly.
Arsenate Reduction.
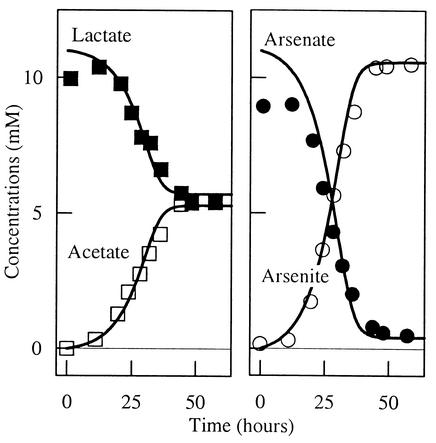
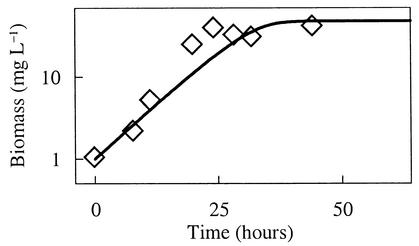
An experimental study of arsenate reduction by Bacillus arsenicoselenatis, reported by Blum et al. (see Fig. 4 of reference 5), provides an example in which thermodynamics controls the respiration rate. In their experiment, B. arsenicoselenatis conserves energy by oxidizing lactate (Lac2−) to acetate (Ac−) and bicarbonate, reducing arsenate to arsenite. The concentrations of acetate and arsenite increase at the expense of lactate and arsenate (Fig. 4), and the biomass concentration increases (Fig. 5) over the course of the experiment. The reaction, however, ceases before either the lactate or arsenate has been completely consumed.
FIG. 4.
Results of an experimental study of arsenate reduction by B. arsenicoselenatis, as reported by Blum et al. (5; their Fig. 4). The plot on the left shows concentrations of species in the electron-donating reaction, and the plot on the right shows those of species in the electron-accepting reaction. Lines are species concentrations predicted by integrating equation 31, using equation 17 to describe biomass growth, as described in the text.
FIG. 5.
Concentration of B. arsenicoselenatis biomass (symbols) in the experiment reported by Blum et al. (5; their Fig. 4). The line is biomass predicted by the rate law integration shown in Fig. 4.
In light of the rate law we propose (equation 16), it is worthwhile considering whether the reaction end point in the experiment represents a thermodynamic balance between the energy available in the environment, ΔG, and that needed to synthesize ATP, mΔGp. In other words, does the reaction in the experiment cease when the thermodynamic drive, f, approaches zero? To test this point, we need to figure values for ΔG, the free energy change of the reaction, and m, the number of ATP molecules synthesized per overall reaction.
The overall reaction
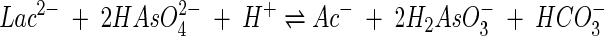 |
(25) |
transfers a total of four electrons per overall reaction; its standard Gibbs free energy change of reaction ΔGo is −140.3 kJ mol−1 (16). The available energy ΔG can be estimated from the concentrations at the end of the experiment of the chemical species in the reaction: arsenite and acetate reach 10.5 and 5.3 mM, respectively, and about 5.2 mM lactate and 0.5 mM arsenate remain to be metabolized. The pH of the growth medium was buffered to 9.8, and we take the bicarbonate concentration to be similar to that of acetate since the two chemical species are produced in stoichiometric proportion. Based on these values, ΔG at the experiment end point is about −130 kJ mol−1. If the thermodynamic drive is indeed in balance, taking ΔGp in equation 6 to be 50 kJ per mol of ATP suggests that the number of ATP molecules produced by the reaction, m, is about 2.6.
Is this value reasonable? We can independently estimate the value of m from a known pathway of lactate oxidation (42) and a pathway proposed recently by Newman et al. (23) for arsenate reduction. Oxidation of lactate to acetate and bicarbonate yields one ATP molecule per lactate through substrate level phosphorylation. The four electrons derived from this process are transferred through the respiratory chain and used to reduce arsenate. According to the speculative model of Newman et al. (23), a single proton is translocated across the cell membrane per electron transferred. Assuming that 3 protons are consumed per ATP synthesized, 1.3 ATP molecules are produced in this step. The total ATP yield, then, is 2.3 ATP molecules per overall reaction, which compares favorably, considering the uncertainties in the calculations, with the value of 2.6 previously determined as representing a thermodynamic balance.
To set up the rate law describing the experiment, we note that proton translocation occurs at a single point in the respiratory chain, once per electron transferred. This observation suggests that the value of χ is 1 per electron transferred, or 4 per reaction 25. Substituting our values for χ, m, and ΔGp into equation 11,
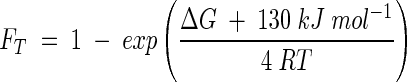 |
(26) |
gives the thermodynamic potential factor, FT.
From the form of the electron-donating half-reaction
 |
(27) |
and taking into account the pH buffering of the growth medium, the kinetic factor, FD (equation 14; see also Appendix), becomes
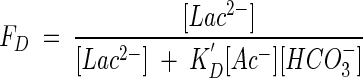 |
(28) |
From the electron accepting half-reaction
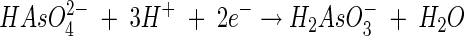 |
(29) |
we can similarly write the kinetic factor, FA (equation 15), in the form
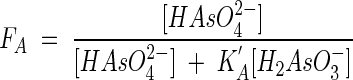 |
(30) |
Substituting these results into equation 16 gives
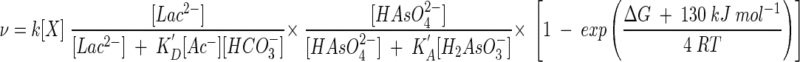 |
(31) |
the rate law applicable to this experiment.
To model biosynthesis over the course of the experiment, as described by equation 17, we can estimate the growth yield, Y, from the net growth in biomass per mole of lactate consumed; this value is about 8.3 g of biomass per mol of lactate. Since biomass did not decrease at any point in the experiment, we neglect decay; i.e., we take the decay constant, D, to be zero.
As such, only three parameters need be estimated to fit the rate law (equation 31) to the results: the rate constant k, which controls the maximum rate, and the constants KD and KA in the kinetic factors, which affect the shape of the curve. We evaluated the rate law using the computer program React (3, 4), taking the following initial concentrations: lactate, 11 mM; acetate, 0 mM; bicarbonate, 0.1 mM; arsenate, 11 mM; and arsenite, 0 mM. Adjusting the program input to reproduce the experimental results, we found best-fit values for k of 4.4 × 10−6 mol s−1 g of biomass−1 and for KD and KA of 10 M−1 and 0.1, respectively. Figures 4 through 6 show the results of this calculation. As can be seen, our rate law predicts variation in the concentrations of the chemical species as well as biomass over the course of the experiment.
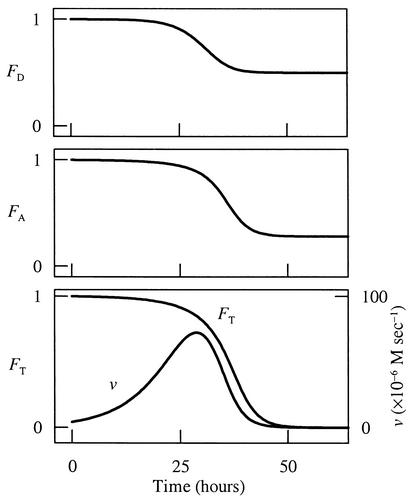
FIG. 6.
Kinetic factors, FD and FA, the thermodynamic potential factor, FT, and the resulting reaction rate, ν, in the rate law integration shown in Fig. 4 and 5. FD and FA are calculated from equations 28 and 30, and FT is calculated from equation 26; ν is the product of the rate constant, k, biomass, [X] (Fig. 5), and the values of FD, FA, and FT.
Having fit the experimental results, we can now interpret them in terms of the factors FD, FA, and FT, as shown in Fig. 6. Initially, these factors are all approximately unity because the reactant species (lactate and arsenate) are present at high concentrations and there is little buildup of the product species (acetate, bicarbonate, and arsenite). The initial rate, therefore, depends only on the product of the rate constant and the biomass concentration. With time, biomass in the experiment increases, allowing the reaction to proceed with increasing rapidity. About 25 h into the experiment, the concentrations of the reactant species have decreased in concentration and the product species have accumulated significantly. Both kinetic and thermodynamic factors, as a result, decrease sharply. The reaction rate reaches a maximum at about 29 h. After this point, the rate begins to fall as the increase in biomass concentration cannot keep pace with the decreasing values of FD, FA, and FT. Continued reaction causes the thermodynamic drive to fall toward zero. About 50 h into the experiment, the thermodynamic drive has essentially disappeared, causing the reaction to nearly cease, even though significant amounts of both lactate and arsenate remain to be metabolized.
Threshold phenomena.
These results bear on the interpretation of threshold phenomena, which are commonly observed in experimental studies of microbial metabolism (8). A threshold is the concentration of an electron donor or acceptor below which the metabolism is observed to not operate. For example, threshold concentrations have been observed for the respiration of dihydrogen by using various electron-accepting processes. The threshold partial pressure of H2 ranges from 10−6 to 10−4 atm when CO2 serves as the electron acceptor (i.e., during methanogenesis) and from 10−6 to 10−5 atm when sulfate is the electron acceptor (17).
Empirical rate laws such as the Monod equation predict that a microbe will continue to metabolize until the concentration of its substrate vanishes. Our rate law, on the other hand, differs from the empirical laws in that it predicts that respiration will cease, even in the presence of finite amounts of substrate, in the absence of a sufficient thermodynamic drive for the cell to conserve energy. In other words, our law gives a thermodynamic explanation for threshold phenomena.
Previous studies (12) have suggested that threshold phenomena represent the minimum substrate concentration or lowest Gibbs free energy needed to activate a metabolic pathway. This is certainly possible, but our study suggests an alternative explanation: the phenomena reflect a microbe's regulation of its respiratory chain under conditions where the energy available in the environment balances that conserved by ADP phosphorylation. As a substrate decreases in concentration, it reaches a point at which the thermodynamic drive approaches zero, i.e., where equilibrium is maintained across the respiratory chain. The cell's regulatory mechanism responds by allowing or causing the activities of its redox enzymes to fall, preventing backward electron transfer from needlessly depleting its ATP stores.
Acknowledgments
We thank Robert Sanford, James Imlay, and Tom Johnson for their interest and generous advice.
This work was supported by Department of Energy grant DE-FG02-02ER15317 and by the generosity of Chevron, ExxonMobil Upstream Research, Idaho National Engineering and Environmental Laboratory, Lawrence Livermore, Sandia, SCK-CEN, Texaco, and the U.S. Geological Survey.
APPENDIX
The discussion in the text of this paper considers that microbial respiration can be represented by a simple electron transfer reaction (reaction 5), although the theory presented, in fact, applies to more complicated reactions (13). We can represent microbial respiration more generally as the reaction
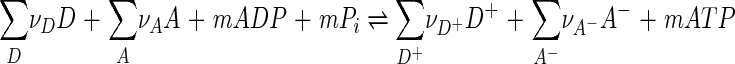 |
(A1) |
In this case, then kinetic factors, FD and FA, are given by
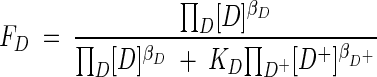 |
(A2) |
and
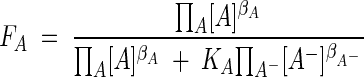 |
(A3) |
and the free energy released by the redox reaction, ΔG, can be calculated by
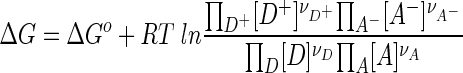 |
(A4) |
Here, the symbols ∑and ∏are summation and multiplicative product operators, i.e., 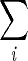 Ai= A1 + A2 + A3 +…, and
Ai= A1 + A2 + A3 +…, and 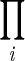 Ai= A1 × A2 × A3 ×… . In evaluating the rate law (equation 16), we use equations A2 and A3 to compute FD and FA and we calculate FT (equation 11) using equation A4.
Ai= A1 × A2 × A3 ×… . In evaluating the rate law (equation 16), we use equations A2 and A3 to compute FD and FA and we calculate FT (equation 11) using equation A4.
REFERENCES
- 1.Bae, W., and B. E. Rittmann. 1996. A stuctured model of dual-limitation kinetics. Biotechnol. Bioeng. 49:683-689. [DOI] [PubMed] [Google Scholar]
- 2.Bekins, B. A., E. Warren, and E. M. Godsy. 1998. A comparision of zero-order, first-order, and Monod biotransformation models. Ground Water 36:261-268. [Google Scholar]
- 3.Bethke, C. M. 1996. Geochemical reaction modeling. Oxford University Press, New York, N.Y.
- 4.Bethke, C. M. 2002. The geochemist's workbench, release 4.0. University of Illinois, Urbana.
- 5.Blum, J. S., A. B. Bindi, J. Buzzelli, J. F. Stolz, and R. S. Oremland. 1998. Bacillus arsenicoselenatis, sp. nov., and Bacillus selenitireducens, sp. nov.: two haloalkaliphiles from Mono Lake, California, that respire oxyanions of selenium and arsenic. Arch. Microbiol. 171:19-30. [DOI] [PubMed] [Google Scholar]
- 6.Boudart, M. 1976. Consistency between kinetics and thermodynamics. J. Phys. Chem. 80:2869-2870. [Google Scholar]
- 7.Characklis, W. G. 1983. Process analysis in microbial systems: biofilms as a case study, p. 171-234. In M. Bazin (ed.), Mathematics in microbiology. Academic Press, Inc., New York, N.Y.
- 8.Conrad, R. 1996. Soil microorganisms as controllers of atmospheric trace gases (H2, CO, CH4, OCS, N2O, and NO). Microbiol. Rev. 60:609-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellis, L. B. M., C. D. Hershberger, E. M. Bryan, and L. P. Wackett. 2001. The University of Minnesota Biocatalysis/Biodegradation Database: emphasizing enzymes. Nucleic Acids Res. 29:340-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erecinska, M., and D. F. Wilson. 1982. Regulation of cellular energy metabolism. J. Membr. Biol. 70:1-14. [DOI] [PubMed] [Google Scholar]
- 11.Frank, V., and B. Kadenbach. 1996. Regulation of the H+/e− stoichiometry of cytochrome c oxidase from bovine heart by intramitochondrial ATP/ADP ratios. FEBS Lett. 382:121-124. [DOI] [PubMed] [Google Scholar]
- 12.Jackson, B. E., and M. J. McInerney. 2002. Anaerobic microbial metabolism can proceed close to thermodynamic limits. Nature 415:454-456. [DOI] [PubMed] [Google Scholar]
- 13.Jin, Q., and C. M. Bethke. 2002. Kinetics of electron transfer through the respiratory chain. Biophys. J. 83:1797-1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joseph-Horne, T., D. W. Hollomon, and P. M. Wood. 2001. Fungal respiration: a fusion of standard and alternative components. Biochimi. Biophys. Acta Bioenergetics 1504:179-195. [DOI] [PubMed] [Google Scholar]
- 15.Koch, A. L. 1998. The Monod model and its alternatives, p. 62-93. In A. L. Koch, J. A. Robinson, and G. A. Milliken (ed.), Mathematical modeling in microbial ecology. Chapman & Hall, New York, N.Y.
- 16.Laverman, A., J. Blum, J. Schaefer, E. Phillips, D. Lovley, and R. Oremland. 1995. Growth of strain SES-3 with arsenate and other diverse electron acceptors. Appl. Environ. Microbiol. 61:3556-3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loffler, F. E., J. M. Tiedje, and R. A. Sanford. 1999. Fraction of electrons consumed in electron acceptor reduction and hydrogen thresholds as indicators of halorespiratory physiology. Appl. Environ. Microbiol. 65:4049-4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Madrid, V. M., G. T. Taylor, M. I. Scranton, and A. Y. Chistoserdov. 2001. Phylogenetic diversity of bacterial and archaeal communities in the anoxic zone of the Cariaco Basin. Appl. Environ. Microbiol. 67:1663-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitchell, P. 1961. Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature 191:144-148. [DOI] [PubMed] [Google Scholar]
- 20.Moser, C. C., J. M. Keske, K. Warncke, R. S. Farid, and P. L. Dutton. 1992. Nature of biological electron transfer. Nature 355:796-802. [DOI] [PubMed] [Google Scholar]
- 21.Neidhardt, F. C., J. L. Ingraham, and M. Schaechter. 1990. Physiology of the bacterial cell: a molecular approach. Sinauer Associates, Inc., Sunderland, Mass.
- 22.Neubauer, S. C., D. Emerson, and J. P. Megonigal. 2002. Life at the energetic edge: kinetics of circumneutral iron oxidation by lithotrophic iron-oxidizing bacteria isolated from the wetland-plant rhizosphere. Appl. Environ. Microbiol. 68:3988-3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newman, D. K., D. Ahmann, and F. M. Morel. 1998. A brief review of microbial arsenate respiration. Geomicrobiol. J. 15:255-268. [Google Scholar]
- 24.Otto, R., B. Klont, and W. N. Konings. 1985. The relation between phosphate potential and growth rate of Streptococcus cremoris. Arch. Microbiol. 142:97-100. [Google Scholar]
- 25.Pao, S. S., I. T. Paulsen, and M. H. Saier, Jr. 1998. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 62:1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pfeiffer, T., S. Schuster, and S. Bonhoeffer. 2001. Cooperation and competition in the evolution of ATP-producing pathways. Science 292:504-507. [DOI] [PubMed] [Google Scholar]
- 27.Richardson, D. J. 2000. Bacterial respiration: a flexible process for a changing environment. Microbiology 146:551-571. [DOI] [PubMed] [Google Scholar]
- 28.Riondet, C., R. Cachon, Y. Waché, G. Alcaraz, and C. Diviès. 1999. Changes in the proton-motive force in Escherichia coli in response to external oxidoreduction potential. Eur. J. Biochem. 262:595-599. [DOI] [PubMed] [Google Scholar]
- 29.Rottenberg, H., and M. Gutman. 1977. Control of the rate of reverse electron transport in submitochondrial particles by the free energy. Biochemistry 16:3220-3227. [DOI] [PubMed] [Google Scholar]
- 30.Schmidt, S. K., S. Simkins, and M. Alexander. 1985. Models for the kinetics of biodegradation of organic compounds not supporting. Appl. Environ. Microbiol. 50:323-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simkins, S., and M. Alexander. 1984. Models for mineralization kinetics with the variables of substrate concentration and population density. Appl. Environ. Microbiol. 47:1299-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simkins, S., and M. Alexander. 1985. Nonlinear estimation of the parameters of Monod kinetics that best describe mineralization of several substrate concentrations by dissimilar bacterial densities. Appl. Environ. Microbiol. 50:816-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spear, J. R., L. A. Figueroa, and B. D. Honeyman. 2000. Modeling reduction of uranium U(VI) under variable sulfate concentrations by sulfate-reducing bacteria. Appl. Environ. Microbiol. 66:3711-3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Temkin, M. I. 1963. Kinetics of stationary reactions. Dokl. Akad. Nauk SSSR 152:156-159. [Google Scholar]
- 35.Teske, A., N. B. Ramsing, K. Habicht, M. Fukui, J. Kuver, B. B. Jorgensen, and Y. Cohen. 1998. Sulfate-reducing bacteria and their activities in cyanobacterial mats of Solar Lake (Sinai, Egypt). Appl. Environ. Microbiol. 64:2943-2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tholosan, O., F. Lamy, J. Garcin, T. Polychronaki, and A. Bianchi. 1999. Biphasic extracellular proteolytic enzyme activity in benthic water and sediment in the northwestern Mediterranean Sea. Appl. Environ. Microbiol. 65:1619-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tomashek, J. J., and W. S. A. Brusilow. 2000. Stoichiometry of energy coupling by proton-translocating ATPase: a history of variability. J. Bioenerg. Biomembr. 32:493-500. [DOI] [PubMed] [Google Scholar]
- 38.Tran, Q. H., and G. Unden. 1998. Changes in the proton potential and the cellular energetics of Escherichia coli during growth by aerobic and anaerobic respiration or by fermentation. Eur. J. Biochem. 251:538-543. [DOI] [PubMed] [Google Scholar]
- 39.Unden, G. 1998. Transcriptional regulation and energetics of alternative respiratory pathways in facultatively anaerobic bacteria. Biochim. Biophys. Acta 1365:220-224. [Google Scholar]
- 40.Unden, G., and J. Bongaerts. 1997. Alternative respiratory pathways of Escherichia coli: energetics and transcriptional regulation in response to electron acceptors. Biochim. Biophys. Acta 1320:217-234. [DOI] [PubMed] [Google Scholar]
- 41.van Bodegom, P., F. Stams, L. Mollema, S. Boeke, and P. Leffelaar. 2001. Methane oxidation and the competition for oxygen in the rice rhizosphere. Appl. Environ. Microbiol. 67:3586-3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van de Pas, B. A., S. Jansen, C. Dijkema, G. Schraa, W. M. de Vos, and A. J. M. Stams. 2001. Energy yield of respiration on chloroaromatic compounds in Desulfitobacterium dehalogenans. Appl. Environ. Microbiol. 67:3958-3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Spanning, R. J., A. P. de Boer, W. N. Reijnders, J. W. De Gier, C. O. Delorme, A. H. Stouthamer, H. V. Westerhoff, N. Harms, and J. van der Oost. 1995. Regulation of oxidative phosphorylation: the flexible respiratory network of Paracoccus denitrificans. J. Bioenerg. Biomembr. 27:499-512. [DOI] [PubMed] [Google Scholar]
- 44.White, D. 1995. The physiology and biochemistry of prokaryotes. Oxford University Press, New York, N.Y.