Abstract
Acetic acid bacteria, especially Gluconobacter species, have been known to catalyze the extensive oxidation of sugar alcohols (polyols) such as d-mannitol, glycerol, d-sorbitol, and so on. Gluconobacter species also oxidize sugars and sugar acids and uniquely accumulate two different keto-d-gluconates, 2-keto-d-gluconate and 5-keto-d-gluconate, in the culture medium by the oxidation of d-gluconate. However, there are still many controversies regarding their enzyme systems, especially on d-sorbitol and also d-gluconate oxidations. Recently, pyrroloquinoline quinone-dependent quinoprotein d-arabitol dehydrogenase and d-sorbitol dehydrogenase have been purified from G. suboxydans, both of which have similar and broad substrate specificity towards several different polyols. In this study, both quinoproteins were shown to be identical based on their immuno-cross-reactivity and also on gene disruption and were suggested to be the same as the previously isolated glycerol dehydrogenase (EC 1.1.99.22). Thus, glycerol dehydrogenase is the major polyol dehydrogenase involved in the oxidation of almost all sugar alcohols in Gluconobacter sp. In addition, the so-called quinoprotein glycerol dehydrogenase was also uniquely shown to oxidize d-gluconate, which was completely different from flavoprotein d-gluconate dehydrogenase (EC 1.1.99.3), which is involved in the production of 2-keto-d-gluconate. The gene disruption experiment and the reconstitution system of the purified enzyme in this study clearly showed that the production of 5-keto-d-gluconate in G. suboxydans is solely dependent on the quinoprotein glycerol dehydrogenase.
Acetic acid bacteria are obligate aerobes well known as vinegar producers and also known to be able to oxidize various sugars and sugar alcohols such as d-glucose, glycerol, d-sorbitol, and so on, in addition to ethanol. Such oxidation reactions are called oxidative fermentation, since they involve incomplete oxidations of such alcohols or sugars accompanied by an accumulation of the corresponding oxidation products in large amounts in the culture medium. Of the two genera of acetic acid bacteria, Gluconobacter species extensively catalyze the oxidation of sugars and sugar alcohols except for ethanol, while Acetobacter species have a high ability to oxidize ethanol to acetic acid. These oxidation reactions of sugars or sugar alcohols seem to be carried out by membrane-bound dehydrogenases linked to the respiratory chain located in the cytoplasmic membrane of the organism (14). Of these oxidative fermentations of acetic acid bacteria, vinegar production from ethanol and 2-keto-d-gluconate (2KGA) production from glucose have each been shown to be carried out by sequential membrane-bound alcohol and aldehyde dehydrogenases and by glucose and gluconate dehydrogenases, respectively (14).
There is still controversy about the mechanism of l-sorbose and 5-keto-d-gluconate (5KGA) production in Gluconobacter species. Three different membrane-bound enzymes have been proposed to be involved in l-sorbose production from d-sorbitol. The first one is a flavoprotein sorbitol dehydrogenase consisting of three subunits, including a cytochrome c subunit which was purified from G. suboxydans IFO 3254 (17); the second enzyme is a quinoprotein sorbitol dehydrogenase purified from G. oxydans ATCC 621, which also consists of three subunits and has a cytochrome c subunit but contains pyrroloquinoline quinone (PQQ) as the prosthetic group instead of flavin adenine dinucleotide in the flavoprotein (7); and the last d-sorbitol dehydrogenase (SLDH) has been recently purified from G. suboxydans IFO 3255, which is a single protein quinoprotein of ∼80 kDa (23). More recently, a gene of the last of these enzymes has been cloned and sequenced (15), and gene disruption led to a defect in l-sorbose production in G. suboxydans IFO 3255 (22), suggesting that the enzyme is extensively involved in l-sorbose production from d-sorbitol in the strain.
Gluconobacter species are unique in accumulating two types of keto-d-gluconate, 5KGA and 2KGA, in the culture medium (18). 2KGA has been shown to be produced by membrane-bound flavoprotein d-gluconate dehydrogenase, as described above, not only in acetic acid bacteria (19) but also in Pseudomonas species and Klebsiella species (13). The enzyme involved in 5KGA production, however, remained unidentified for a long time. Klasen et al. have purified d-gluconate:NADP 5-oxidoreductase (called 5-keto-d-gluconate reductase, 5KGR) from Gluconobacter oxydans and sequenced the gene, and tried to express 5KGR in G. oxydans for 5KGA production (10). However, recently, Shinagawa et al. (20) have shown that the membrane suspension from G. suboxydans can produce 5KGA as well as 2KGA under acidic conditions in the absence of NADP, and also that exogenous addition of PQQ and CaCl2 stimulate 5KGA formation, thus suggesting that the accumulation of 5KGA involves a membrane-bound quinoprotein. However, such a quinoprotein has never been isolated, and thus the enzyme involved in 5-ketogluconic acid production remains to be solved.
Recently, membrane-bound quinoprotein d-arabitol dehydrogenase (ARDH) purified from G. suboxydans IFO 3257 showed the oxidation activity of d-gluconate in addition to sugar alcohols such as d-arabitol, d-mannitol, d-sorbitol, glycerol and so on (2). Furthermore, a single protein PQQ-dependent SLDH purified from G. suboxydans IFO 3255 (23) described above has also been shown to have a substrate specificity similar to that of ARDH and to have some d-gluconate oxidation activity.
Thus, in this study, the relationship between both quinoproteins, ARDH and SLDH, was examined at first, and then we examined whether these enzymes are able to produce 5KGA or 2KGA. It was clearly shown that both enzymes are identical quinoproteins able to catalyze d-gluconate oxidation to 5KGA.
MATERIALS AND METHODS
Microorganisms and cultivation.
G. suboxydans IFO 3257 and G. suboxydans IFO 3255 were supplied by the Institute for Fermentation, Osaka (IFO), Japan, and used throughout this work. Gluconobacter strains and the SLDH disruptants were grown in either glycerol, arabitol, or glucose-gluconate medium. Glycerol medium consisted of 10 g of glycerol, 0.5 g of d-arabitol, 3 g of yeast extract, and 3 g of Polypeptone in 1 liter of tap water; arabitol medium consisted of 1% d-arabitol, 3 g of yeast extract, and 3 g of Polypeptone in 1 liter of tap water; and glucose-gluconate medium consisted of 10 g of d-glucose, 10 g of sodium d-gluconate, 2 g of yeast extract, and 2 g of Polypeptone in 1 liter of tap water. For the preculture of the Gluconobacter strains, potato medium was used (11). Cultivation was carried out aerobically with rotary shaking at 30°C. Escherichia coli transformant was cultivated at 37°C in Luria-Bertani (LB) medium containing kanamycin (50 μg/ml).
Construction of SLDH-defective mutant from G. suboxydans IFO 3257.
SLDH-disrupted mutant strain of G. suboxydans IFO 3255 was prepared in the previous study (22), and that of G. suboxydans IFO 3257 was also prepared by transconjugation with pSUP202 sldA::Km, essentially by the same method as described in the previous study (22). E. coli HB101 harboring pSUP202 sldA::Km and E. coli HB101 harboring pRK2013 grown on LB medium containing kanamycin (50 μg/ml) at 30°C were mixed with G. suboxydans IFO 3257 grown on potato medium at 30°C when the growth of these strains reached the mid-exponential phase. The cell suspension was precipitated by centrifugation and washed with fresh potato medium, and the precipitate was suspended with a small volume of potato medium and spotted on a potato plate. After being incubated for 4 h at 30°C, the cells were removed from the agar plate, suspended in a small volume of potato medium, and spread on a potato agar plate containing kanamycin (50 μg/ml) and nalidixic acid (25 μg/ml), which prevented the growth of E. coli (9).
Colonies on the plate were isolated, and the gene disruption was confirmed by PCR. After isolation of chromosomal DNAs from the mutant strain, the insertion of a Km cassette to the SLDH-encoded gene was checked by agarose gel electrophoresis after PCR (data not shown). The primers ACGCGATACTCTCCGCTTTC and CGGTACGACGGTCAACACG were used for the PCR as 5′- and 3′-primers, respectively.
Preparation of resting cells.
G. suboxydans wild-type and mutant strains were cultivated for 24 h (stationary phase) in glycerol medium at 30°C. The cells were harvested by centrifugation at 9,000 × g and washed twice with 10 mM potassium phosphate buffer (KPB), pH 6.0. The cells were suspended with the same buffer and the cell suspension was used as the resting cell solution.
Preparation of membrane fraction.
The cells were harvested by centrifugation at 10,000 × g for 10 min and washed with distilled water. The cell paste was suspended in 10 mM KPB, pH 6.0, and passed through a French pressure cell at 16,000 lb/in2. After centrifugation to remove intact cells, the supernatant was centrifuged at 100,000 × g for 60 min. The resultant precipitate was resuspended in the same buffer and used as the membrane fraction.
Enzyme assay.
Enzyme activity was measured spectrophotometrically by the reduction of 2,6-dichlorophenol indophenol (DCIP) at 600 nm coupling with phenazine methosulfate (PMS) at 25°C. The reaction mixture contained enzyme solution, buffer (McIlvaine buffer [McB] or 0.2 M glycine-NaOH buffer), substrate, 0.2 mM PMS, 0.11 mM DCIP, and 3.0 mM sodium azide in a total volume of 1 ml. One unit of enzyme activity was defined as the amount of enzyme catalyzing the oxidation of 1 μmol of substrate per min, which was calculated using a millimolar extinction coefficient of DCIP, 3.43 at pH 5.0 and 15.6 at pH 9.0.
Detection of ketoses and keto-d-gluconates.
2KGA and 5KGA were measured enzymatically with 2KGA reductase (4) and 5KGA reductase (3), respectively, by a routine method used for common NAD(P)H-linked enzymes where the enzyme activities were measured by monitoring the rate of decrease of NADPH at 340 nm at 25°C in 100 mM KPB (pH 6.5). d-Xylulose and l-sorbose contents were measured with a resorcinol-dependent color reaction. The reaction was performed by mixing 0.2 ml of the sample solution, culture medium, or reaction mixture with 1 ml of a resorcinol reagent and incubating at 80°C for 20 min. The resorcinol reagent consisted of 3.5 ml of 0.5% resorcinol solution, 12 ml of concentrated HCl, and 19.5 ml of distilled water. After the reaction mixture had been cooled in an ice bath, the color intensity was measured at 530 nm (ketohexose) and 600 nm (ketopentose). These ketose contents were calculated from a calibration curve prepared at the same time with an authentic chemical.
Determination of the reaction product with the purified ARDH.
The reaction mixture consisted of McB (pH 5.0), 100 mM d-gluconate, 30 μM ubiquinone-2 (Q2), 0.025% Tween 20, 10 U of ubiquinol oxidase (cytochrome bo oxidase) purified from Acetobacter aceti (12), and 5 U of the purified ARDH (2). The reaction was performed at 25°C overnight and then terminated by heating at 90°C for 1 min. The reaction product was identified by enzymatic measurement with 2KGA reductase and 5KGA reductase as described above.
DNA techniques.
Chromosomal DNAs of G. suboxydans IFO 3257 and of the SLDH disruptants were isolated from cells grown to a late-log phase in a potato medium. PCR was performed using a Ready To Go/PCR beads kit (Pharmacia) using about 0.1 μg of a template chromosomal DNA in the mixture in a final volume of 25 μl. The reaction was subjected to 30 cycles of amplification in a GeneAmp PCR System 2400 (Perkin-Elmer) under conditions including denaturation for 1 min at 95°C, annealing for 1 min at 58°C, and extension for 2 min at 72°C.
Immunoblot analysis.
After sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed, proteins in the gel were transferred electrophoretically onto a polyvinylidene fluoride (PVDF) membrane (Millipore) at 100 mA for 4 h. After blocking with 3% gelatin and washing with 20 mM Tris-HCl buffer (pH 7.5) containing 500 mM NaCl (TBS), the membrane was incubated with anti-SLDH and also with anti-ARDH antibodies in TBS containing 1% gelatin for 2 h. Then, the membrane was washed again and incubated with TBS containing protein A-peroxidase for 2 h. After washing the membrane with TBS containing 0.05% Tween 20, the enzyme band was visualized by the addition of color reagents and H2O2. Prestained marker proteins (Bio-Rad) were used to estimate the relative molecular weight. SLDH-specific antibody was obtained against SLDH purified from G. suboxydans IFO 3255 in the previous study (4), while ARDH-specific antibody was prepared with 1.5 mg of ARDH purified from G. suboxydans IFO 3257 (2) in this study.
Determination of N-terminal amino acid sequence.
Purified ARDH was subjected to SDS-PAGE. Then, proteins in the gel were electrophoretically blotted onto a PVDF membrane at 100 mA for 4 h in 10 mM CAPS (N-cyclohexyl-3-aminopropanesulfonic acid; pH 11.0) containing 10% methanol. The transferred membrane was stained with Coomassie brilliant blue R-250 for 1 min and destained with 50% methanol. A single band was cut out, and N-terminal amino acid sequencing was performed by automated Edman degradation on a peptide sequence analyzer (Shimadzu).
Determination of protein.
Protein concentration was determined according to the modified Lowry method with bovine serum albumin as the standard (8).
RESULTS
Relation between ARDH and SLDH.
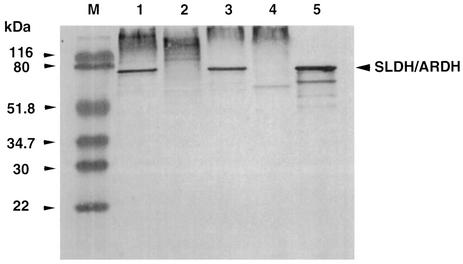
As described in the introduction, ARDH purified from G. suboxydans IFO 3257 has a similar substrate specificity to SLDH purified from G. suboxydans IFO 3255. However, their subunit structures were different from each other; ARDH has two peptides of 82 and 14 kDa (2), while SLDH exhibited a single peptide of 80 kDa (23). In order to confirm the relationship between both enzymes, immunoblotting analysis of ARDH was performed with antibody prepared against SLDH purified from G. suboxydans IFO 3255 (Fig. 1). As shown, there was a single cross-reactive band with anti-SLDH in the membranes of strain IFO 3257 as well as of strain IFO 3255 and, furthermore, the 82-kDa peptide of purified ARDH was cross-reacted with anti-SLDH. Although data are not shown, the same immunoblotting pattern was obtained with the membranes of both strains and with the purified ARDH, even when anti-ARDH was used instead of anti-SLDH.
FIG. 1.
Immunoblotting analysis of the membrane fractions of G. suboxydans IFO 3257 and 3255 strains and their SLDH disruptants and of the purified ARDH with anti-SLDH. Protein bands in the gel of SDS-PAGE were transferred electrophoretically onto PVDF membrane, and the enzyme band was visualized as described in Materials and Methods. Lane M: prestained marker proteins; lanes 1 to 4: membrane fractions (2 μg of protein each) of G. suboxydans strain 3255, 3255sldA::Km, 3275, and 3275sldA::Km, respectively; lane 5: ARDH (0.5 μg of protein) purified from G. suboxydans IFO 3257.
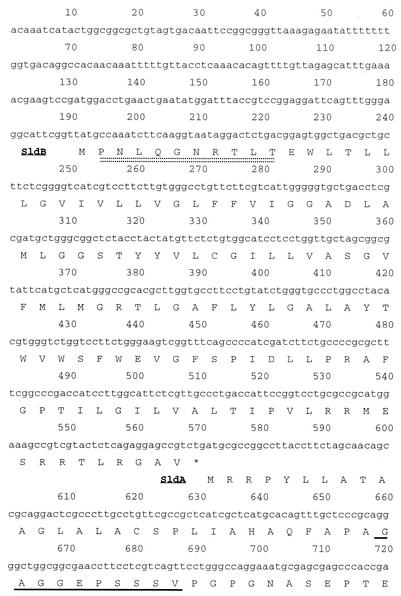
Furthermore, the N-terminal amino acid sequences in the 82- and 14-kDa peptides of ARDH were analyzed to confirm the relationship between both enzymes. The N-terminal amino acid sequences were shown to be GAGGEPSSSV in the large subunit, while in the small subunit fraction two amino acid residues were found in each cycle from the 1st to 10th cycles, of which the residues were E or P, T or N, I or L, H or Q, L or G, F or N, G or R, P or T, F or L, and K or T, respectively. Since the gene(s) encoding SLDH of G. suboxydans IFO 3255 has been cloned and sequenced (5), these sequences of the large and small subunits were compared with the deduced amino acid sequence of the SLDH gene(s). The sequence of GAGGEPSSSV was shown to correspond to that after the 29 amino acid residues in the N-terminal of SldA, the structural gene product of SLDH (Fig. 2). Furthermore, the N-terminal sequence of SldB, which is the product of a gene present just upstream of sldA, was shown to be identical to one of the two sets of amino acid sequences, PNLQGNRTLT (Fig. 2). The rest of the sequence, ETIHLFGPFK, found in the 14-kDa band fraction seemed to be contaminated in the purified enzyme, and the sequence was shown to have no significant homology to any other peptides in a database. Thus, the immunoblotting result and the N-terminal amino acid sequences of ARDH suggested that ARDH consists of two peptides corresponding to SldA and SldB of SLDH.
FIG. 2.
Part of the amino acid sequence alignment of SldA and SldB and the N-terminal sequence of the purified ARDH large and small subunits. The N-terminal sequence of ARDH large subunit (underlined with a solid underline) was identical to residues 30 to 39 of the N-terminal amino acid residues deduced from sldA, while the N-terminal sequence of ARDH small subunit (underlined with a double dotted line) was identical to residues 2 to 11 in the N-terminal amino acid residues deduced from sldB.
Detection of d-gluconate oxidation product catalyzed by ARDH.
As shown in a previous work(2), the substrate specificity of ARDH was highly broad, and thus many kinds of substrates, not only pentitols but also other polyhydroxy alcohols, were oxidized. In addition, there was an interesting finding that ARDH also catalyzes the oxidation of one sugar acid, d-gluconate, although the affinity to d-gluconate is rather low (Km, 420 mM) compared with those of other sugar alcohols. As described in the introduction, either of two different reaction products, 2KGA or 5KGA, is possibly produced by the oxidation of d-gluconate, which is related to the reaction mechanism of the enzyme.
Therefore, in this study, the reaction product of d-gluconate oxidation with the purified enzyme was examined by a reaction system where the purified enzyme was mixed with ubiquinol oxidase purified from A. aceti (12) and also with Q2 to reproduce the d-gluconate oxidase system, as described in Materials and Methods. In the reaction system, a continuous electron flow from d-gluconate to oxygen occurred by both enzymes, ARDH and the ubiquinol oxidase via Q2. Thus, the reaction product of d-gluconate oxidation by this system was determined to be 5KGA but not 2KGA (data not shown).
SLDH disruption in G. suboxydans IFO 3257.
In order to confirm the reaction product of ARDH and also to confirm the relationship between ARDH and SLDH, disruption of ARDH in strain IFO 3257 was attempted using a suicide vector harboring the SLDH gene (sldA), which was inserted by a kanamycin cassette in the middle of the gene, pSUP202-sldA::Km, as shown in the previous study (22). Colonies having a phenotype of Kmr Tcs were isolated after conjugation with the pSUP202-sld::Km. The colonies isolated were further analyzed based on the inability to produce d-xylulose from d-arabitol, which was performed by the resorcinol test after being cultivated on a medium containing 1% d-arabitol, 1% glycerol, 0.3% yeast extract, and 0.3% Polypeptone. Since the presence of d-xylulose in the broth is shown as a green color, a strain which did not exhibit any color by the resorcinol test was selected. The gene disruption of the cells exhibiting no resorcinol reaction was also confirmed by PCR (data not shown). As shown in Fig. 1, the disruptant obtained in strain IFO 3257 (3257sldA::Km) as well as in strain IFO 3255 strain (3255sldA::Km) (22) showed no protein bands cross-reactive to anti-SLDH by immunoblotting analysis. The same result was obtained when anti-ARDH was used as the antibody (data not shown). Thus, the data also suggest that the membranes of both strains of IFO 3257 and IFO 3255 contained only a single protein responsible for both SLDH and ARDH.
When the growth of the disruptant 3257sldA::Km was compared with that of the wild strain IFO 3257, a prolonged lag time of the disruptant was observed in arabitol medium, where the mid-log time was ∼15 and ∼44 h in the wild and disruptant strains, respectively, but not in glycerol medium (data not shown).
In order to examine the effect of SLDH disruption on polyol oxidation, the enzyme activities for several polyols were measured with membranes prepared from the wild strain and 3257sldA::Km strain (Table 1). As shown, all the oxidation activities detected in the purified ARDH (2) were diminished, including d-gluconate oxidation, but the oxidation activities for d-glucose and acetaldehyde were retained, which come from different enzymes: d-glucose dehydrogenase and acetaldehyde dehydrogenase, respectively. It should be noted that significant enzyme activity for d-gluconate oxidation measured at pH 5.0 was detected in the disruptant, about 16% of that observed in the wild strain.
TABLE 1.
Effects of SLDH disruption on polyol dehydrogenase activities in both acidic and alkaline pHsa
| Substrate | Polyol dehydrogenase activity (U/mg) at:
|
|||
|---|---|---|---|---|
| pH 5.0
|
pH 9.0
|
|||
| Wild strain | 3257sldA::Km | Wild strain | 3257sldA::Km | |
| d-Arabitol | 1.94 | NDb | 0.96 | 0.04 |
| d-Sorbitol | 1.64 | ND | 0.75 | 0.04 |
| d-Mannitol | 1.12 | ND | 0.46 | ND |
| Ribitol | 1.17 | ND | 0.62 | 0.13 |
| meso-Erythritol | 1.56 | ND | 0.97 | ND |
| Glycerol | 1.38 | ND | 0.92 | ND |
| d-Gluconate | 0.73 | 0.12 | 0.06 | ND |
| d-Glucose | 0.99 | 0.68 | —c | — |
| Acetaldehyde | 1.99 | 2.37 | — | — |
G. suboxydans IFO 3257 and 3257sldA::Km were cultivated for 24 h on a glycerol medium, and the membrane fractions were prepared from these cells. Enzyme activity was measured by PMS-DCIP assay in the presence of 100 mM substrate in McB, pH 5.0, and in 0.2 M glycine-NaOH buffer, pH 9.0.
ND, not detected.
—, not determined.
In the purified ARDH, significant enzyme activity has also been detected at alkaline pH such as pH 8 or 9 (2). Furthermore, in the membranes of G. suboxydans, ribitol oxidase activity, which is also expected to be dependent on ARDH, has also been detected at alkaline pH (1). Thus, in this study, several polyol dehydrogenase activities were also measured at the alkaline pH of 9.0. As shown in Table 1, all the polyol dehydrogenase activities detected at pH 5.0 were also detected at pH 9.0. Furthermore, almost all the polyol dehydrogenase activities detected at the alkaline pH were also diminished in the membranes of the SLDH disruptant. The only exception was the ribitol dehydrogenase activity, where the activity at pH 9.0 was not completely diminished by the disruption, the reason for which remains unclear at present.
Keto-d-gluconate production by wild-type and SLDH-defective strains.
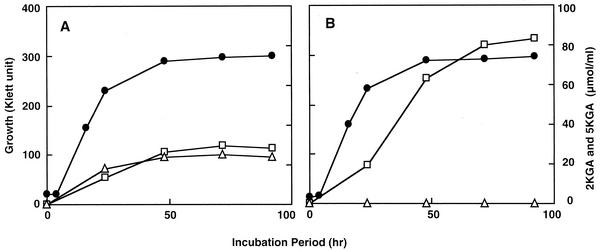
To examine the role of SLDH or ARDH on the oxidative 5KGA fermentation, wild-type and SLDH-defective mutants of G. suboxydans were cultivated in glucose-gluconate medium containing d-glucose and d-gluconate as the carbon source at 30°C with rotary shaking. No difference in the growth was detected between strains 3257 and 3257sldA::Km, unlike in the arabitol medium. In the culture conditions, as shown in Fig. 3, the wild-type strain could accumulate both 2KGA and 5KGA in the culture broth, while the mutant strain could produce only 2KGA. Thus, it was shown clearly that SLDH disruptant is unable to produce 5KGA. In addition, it should be noted that the mutant strain showed a higher yield of 2KGA than the wild-type strain, suggesting that 2KGA and 5KGA production from d-gluconate could compete in vivo.
FIG. 3.
Effects of SLDH disruption on keto-d-gluconate production in G. suboxydans IFO 3257. G. suboxydans IFO 3257 wild-type strain (A) and SLDH-defective mutant 3257sldA::Km (B) were cultivated in 100 ml of glucose-gluconate medium. During the cultivation, samples were taken from these cultures aseptically, and ketogluconates, 2KGA (□) and 5KGA (▵), were measured as described in Materials and Methods. The growth of both strains (•) was also measured.
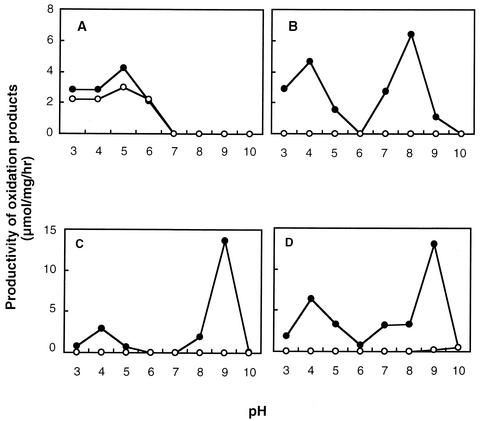
Keto-d-gluconate production was also examined in the resting cells of wild-type and the mutant strain at different pHs (Fig. 4 A and B). Similar to the case in the growing cells, the wild-type strain could produce both 2KGA and 5KGA, whereas 2KGA production was detected only at an acidic pH but 5KGA was produced at both acidic and alkaline pHs. Interestingly, a considerable 5KGA production was observed at pH 9.0. However, the disruption of gene-encoded SLDH led to a complete loss of 5KGA productivity not only in acidic range but also in alkaline range, while 2KGA production remained constant. Although data are not shown here, the same results were obtained from the SLDH disruption in G. suboxydans IFO 3255 strain.
FIG. 4.
Effects of SLDH disruption on keto-d-gluconates, l-sorbose and d-xylulose production with the resting cells of G. suboxydans IFO 3257 at acidic and alkaline pHs. G. suboxydans IFO 3257 wild-type strain and SLDH-defective mutant 3257sldA::Km were cultivated for 24 h on a glycerol medium and washed with buffer to prepare the resting cells, as described in Materials and Methods. The resting cell reaction was performed for 3 h at 30°C in a reaction mixture consisting of 100 μl of 1 M substrate, 100 μl of the resting cell suspension of 3257 (protein contents, 14.5 mg/ml [•]) and of 3257sldA::Km (13.8 mg/ml [○]), and 800 μl of buffer (pH 3.0 to 8.0; McB, pH 9.0 to 10.0; 0.2 M glycine-NaOH buffer). 2KGA (A) and 5KGA (B) concentrations were measured enzymatically while l-sorbose (C) and d-xylulose (D) concentrations were measured by the resorcinol reagent as described in Materials and Methods.
The conversion of d-arabitol or d-sorbitol to d-xylulose or l-sorbose was also confirmed by the same resting cells of G. suboxydans IFO 3257 and the SLDH disruptant. As shown in Fig. 4C and D, both the substrates were also oxidized at both acidic and alkaline pHs, and the production of both d-xylulose and l-sorbose at both pHs was completely diminished by the SLDH disruption, as in the case of 5KGA production. In G. suboxydans IFO 3255 strain and the disruptant, the same results were also observed (data not shown).
DISCUSSION
ARDH or SLDH is a major polyol dehydrogenase in Gluconobacter sp.
Recently, PQQ-dependent, membrane-bound quinoproteins ARDH and SLDH have been purified back to back from G. suboxydans IFO 3257 (2) and G. suboxydans IFO 3255 (23), respectively. And also the SLDH gene has been cloned and expressed in E. coli (15). Both enzymes have been shown to possess very similar substrate specificities on several sugar alcohols such as d-sorbitol, d-mannitol, d-arabitol, meso-erythritol, and glycerol, including d-gluconate. ARDH has been shown to contain PQQ in the enzyme molecule (2). SLDH has also been suggested to have PQQ based on the gene sequence showing homology to a typical quinoprotein glucose dehydrogenase (15), although the presence of PQQ could not be shown directly in the purified enzyme (23). The differences between both enzymes, described in the published papers (2, 23), are in their subunit structure, their optimum pH, and their pH stability, as follows. (i) SLDH consists of a single peptide of 80 kDa, while ARDH has two peptides of 82 and 14 kDa. (ii) Their optimum pH in PMS-DCIP assay was 6.0 in SLDH, while two optimum pHs, high and low activities, at pH 5.0 and 8.0, were observed in ARDH. (iii) SLDH is stable at alkaline pH around 8.0, while ARDH is stable below pH 5.0.
In this study, however, both enzymes were shown to be identical by the following criteria: (i) the large subunit of both enzymes was cross-reacted each other with antibody prepared from both enzymes (Fig. 1); (ii) the N-terminal sequence of the ARDH large subunit was identical to that after the putative signal sequence in the deduced amino acid sequence of the sldA gene encoding the SLDH large subunit (Fig. 2); and (iii) the disruption of the sldA gene led to no protein (Fig. 1) and also no enzyme activities corresponding to ARDH and also to SLDH (Table 1). Since the small subunit found in ARDH was shown to be the product of sldB present just in front of sldA, it would be essential for the enzyme activity or for producing the active large subunit (15). Thus, the lack of a small subunit in the purified SLDH indicates it is either detached from the large subunit during the purification or invisible in the SDS-PAGE due to its small size and thus resulting in a weak staining band. One other finding that should be noted in this study is the signal sequence of SldA. Although a previous study suggested that the signal sequence is the first 24 residues of SldA (15), 29 amino acid residues instead of 24 would be the signal sequence since the N-terminal of ARDH started from the 30th residue, G, just after APA.
Regarding the difference in their optimum pH, it seems to be due to the molecular extinction coefficient used of DCIP, which varies greatly depending on pH, and the value is especially low at lower pHs. The difference in pH stability and in the activity at alkaline pH seem to be related to holoenzyme formation with PQQ and divalent cations. We have some evidence showing different activity at alkaline pH when holoenzyme is constructed with different divalent cations such as calcium, magnesium or nickel (unpublished information). Since ARDH and SLDH have been isolated from different strains grown in different culture media (2, 23), both the holoenzymes may have different cations as the counter ion to bind to PQQ, which may affect their enzyme activity and also their stability.
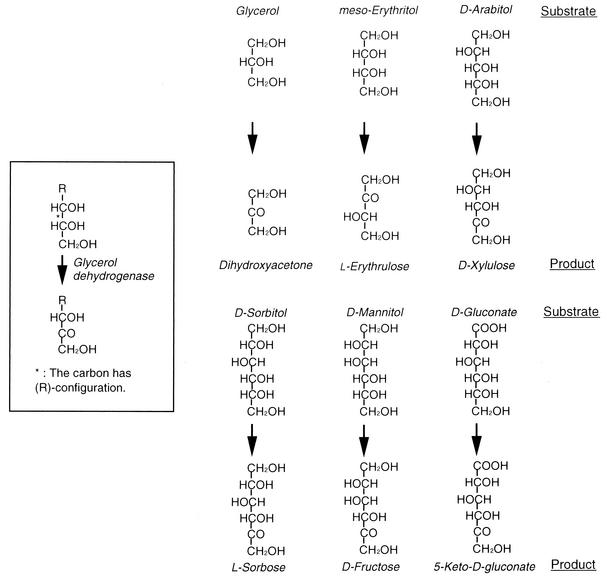
Thus, ARDH and SLDH are considered to be identical quinoproteins responsible for almost all sugar alcohol oxidation in Gluconobacter species, and glycerol dehydrogenase (5), polyol dehydrogenase (6), and mannitol dehydrogenase (16) previously isolated from several Gluconobacter sp. all seem to be the same major polyol dehydrogenase as SLDH or ARDH. As shown in Fig. 5, SLDH and ARDH including other so-called polyol dehydrogenases are able to recognize secondary alcohols with R-configuration in the sugar alcohols, in which d-gluconate is also included. ARDH is actually able to oxidize secondary alcohols such as 2-butanol, 3-pentanol, or 2,3-butanediol (2). The least unit that fulfills the essential polyol structure is glycerol in such sugar alcohols. Thus, the quinoprotein major polyol dehydrogenase, so far called as SLDH, ARDH, mannitol dehydrogenase, or polyol dehydrogenase, would be better call glycerol dehydrogenase (EC 1.1.99.22).
FIG. 5.
Reaction mechanism of quinoprotein glycerol dehydrogenase.
Major polyol dehydrogenase, glycerol dehydrogenase, is involved in the production of 5KGA.
It has been suggested that membrane-bound 5KGA-forming d-gluconate dehydrogenase is a quinoprotein in the genus Gluconobacter (20). In this study, it was shown in the purified enzyme reconstituted with Q2 and ubiquinol oxidase that ARDH is able to produce 5KGA by oxidation of d-gluconate during the turnover. The reaction, oxidation of d-gluconate to 5KGA, in the enzyme is likely performed by the same mechanism for other polyol oxidations, as shown in Fig. 5. Furthermore, the gene disruption of SLDH also inhibited the ability to produce 5KGA (Fig. 4). These data, taken together, clearly indicate that the oxidative fermentation of 5KGA was attributed to SLDH or ARDH, now called glycerol dehydrogenase, in Gluconobacter species.
The 5KGA production was observed in the resting cells not only at acidic but also at alkaline pH, both of which are diminished by the gene disruption (Fig. 4). However, only weak activity of d-gluconate oxidation at alkaline pH was observed in the purified enzyme (2) and even in the membrane fraction (20). Therefore, the 5KGA production at alkaline pH in the resting cells may be considered to occur by the function of a cytoplasmic 5KGR which has an alkaline optimum pH of 10.0 (10). However, as demonstrated in the case of NADPH-dependent l-sorbose reductase in G. suboxydans IFO 3291 (21), these cytoplasmic reductases, such as l-sorbose reductase, 5KGR, or 2KGR, mainly work for the assimilation of oxidation products such as l-sorbose, 5KGA, or 2KGA in the cytoplasm by being taken up from the periplasm; converted to the corresponding sugar alcohol, d-sorbitol or d-gluconate, in the presence of NADPH; and further metabolized in the pentose-phosphate pathway. Therefore, the inconsistency between 5KGA production with the resting cells at the alkaline pH and the weak alkaline activity of the so-called glycerol dehydrogenase may be related to the difference in the situation of holoenzyme formation between the intact cells and the purified enzyme, as described above. This phenomenon is now under investigation.
Thus, it can be concluded that d-gluconate was oxidized to 5KGA by a quinoprotein glycerol dehydrogenase, which is in agreement with the previous observation that membrane-bound quinoprotein dehydrogenase is concerned with 5KGA production (20). Gluconobacter has two types of d-gluconate dehydrogenase, 2KGA-producing flavoprotein (18) and quinoprotein glycerol dehydrogenase, as shown in this study, in the cytoplasmic membrane, which seem to compete with each other to oxidize d-gluconate to keto-d-gluconates in the periplasmic space, as shown in Fig. 3. Thus, selective expression of either dehydrogenase in Gluconobacter sp. is important to produce either of the keto-d-gluconates, which was actually shown in the selective production of 2KGA in the quinoprotein-disrupted strain (Fig. 3)
REFERENCES
- 1.Adachi, O., Y. Fujii, Y. Ano, D. Moonmangmee, H. Toyama, E. Shinagawa, G. Theeragool, N. Lotong, and K. Matsushita. 2001. Membrane-bound sugar alcohol dehydrogenase in acetic acid bacteria catalyzes L-ribulose formation and NAD-dependent ribitol dehydrogenase is independent of the oxidative fermentation. Biosci. Biotechnol. Biochem. 65:115-125. [DOI] [PubMed] [Google Scholar]
- 2.Adachi, O., Y. Fujii, M. F. Ghaly, H. Toyama, E. Shinagawa, and K. Matsushita. 2001. Membrane-bound quinoprotein D-arabitol dehydrogenase of Gluconobacter suboxydans IFO 3257: a versatile enzyme for the oxidative fermentation of various ketoses. Biosci. Biotechnol. Biochem. 65:2755-2762. [DOI] [PubMed] [Google Scholar]
- 3.Ameyama, M., and O. Adachi. 1982. 5-Keto-D-gluconate Reductase from Gluconobacter suboxydans. Methods Enzymol. 89:198-202. [Google Scholar]
- 4.Ameyama, M., and O. Adachi. 1982. 2-Keto-D-gluconate reductase from acetic acid bacteria. Methods Enzymol. 89:203-210. [DOI] [PubMed] [Google Scholar]
- 5.Ameyama, M., E. Shinagawa, K. Matsushita, and O. Adachi. 1985. Solubilization, purification and properties of membrane-bound glycerol dehydrogenase from Gluconobacter industrius. Agric. Biol. Chem. 49:1001-1010. [Google Scholar]
- 6.Cho, N. C., K. Kim, and D.-Y. Jhon. 1990. Purification and characterization of polyol dehydrogenase from Gluconobacter melanogenus. Korean Biochem. J. 23:172-178. [Google Scholar]
- 7.Choi, E.-S., E.-H. Lee, and S.-K. Rhee. 1995. Purification of a membrane-bound sorbitol dehydrogenase from Gluconobacter suboxydans. FEMS Microbiol. Lett. 125:45-50. [Google Scholar]
- 8.Dully, J. R., and P. A. Grieve. 1975. A simple technique for eliminating interference by detergents in the Lowry method of protein determination. Anal. Biochem. 64:136-141. [DOI] [PubMed] [Google Scholar]
- 9.Gupta, A., V. Verma, and G. N. Qazi. 1997. Transposon induced mutation in Gluconobacter oxydans with special reference to its direct-glucose oxidation metabolism. FEMS Microbiol. Lett. 147:181-188. [DOI] [PubMed] [Google Scholar]
- 10.Klasen, R., S. Bringer-Meyer, and H. Sahm. 1995. Biochemical characterization and sequence analysis of the gluconate:NADP 5-oxidoreductase gene from Gluconobacter oxydans. J. Bacteriol. 177:2637-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsushita, K., and M. Ameyama. 1982. D-Glucose dehydrogenase from Pseudomonas fluorescens, membrane-bound. Methods Enzymol. 89:149-153. [DOI] [PubMed] [Google Scholar]
- 12.Matsushita, K., H. Ebisuya, and O. Adachi. 1992. Homology in the structure and the prosthetic groups between two different terminal ubiquinol oxidases, cytochrome a1 and cytochrome o, of Acetobacter aceti. J. Biol. Chem. 267:24748-24753. [PubMed] [Google Scholar]
- 13.Matsushita, K., E. Shinagawa, and M. Ameyama. 1982. D-Gluconate dehydrogenase from bacteria, 2-keto-D-gluconate-yielding, membrane-bound. Methods Enzymol. 89:187-193. [DOI] [PubMed] [Google Scholar]
- 14.Matsushita, K., H. Toyama, and O. Adachi. 1994. Respiratory chains and bioenergetics of acetic acid bacteria. Adv. Microb. Physiol. 36:247-301. [DOI] [PubMed]
- 15.Miyazaki, T., N. Tomiyama, M. Shinjoh, and T. Hoshino. 2002. Molecular cloning and functional expression of D-sorbitol dehydrogenase from Gluconobacter suboxydans IFO 3255, which requires pyrroloquinoline quinone and hydrophobic protein SldB for the activity development in Escherichia coli. Biosci. Biotechnol. Biochem. 66:262-270. [DOI] [PubMed] [Google Scholar]
- 16.Oikawa, T., J. Nakai, Y. Tsukagawa, and K. Soda. 1997. A novel type of D-mannitol dehydrogenase from Acetobacter xylinum: occurrence, purification, and basic properties. Biosci. Biotechnol. Biochem. 61:1778-1782. [DOI] [PubMed] [Google Scholar]
- 17.Shinagawa, E., K. Matsushita, O. Adachi, and M. Ameyama. 1982. Purification and characterization of D-sorbitol dehydrogenase from membrane of Gluconobacter suboxydans var.α. Agric. Biol. Chem. 46:135-141. [Google Scholar]
- 18.Shinagawa, E., K. Matsushita, O. Adachi, and M. Ameyama. 1983. Selective production of 5-keto-D-gluconate by Gluconobacter strains. J. Ferment. Technol. 61:359-363. [Google Scholar]
- 19.Shinagawa, E., K. Matsushita, O. Adachi, and M. Ameyama. 1984. D-Gluconate dehydrogenase, 2-keto-D-gluconate yielding, from Gluconobacter dioxyacetonicus: purification and characterization. Agric. Biol. Chem. 48:1517-1522. [Google Scholar]
- 20.Shinagawa, E., K. Matsushita, M. Ameyama, and O. Adachi. 1999. Production of 5-keto-D-gluconate by acetic acid bacteria is catalyzed by pyrroloquinoline quinone (PQQ)-dependent membrane-bound D-gluconate dehydrogenase. J. Mol. Catal. B Enzyme 6:341-350. [Google Scholar]
- 21.Shinjoh, M., M. Tazoe, and T. Hoshino. 2002. NADPH-dependent l-sorbose reductase is responsible for l-sorbose assimilation in Gluconobacter suboxydans IFO 3291. J. Bacteriol. 184:861-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shinjoh, M., N. Tomiyama, T. Miyazaki, and T. Hoshino. 2002. Main polyol dehydrogenase of Gluconobacter suboxydans IFO 3255, membrane-bound D-sorbitol dehydrogenase, that needs product of upstream gene. sldB, for activity. Biosci. Biotechnol. Biochem. 66:2314-2322. [DOI] [PubMed] [Google Scholar]
- 23.Sugisawa, T., and T. Hoshino. 2002. Purification and properties of membrane-bound D-sorbitol dehydrogenase from Gluconobacter suboxydans IFO 3255. Biosci. Biotechnol. Biochem. 66:57-64. [DOI] [PubMed] [Google Scholar]