Abstract
Populations of Rhizobium leguminosarum biovar viciae were sampled from two bulk soils, rhizosphere, and nodules of host legumes, fava bean (Vicia faba) and pea (Pisum sativum) grown in the same soils. Additional populations nodulating peas, fava beans, and vetches (Vicia sativa) grown in other soils and fava bean-nodulating strains from various geographic sites were also analyzed. The rhizobia were characterized by repetitive extragenomic palindromic-PCR fingerprinting and/or PCR-restriction fragment length polymorphism (RFLP) of 16S-23S ribosomal DNA intergenic spacers as markers of the genomic background and PCR-RFLP of a nodulation gene region, nodD, as a marker of the symbiotic component of the genome. Pairwise comparisons showed differences among the genetic structures of the bulk soil, rhizosphere, and nodule populations and in the degree of host specificity within the Vicieae cross-inoculation group. With fava bean, the symbiotic genotype appeared to be the preponderant determinant of the success in nodule occupancy of rhizobial genotypes independently of the associated genomic background, the plant genotype, and the soil sampled. The interaction between one particular rhizobial symbiotic genotype and fava bean seems to be highly specific for nodulation and linked to the efficiency of nitrogen fixation. By contrast with bulk soil and fava bean-nodulating populations, the analysis of pea-nodulating populations showed preferential associations between genomic backgrounds and symbiotic genotypes. Both components of the rhizobial genome may influence competitiveness for nodulation of pea, and rhizosphere colonization may be a decisive step in competition for nodule occupancy.
Rhizobia are soil bacteria which have the ability to induce nitrogen-fixing nodules on the roots or stems of legume plants. There have been extensive data showing that rhizobial strains differ in their nodulating competitiveness, as estimated by the percentages of nodules formed when host legumes are inoculated with a mixture of strains or when they are applied as a single inoculant in soil containing indigenous rhizobial populations (2, 10, 39, 40). The genetic basis of competitiveness for nodule formation is not fully understood yet. Efficient nodulation is controlled by rhizobial nodulation genes, cell surface determinants, genes controlling catabolism of legume or bacterial metabolites, and other rhizobial genes the functions of which have not been elucidated (see the review by Vlassak and Vanderleyden [46]).
Dominance of particular rhizobial genotypes in nodules may result from a higher abundance in soil rather than a greater nodulation competitiveness, as shown for certain genotypes within Sinorhizobium meliloti indigenous populations (6, 16, 43). This may explain the finding that nodule-dominant genotypes from soil populations do not necessarily show superior competitiveness for nodulation compared to minor occupants when evaluated under nonsoil conditions (7, 27, 31). Prior to competition for root infection and nodule formation, rhizobia should survive and grow in the soil in the absence of the host plant and be able to colonize the host plant rhizosphere. The responses of indigenous rhizobia to abiotic and biotic environmental factors influence their saprophytic and rhizosphere competence and finally the issue of nodule occupancy by individual genotypes (46). There have been few studies examining the selectivity of host rhizosphere towards indigenous rhizobial genotypes and the effect of this selectivity on the outcome of nodulation (8, 28, 32, 37). A selective enrichment of rhizobial types in the clover rhizosphere was reported previously (28), but the relative abundance of different rhizobial types in the host rhizosphere did not correlate with their relative abundance in nodule populations (28, 32, 37).
The breadth of host range varies among rhizobial species, and thus host species and genotype within species may be major factors that influence the outcome of competition for nodulation. Rhizobium leguminosarum bv. viciae is the specific symbiont of the legumes of the tribe Vicieae which comprises the genera Vicia, Pisum, Lens, and Lathyrus. Differences in host plant preference for specific rhizobial genotypes within natural populations have been previously reported among these legume genera and species (12, 15, 22, 23). In particular, peas (Pisum sativum) and field beans (Vicia faba) showed contrast in the selection of indigenous rhizobial genotypes (22) as revealed by plasmid profiling.
Genetic determinants of nodulation competitiveness have been localized on plasmids in R. leguminosarum including the symbiotic (Sym) plasmid, which carries genes essential for nodulation and nitrogen fixation (5, 33, 35, 49). A previous study has shown that associations constructed by introduction of the same Sym plasmid in different recipient strains varied in competitiveness for nodulation of pea plants, indicating that both components of the genome were involved (5). Several reports have shown that the Sym plasmid is not strictly associated with the chromosomal background in natural populations of R. leguminosarum (14, 23, 25, 30, 38, 50, 52). The analysis of the genetic structure of populations of R. leguminosarum bv. viciae isolated directly from bulk soil (30) suggests that the diversity of associations of genomic backgrounds and Sym genotypes may be greater in soils than in nodules, which are formed only by the most competitive genotypes for a given host plant.
In this study, our aims were (i) to investigate in more detail the contribution of the two components of the rhizobial genotype (genomic background and Sym genotype) to the outcome of competition for nodulation by studying natural populations of R. leguminosarum bv. viciae and using different host plants, (ii) to determine if nodulation-competitive genotypes are selected at the stage of rhizosphere colonization, and (iii) to evaluate the influence of environmental factors on the nodulating populations. We have previously developed a procedure to isolate free-living populations of R. leguminosarum bv. viciae from soil samples (29) and described PCR-based molecular methods for typing genomic background and symbiotic genotypes (24). At the same time as natural populations of R. leguminosarum bv. viciae were directly isolated from two soils (30), pea and fava bean plants were cultivated in these two soils. In the present study, we report the results of comparisons of the relative abundance of indigenous rhizobial genotypes isolated from bulk soils, host rhizospheres, and nodules. The compositions of pea, fava bean, and vetch nodule populations from soils collected at another geographic location were also analyzed, as well as a collection of rhizobial strains isolated from nodules of diverse varieties of fava beans grown at various geographic sites.
MATERIALS AND METHODS
Soil samples.
Soils P1 and F5 were collected at two 500-m-distant fields at the Institut National de la Recherche Agronomique (INRA) Experiment Station, Bretenières (Côte d'Or, France). They are clay loam with pHs of 8 and 7.5 (in water), respectively. The characteristics of these soils and the sampling procedure have been previously described (30). Most probable numbers (MPN) (45) of R. leguminosarum bv. viciae cells were in the range of 104 to 105/g of soil (30). The cropping history over the 10 to 12 years before the sampling dates, March 1993 and April 1994 for soils P1 and F5, respectively, included the cultivation of various nonlegume crops and host legumes for R. leguminosarum bv. viciae, which were peas (field P1 in 1982 and field F5 in 1984, 1986, and 1987), lentils (field F5 in 1986 and 1987), and fava beans (field F5 in 1989).
Soils TTF4 and M62 were collected at the field site of the INRA Experimental Station of Grignon (Yvelines, France), which is about 350 km distant from Bretenières. These soils are also clay loam with a pH of 7.8 (in water). Their characteristics were previously described by Barrusio and Houot (4). From 1973 until the sampling date in March 1996, the TTF4 field was a randomized block design (three blocks) including two treatments, A and C. Treatment A was a wheat-maize rotation cultivated with maize during the 1995-1996 cropping season. Treatment C was a wheat monoculture. Soil samples were collected in each of the A and C plots (three samplings per plot). The soil was sieved at 5 mm, and the three subsamples from each plot were mixed together before use. MPN of R. leguminosarum bv. viciae cells for TTF4A and TTF4C plots were within the range of 3 × 102 to 5 × 103 and 1.4 × 103 to 2 × 104/g of soil, respectively. Field M62 was about 150 m distant from TTF4. From 1962 to the sampling date in March 1998, this field was continuously cultivated with maize. The sampling procedure was the same as that for TTF4.
R. leguminosarum bv. viciae isolation.
Rhizobia were directly isolated from soil by a procedure previously described (29) which involves the use of a semiselective medium (including antibiotics to inhibit the growth of gram-positive bacteria and fungicides), a counterselection step (inability to grow on a NaCl-rich agar medium), and a specific identification method (colony blot hybridization with a biovar-specific DNA probe). At the same time as bacteria were isolated from bulk soils (30), samples of soils P1 and F5 were used to cultivate peas (P. sativum cv. Solara) and fava beans (V. faba L. Ad23MS, a sterile male line obtained from G. Duc, Plant Breeding and Genetic Department of INRA). Seeds were surface sterilized and sown in pots filled with Terragreen (1:5 [vol/vol]) in the bottom and 1.8 kg of soil in pots of 2.5 liters (soil P1) or 2.5 kg of soil in pots of 5 liters (soil F5). Six replicate pots (four plants per pot) were cultivated in a greenhouse. After 3 weeks, plants with the soil adhering to the roots were removed from three replicate pots and allowed to dry for 12 h at 28°C. The non-closely adhering soil was then removed by vigorous manual shaking of roots, and the adhering soil, considered to be rhizosphere soil, was collected with a brush. MPN of R. leguminosarum bv. viciae cells in the rhizosphere soils were within the range of 0.5 × 105 to 1.4 × 106/g of soil, i.e., about 10 times more than in the bulk soils. Direct isolation of R. leguminosarum bv. viciae from the rhizosphere soil samples was performed as for the bulk soil samples. After 6 weeks of growth, 60 nodules per pot (15 per plant) were collected from the remaining pots (three replicates). The sampling was representative of the proportion of nodules on the principal and the lateral roots. For soils TTF4 and M62, R. leguminosarum bv. viciae bacteria were isolated from nodules of pea (P. sativum cv. Solara), fava bean (V. faba cv. Divine), and vetch (Vicia sativa cv. Cristal) plants grown in pots for 4 to 5 weeks under greenhouse conditions as described above. Three replicates were made for each plant species and each plot of field TTF4 and from field M62, and 30 nodules per pot were collected. The procedure of isolation of R. leguminosarum bv. viciae from nodules was as described by Vincent (45). The isolates were maintained on MGY agar medium (29) at 4°C.
Additional R. leguminosarum bv. viciae strains.
A collection of 53 bacterial strains isolated from nodules of various cultivars of V. faba grown under field conditions was also included in this study. Forty-eight of them were isolated at various sites all over France (3). Four strains, USDA2497, USDA2498, USDA2501, and USDA2508, originated from Spain and were kindly provided by P. van Berkum (Agricultural Research Service, U.S. Department of Agriculture, Beltsville, Md.), and one strain (MSDJ0545) originated from Poland.
Characterization of R. leguminosarum bv. viciae by plasmid profiling and PCR fingerprinting.
The plasmid content of all isolates except those from soil M62 was analyzed for a preliminary classification as previously described (30). The isolates from the rhizospheres and nodules of plants grown in soil P1 were further characterized by repetitive extragenomic palindromic PCR (REP-PCR) and classified in REP groups as previously described (24, 30). Since isolates sharing identical plasmid profiles were always grouped by REP-PCR fingerprinting, only one representative of each plasmid profile was typed by REP-PCR within the pea and fava bean populations from soil F5. REP-PCR classification was checked by PCR-restriction fragment length polymorphism (RFLP) of the intergenic spacer (IGS) region between 16S and 23S ribosomal DNA (rDNA) as previously described (24, 30) for a representative subsample of isolates (23%) from soils P1 and F5. In the subsequent experiments, we abandoned the REP-PCR method because of the lack of reproducibility of the fingerprints between independent experiments made at different periods with changes in products (primers and enzymes). A subsample of isolates (73%) from the TTF4 plots and all isolates from soil M62 were further characterized by PCR-RFLP of 16S-23S rDNA IGS. The Sym genotype of each isolate and strain used in this study was also characterized by PCR-RFLP of the nodulation gene region nodD-F as previously described for the populations from the P1 and F5 bulk soil samples (30). Primer pair NBA12 and NBF12 were used to amplify the nodD-F gene region, which includes a part of the IGS region between nodA and nodD genes, the nodD gene, and the IGS region between nodD and nodF genes. Primers NODD2PH678 and NODDRL2′ were also used to amplify an 876-bp internal fragment of the nodD gene. Primer nucleotide sequences and locations were previously described (24). Amplification of this nodD gene fragment by nested PCR from the NBA12-NBF12 PCR products was used to authenticate the nodD-F gene region. Sequence divergence between nodD genes was estimated by mapping restriction sites, computing a similarity matrix, and clustering as previously described (24). The following restriction enzymes were used: AluI, CfoI, DdeI, HaeIII, MspI, and NdeII.
Plant tests.
The ability of rhizobial strains to form nodules on fava beans (line Ad23MS and the commercial cultivar Divine) and peas (cultivar Solara) was tested on plants grown in pots (four replicates) filled with perlite as previously described (23). After 7 weeks, plants were harvested, shoot dry matter was measured, numbers of nodules were recorded, and 18 nodules per strain were excised. The rhizobia were reisolated from nodules and genetically characterized, eliminating the possibility that contaminant strains might have formed nodules.
Data analysis.
The distribution of genotypes was statistically compared among populations by analysis of molecular variance (AMOVA version 1.55, 1995) (13). R × C tests of independence, R and C standing for the numbers of rows (e.g., REP groups) and columns (e.g., frequency of each nod type) in unordered contingency tables, respectively, were used to test whether the nod types were randomly distributed in REP groups and whether the distribution of the rhizobial genotypes was independent of the plant cultivar. The exact value of probability to accept or reject the null hypothesis was computed with the StatXact program (version 3 for Windows, 1997; Cytel Software Corporation). To estimate the number of dominant types, we calculated the Simpson inverse diversity index, 1/D = 1/Σ[ni(ni − 1)/N(N − 1)], where ni is the number of the ith type and N is the number of individuals in the population (19). Rarefaction analysis was done to check that the size of our sampling (the numbers of isolates) was high enough to allow direct comparisons of genotype richness (number of types) between populations with the EstimateS program (http://viceroy.eec.uconn.edu/estimates, version 6.0b1 for Windows, 2000). Analysis of variance was performed on the weights of shoot dry matter obtained from plant tests.
RESULTS
Characterization of R. leguminosarum bv. viciae populations.
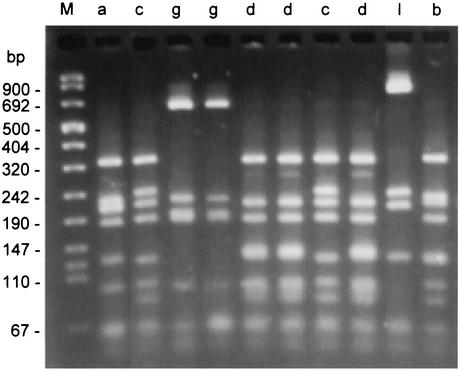
The nodD-F gene region of the Sym plasmid could be amplified by PCR for 463 R. leguminosarum bv. viciae isolates from Bretenières soils. Eleven distinct nod types (HaeIII restriction patterns) were detected (Table 1; Fig. 1). The background genotypes of the 481 R. leguminosarum bv. viciae isolates from Bretenières were classified in 33 REP groups (Table 2). A total of 46 composite genotypes defined by the REP group-nod type associations were recorded among the Bretenières isolates (Table 1). The classification in nod types was independent of the classification in REP groups (P > 0.05) except for the pea nodule populations (P < 0.01), in which the frequency of the association of REP group A and nod type g was two to three times lower than expected. Noteworthy is the fact that the predominant nod type g (63% of isolates) was found to be associated with most of the REP groups (24 out of 33).
TABLE 1.
Distribution of R. leguminosarum bv. viciae isolates from bulk soil, rhizosphere, and nodules in associations of nod types and REP groups
| nod type | REP group | % of isolates from population (no. of isolates)a
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| PISNb (59) | PIRF (52) | PIRP (56) | PINF (67) | PINP (62) | F5SNb (54) | F5NF (55) | F5NP (58) | Total (463) | ||
| a | A | 10 | 8 | 11 | 14 | 2 | 5.6 | |||
| a | G | 2 | 2 | 9 | 48 | 7.6 | ||||
| b | G | 9 | 1.5 | |||||||
| d | A | 7 | 6 | 39 | 19 | 8.9 | ||||
| g | A | 46 | 56 | 25 | 49 | 15 | 33 | 25 | 3 | 31.5 |
| g | B | 10 | 2 | 5 | 13 | 11 | 7 | 4 | 6.9 | |
| g | D | 10 | 4 | 2 | 9 | 3 | 20 | 16 | 3 | 8.4 |
| g | E | 2 | 11 | 1.5 | ||||||
| g | F | 2 | 7 | 5 | 1.9 | |||||
| g | G | 2 | 2 | 6 | 22 | 16 | 5.6 | |||
| g | T | 2 | 9 | 5 | 1.9 | |||||
| g′ | A | 5 | 8 | 2 | 18 | 3 | 4 | 4 | 5.6 | |
| j | A | 2 | 4 | 4 | 19 | 3.7 | ||||
| Variousc | Variousc | 9 | 10 | 11 | 7 | 3 | 19 | 9 | 16 | 9.7 |
P1, soil P1; F5, soil F5; SN, bulk soil; RF, fava bean rhizosphere soil; RP, pea rhizosphere soil; NF, fava bean nodules; NP, pea nodules.
The results have been previously published (30).
The results were pooled for 33 associations of nine nod types and 30 REP groups represented by fewer than five isolates within each subpopulation.
FIG. 1.
Restriction patterns of PCR-amplified nodD-F gene fragments digested with HaeIII. The lane assignments correspond to restriction pattern types (nod types) which are given in Tables 1 and 3. Lane M, molecular size marker VIII (Boehringer Mannheim).
TABLE 2.
Distribution of R. leguminosarum bv. viciae isolates from soils P1 and F5 in REP groups
| Populationa | % of isolates in REP group:
|
No. of isolates | Diversity indexd | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| A | G | D | B | T | F | E | Othersc | |||
| PISNb | 69 | 2 | 11 | 13 | 5 | 62 | 2.0 | |||
| PIRF | 80 | 2 | 4 | 2 | 2 | 10 | 56 | 1.6 | ||
| PIRP | 81 | 2 | 2 | 5 | 2 | 8 | 58 | 1.5 | ||
| PINF | 68 | 9 | 13 | 7 | 3 | 68 | 2.1 | |||
| PINP | 71 | 3 | 5 | 11 | 2 | 5 | 3 | 63 | 1.9 | |
| Total P1 | 73.6 | 1.6 | 6.2 | 9.1 | 0.3 | 3.3 | 15.0 | 307 | 1.8 | |
| F5SNb | 39 | 14 | 19 | 7 | 2 | 4 | 15 | 57 | 4.9 | |
| F5NF | 30 | 21 | 16 | 4 | 9 | 10 | 10 | 57 | 6.0 | |
| F5NP | 3 | 82 | 3 | 7 | 5 | 60 | 1.5 | |||
| Total F5 | 23.6 | 39.7 | 12.6 | 3.4 | 5.7 | 4.6 | 10.4 | 174 | 4.3 | |
P1, soil P1; F5, soil F5; SN, bulk soil; RF, fava bean rhizosphere soil; RP, pea rhizosphere soil; NF, fava bean nodules; NP, pea nodules.
The results have been previously published (30).
The results from 26 REP groups represented by fewer than five isolates within each population were pooled.
Simpson inverse index.
The nodD-F gene region was also PCR amplified for 666 isolates of R. leguminosarum bv. viciae from other geographical sites. Eleven nod types were detected by RFLP analysis (Table 3). Seven of them, which grouped 95% of isolates, were similar to those found at Bretenières. Again, the nod type g largely predominated in this collection, representing 76% of the isolates.
TABLE 3.
Diversity of nod types among R. leguminosarum bv. viciae isolates from nodules of fava bean, pea, and vetch plants cultivated in soils of various origins
| Host plant of origin | Soil | % of isolates in nod type:
|
No. of isolates | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| a | c | d | g | g" | h | l | Othersb | |||
| Fava bean | Variousa | 6 | 11 | 77 | 2 | 4 | 53 | |||
| Fava bean | TTF4C | 96 | 2 | 1 | 84 | |||||
| Fava bean | TTF4A | 96 | 4 | 83 | ||||||
| Fava bean | M62 | 100 | 90 | |||||||
| Pea | TTF4C | 4 | 69 | 7 | 12 | 4 | 2 | 88 | ||
| Pea | TTF4A | 18 | 64 | 1 | 4 | 6 | 2 | 89 | ||
| Pea | M62 | 8 | 63 | 29 | 90 | |||||
| Vetch | M62 | 6 | 5 | 78 | 9 | 1 | 2 | 89 | ||
Collection of isolates from plants grown under field conditions at various geographical sites.
The results were pooled for four nod types represented by fewer than five isolates within each population.
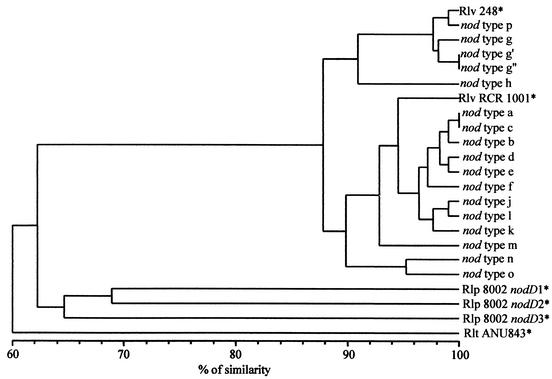
The size of the nodD-F PCR products varied from 1,350 to 1,500 bp except for isolates characterized by nod types g′ and g", which gave a fragment of approximately 2,200 bp. Representatives of each nod type were further analyzed by PCR with primer pair NBA12-NODDRL2′, which targets 876 bp of the nodD gene plus 35 bp of the IGS region between nodA and nodD. All the PCR products had the same expected size of approximately 960 bp. We deduced from this result that the length polymorphism of the nodD-F gene region was located within the IGS between nodD and nodF. By using the primer pair NODD2PH678-NODDRL2′, an internal fragment of 875 bp of the the nodD gene was amplified to estimate sequence divergence between the various nod types. RFLP analyses with six restriction enzymes were performed, and the map location of 108 restriction sites could be inferred from the available nodD gene sequences as previously described (24). Five groups including two main clusters could be delineated at the restriction site similarity level of 95% (Fig. 2). One cluster included nine nod types and the nodD gene of the reference strain R. leguminosarum bv. viciae RCR 1001. The second main cluster was formed by nod types g, g′, g", and p and included the nodD gene of the reference strain R. leguminosarum bv. viciae 248, which also had the HaeIII restriction pattern g by PCR-RFLP of the nodD-F gene region.
FIG. 2.
Dendrogram (unweighted pair group method with arithmetic mean) of similarities between nod types based on mapped restriction site analysis of nodD gene fragments (875 bp) amplified by PCR. The nod types of R. leguminosarum bv. viciae (Rlv), bv. trifolii (Rlt), and bv. phaseoli (Rlp) strains that are designated by an asterisk were included by inferring their restriction site polymorphism from the nodD sequences available in GenBank (accession numbers Y00548, J03671, X03721, X54214, and X54215).
Comparison of the Bretenières R. leguminosarum bv. viciae populations isolated from bulk soils, rhizosphere soils, and nodules.
Four to seven composite REP-nod genotypes were represented by at least three isolates (5%) in each of the eight populations from Bretenières soils P1 and F5 (Table 1). Pairwise comparisons indicated that the distribution of the composite genotypes significantly differed (generally P < 0.001) between the populations, except in three cases. The soil P1 populations from bulk soil (P1SN) and from fava bean rhizosphere (P1RF), P1RF and P1NF from fava bean nodules, and P1RP from pea rhizosphere and P1NP from pea nodules had pairwise similar compositions (P > 0.1). In P1RP and P1NP, the association of REP group A with nod type d was one of the most abundant, including 20 to 40% of isolates. In the other populations from soil P1, the REP A-nod g association largely predominated (>45% of isolates). In soil F5, other composite genotypes codominated with REP A-nod g in the bulk soil (F5SN) and the fava bean (F5NF) populations. The pea nodule population (F5NP) was markedly different from F5SN and F5NF. The REP A-nod g association was scarcely represented in F5NP, while the REP G-nod a association largely predominated (48% of isolates).
The distributions of the REP groups and the nod types were then separately analyzed. No significant differences (P > 0.15) were found between the distribution of the REP groups among the populations from soil P1 by pairwise comparisons, except for P1RF and P1NF and for P1RF and P1NP (Table 2). Differences between P1RF and the nodule populations may be explained by the greater richness in genotypes detected in P1RF (11 REP groups) compared to P1NF and P1NP (six and eight REP groups, respectively) and also differences in evenness. A single REP group, A, largely predominated in these five populations (68 to 81% of the isolates). AMOVA indicated that the two F5 subpopulations from bulk soil (F5SN) and fava bean nodules (F5NF) significantly differed (P < 0.001), but most of the diversity of the REP groups was found within each subpopulation, while the differences between the two populations were small (0.84%). Both populations markedly differed from the pea nodule population F5NP (P < 0.001) with estimated variances among populations of 28 and 36% for F5SN-F5NP and F5NF-F5NP, respectively. Three REP groups, A, D, and G, were prevalent (14 to 39% of isolates) within F5SN and F5NF, while REP group G included more than 80% of the isolates from F5NP (Table 2). We have previously reported that the distribution of REP groups significantly differed between the two bulk soil populations (30). Differences were also found between the two fava bean nodule populations and the two pea nodule populations. Five REP groups were common to both soils, including 91 and 85% of the isolates from soils P1 and F5, respectively. However, the diversity of REP groups estimated by the Simpson inverse index was higher in the bulk soil and fava bean populations from soil F5 than in the other populations (Table 2). Additionally, REP group G, which was abundant in soil F5, and REP group A, which was more predominant in soil P1, were found to be distantly related genotypes within the species R. leguminosarum based on the comparison of various typing methods (24).
The nod type g largely predominated in Bretenières populations except in those isolated from the rhizosphere and the nodules of pea plants (Table 1). We have previously reported that the distributions of the nod types were not significantly different between the two bulk soil populations P1SN and F5SN based on a Pearson chi-square statistical approach (30). The statistical analysis used in the present study, AMOVA, has the advantage of taking into account the variability within treatment. AMOVA performed on the same set of data confirmed the previous result. The frequencies of nod types were also similar (P > 0.3) between P1SN and P1RF (fava bean rhizosphere) and between P1RP (pea rhizosphere) and P1NP (pea nodules). The distributions of the nod types were significantly different (P < 0.001) between the other populations from soil P1 and also among the three populations from soil F5. All isolates excepted one from nodules of fava beans grown in soils P1 and F5 harbored nod type g or nod type g′, two closely related genotypes. By contrast, a diversity of nod types were detected in populations isolated from the rhizosphere and from the nodules of pea plants. Two to three other types predominated with nod type g. The frequency of nod type d in P1RP and P1NP was notably higher than in the other P1 populations. Similarly, the frequency of nod type a was higher in F5NP than in the other F5 populations. The nod type j was abundant only in pea nodules from both soils. The distributions of the nod types were significantly different between the pea nodule populations from soils P1 and F5, mainly due to the absence of detection of nod type d in soil F5.
Comparison of fava bean, pea, and vetch nodule populations from different geographic sites.
The distribution of nod types was investigated in a collection of 53 strains of R. leguminosarum bv. viciae isolated from fava beans grown at various geographic sites, mainly in France. The nod type g was again found to be the most frequently occurring genotype in fava bean nodules (Table 3). The frequency of nod types was independent of the fava bean genotype or cultivar (P > 0.05). This result was fully corroborated by the analysis of the fava bean populations from three different plots at the Grignon site. The nod type g represented 96 to 100% of the isolates, as in the Bretenières populations isolated from fava bean nodules (Table 3).
The analysis of three pea nodule populations from Grignon also corroborated the results obtained from the Bretenières populations. Nine nod types were detected in addition to type g (Table 3). As in the pea population from soil P1, the frequencies of nod type d were high in Grignon plots TTF4A (18% of isolates) and M62 (63% of isolates), and nod type a was also present in M62 soil.
The diversity of nod types was higher in the vetch population from Grignon plot M62 (seven nod types) than in the fava bean and pea nodule populations from the same plot (one and three nod types, respectively). The nod type g predominated in the vetch population (77.5% of isolates). Similarly, the diversity of background genotypes was higher in the vetch population than in the fava bean and pea populations, in which 16, 5, and 1 genotype were detected, respectively, by PCR-RFLP of 16S-23S rDNA IGS (data not shown).
The rDNA type that was associated with REP group A in P1 and F5 populations was also frequent in the various Grignon populations (13 to 100% of isolates). The association of this genotype with nod type g included 13 to 72% of the fava bean isolates, 48% of the vetch isolates, and 10 to 39% of the pea isolates; as in P1NP, the association of this genotype with nod type d was abundant in the pea populations from Grignon TTF4A and M62 fields, including 18 and 63% of isolates, respectively.
Nodule formation and nitrogen fixation effectiveness of R. leguminosarum bv. viciae genotypes with fava beans.
Seven strains representative of associations of REP groups and nod types (A/d, A/a, A/j, G/a, G/b, G/j, and G/g) were selected from the rhizosphere and nodule populations from pea. The reference strain MSDJ0822, isolated from a fava bean nodule, was included as a positive control for nitrogen fixation effectiveness with both peas and fava beans (1). Strain MSDJ0822 was characterized by the composite genotype A/g.
All the strains formed numerous nodules with pea plants and were as effective in nitrogen fixation as was the positive-control strain. All these strains were also able to form nodules with fava bean plants, but the number, size, color, and shape of nodules varied according to the strain inoculated. The strain characterized by the composite genotype A/d formed few nodules (<10 per plant). The highest numbers of nodules (>200 nodules per plant) were obtained with the strains characterized by the composite genotypes G/g and A/g. Only these two strains plus one other strain (genotype G/a) were significantly effective for nitrogen fixation with fava beans (15 to 18 g of shoot dry matter/pot) by comparison with the uninoculated control pots (3 g of shoot dry matter/pot). The results were similar for the V. faba line Ad23MS and the V. faba commercial cultivar Divine.
DISCUSSION
Comparison of the diversities of R. leguminosarum bv. viciae populations.
The richness of combined genotypes detected in nodules when the results from fava beans and peas were pooled was as high as that detected in bulk soils (10 to 12 genotypes based on rarefaction analysis of samples with identical numbers of isolates). Most genotypes were shared by soil and nodule populations, and the shared genotypes represented 80 to 90% of each population.
The richness of nod types was similar in fields from two distinct geographical sites, and about half of the nod types identified in this study were found at both sites. This shows that the levels of diversity of this functional gene were similar in arable soils which had roughly comparable physicochemical characteristics. However, the estimation of the nodD gene diversity might have been biased by the cultivation-based approach that we used. We are currently developing specific molecular tools to quantify and to describe the diversity of this gene by direct PCR amplification from total community DNA.
Prevalence of a single Sym genotype in several arable fields.
The nod type g largely predominated in the Bretenières soils, and also probably in the Grignon soils, because the frequency of this nod type in nodule populations was as high as, or higher than, that in Bretenières nodule populations. The fields sampled in this study have been subjected to various crop management plans including, for two of them, long-term monoculturing of nonlegume plants. Therefore, the prevalence in soils of the Sym genotype characterized by nod type g cannot be related to the cropping history and the presence of its compatible host plant. This Sym genotype has a notably wide host range for the genomic backgrounds associated with it, suggesting that the Sym plasmid that carries it can be transferred by conjugation and stably maintained in soil conditions in a wide range of R. leguminosarum bv. viciae genotypes. The apparent superior saprophytic ability of nod type g may thus be due to the breadth of diversity of genomic backgrounds that harbor it, which enables the species to adapt to various environmental conditions. Another possible explanation might be that genes localized on the Sym plasmids with nod type g are directly involved in the selective maintenance of this plasmid type or confer a selective advantage on the strains for saprophytic competition. The production of bacteriocin may play this role. The Sym plasmid pRL1J1 (nod type g) from R. leguminosarum bv. viciae strain 248 contains genes for bacteriocin production (21). Although bacteriocin production is not specific to this strain, it is restricted to a limited number of strains and various R. leguminosarum bv. viciae strains were found to be sensitive to the bacteriocins produced by strain 248 (20, 36). However, according to the work of Wilson et al. (48), the prevalence of a bacteriocin-producing strain in one field may have led to the subsequent proliferation of resistant strains the following year.
Host selection of R. leguminosarum bv. viciae genotypes from soil free-living populations.
Both fava bean and pea plants are able to discriminate among the diversity of rhizobial genotypes that are present in soil. Similar results were reported previously for other legumes (6, 16, 51). Furthermore, relatively small differences in frequencies of R. leguminosarum bv. viciae genotypes between soils appear to strongly influence the distribution of R. leguminosarum bv. viciae genotypes in nodules. For example, genotype G/a was not detected in nodules of plants grown in soil P1 but represented 48% of nodules of pea plants grown in soil F5. This genotype also predominated in nodules of pea plants grown in field conditions at the same site 8 years before the sampling of soil (25), which suggests the stability of the R. leguminosarum bv. viciae population over several seasons.
Differences in host preference for R. leguminosarum bv. viciae genotypes.
The frequencies of the various R. leguminosarum bv. viciae genotypes isolated from nodules varied with the host of isolation, which confirmed previous studies (22, 23, 25). We found that fava beans were almost exclusively nodulated by rhizobial strains harboring nod type g, independently of the rhizobial genomic background, the plant genotype, the geographical location, and the cropping history of the soil of origin. Conversely, the frequency of nod type g was significantly lower in pea nodules than in the soil populations, which suggests that strains harboring this genotype are not competitive for nodule formation on pea plants. van Berkum et al. (41) also observed differences in nod gene fingerprints between pea and fava bean R. leguminosarum bv. viciae strains from various geographical origins. We found that the fava bean strains that they studied had the nod type g. This genotype was found to be distantly related to most of the other nod types. The nodD gene products of R. leguminosarum bv. viciae strain 248 (nod type g) and strain RCR 1001 (nod type closely related to several other nod types characterized in this study) have 22 differences in amino acid sequences (92.7% similarity), which may potentially influence the regulatory activity of NodD. However, polymorphism in the nodD gene also corresponded to polymorphism in other nod genes (24, 26) which encode Nod factor biosynthesis and are involved in host specificity.
The nod type g seems not to be more competitive than the other Sym genotypes available in the bulk soil for fava bean rhizosphere colonization. Its selection in fava bean nodules should occur later on at the stage of infection of roots or in the infection threads. It is possible that this nod gene type shows a specific or superior response to signal molecules (such as flavonoids) produced by fava bean roots, which act as inducers of nod gene expression and Nod factor production (9). Furthermore, 60% of the fava bean rhizobia isolated from various French soils and included in our study were previously examined for effectiveness of nitrogen fixation with fava bean (3). The quasitotality of them were effective, and we found that 91% of the strains that were effective had the nod type g. Conversely, several other nod types isolated from pea nodules were ineffective in nitrogen fixation with fava beans independently of the plant genotype. Collectively, these results give additional evidence of the specificity between particular genotypes of R. leguminosarum bv. viciae and fava beans and suggest that specific Sym plasmid-encoded genes should play a significant role in competition for nodulation of fava beans and probably in nitrogen-fixing effectiveness. The possible correlation between Sym genotypes and effectiveness of nitrogen fixation with fava bean needs to be further investigated by testing a larger sample of rhizobial strains with various nod types.
Peas and vetches showed less specificity than did fava beans for R. leguminosarum bv. viciae nod types. Additionally, vetches were less selective than were peas and fava beans for genomic backgrounds. We also found meaningful differences between peas and vetches in selection of nod types. Similar differences have been previously observed between lentils and peas (23).
In contrast to fava beans, peas discriminated also among rhizobial genomic backgrounds. We observed preferential associations between REP groups and nod types in pea nodule populations, while in bulk soils and in fava bean nodules, the Sym genotypes were randomly distributed in genomic backgrounds. Correlations between chromosomal and Sym plasmid genotypes have been previously observed within pea R. leguminosarum bv. viciae populations (11, 23, 50, 52). This suggests that both components of the rhizobial genome are involved in competitiveness for nodule formation with peas. On the basis of coinoculation experiments with various recipient strains lacking Sym plasmids and introduced Sym plasmids, Brewin et al. (5) concluded that the two genetic components were equally involved in competitiveness for nodule formation, while the competitiveness for growth in the rhizosphere (defined by the root surface) was due only to the genetic backgrounds. Our study did not provide clear evidence that the predominant genomic background in pea nodules, REP group A, was selected in the rhizosphere, but this might be hidden due to its high frequency in the bulk soil. However, the combined genotype A/d was significantly more frequent in the rhizosphere and in nodules than in the bulk soil, by contrast to type A/g, which was more or less counterselected in the rhizosphere. These results suggest that the Sym plasmid may contribute to the competitive growth in the rhizosphere of peas.
Various factors may be involved in competitive growth in the rhizosphere and in competition for root surface colonization. Among these factors is competition for nutrient sources. In some R. leguminosarum bv. viciae strains, the Sym plasmid carries genetic determinants for the catabolism of plant-associated organic compounds which may be produced or exudated in the rhizosphere of peas (34, 42). In particular, homoserine is an amino acid abundantly exuded by pea roots (42), and R. leguminosarum bv. viciae strains that were able to use homoserine as C and N sources were found to be prevalent in pea nodules while strains prevalent in fava bean nodules were not able to use homoserine in that way (22). The Sym plasmids of some R. leguminosarum bv. viciae strains also carry genes for the synthesis and the catabolism of rhizopines, which are organic compounds synthesized by rhizobia in the nodule formed with pea plants (34, 47) and, possibly, in the rhizosphere (18). However, although it has been shown elsewhere that the ability to catabolize the rhizopine confers a competitive advantage for nodule formation, there is no clear evidence of the role of rhizopines in competitive growth in the rhizosphere (17, 18).
The genetic background of R. leguminosarum bv. viciae strains includes non-Sym plasmids which are stably associated with the chromosomal backgrounds in the species R. leguminosarum, whatever the biovar and the Sym plasmid type (23). Chemotaxis and motility as well as bacteriocin production are other factors that may play a role in competition for rhizosphere and root colonization, and genes involved in chemotaxis and bacteriocin production have been identified and localized on non-Sym plasmids among R. leguminosarum bv. viciae strains (21, 36, 44, 49).
Collectively, these results emphasize the influence of the host plant on the diversity and the genetic structure of R. leguminosarum bv. viciae populations and reflect differences in the degree of host specificity within the Vicieae cross-inoculation group. Frequently isolated genotypes in nodule populations may not be highly competitive for nodule occupancy, and their success in nodule formation may just reflect their prevalence in soil possibly due to their ability to survive in the soil environment and/or their strong competitive saprophytic abilities. Conversely, dominant genotypes in nodules, especially of pea plants, are not necessarily dominant in bulk soil. Further work is needed to confirm the correlation among R. leguminosarum bv. viciae genotypes, rhizosphere competence, and competition for nodulation and subsequently to investigate the determinants of these processes.
Acknowledgments
This work was funded by INRA as part of AIP EcoSol and was also financially supported by a grant from the Conseil Régional de Bourgogne.
We thank V. Macheret, M.-C. Breuil, F. Revoy, and J. Sommer for technical assistance and Fabrice Dessaint for his helpful advice in statistical analyses.
REFERENCES
- 1.Amarger, N. 1981. Selection of Rhizobium strains on their competitive ability for nodulation. Soil. Biol. Biochem. 13:481-486. [Google Scholar]
- 2.Amarger, N. 1984. Evaluation of competition in Rhizobium spp., p. 300-305. In M. J. Klug and C. A. Reddy (ed.), Current perspectives in microbial ecology. American Society for Microbiology, Washington, D.C.
- 3.Amarger, N. 1988. The microbial aspects of faba bean culture, p. 173-178. In D. P. Beck and L. A. Materon (ed.), Nitrogen fixation by legumes in Mediterranean agriculture. International Center for Agricultural Research in the Dry Areas. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 4.Barrusio, E., and S. Houot. 1996. Rapid mineralization of the S-triazine ring of atrazine in soils in relation to soil management. Soil Biol. Biochem. 28:1341-1348. [Google Scholar]
- 5.Brewin, N. J., E. A. Wood, and J. P. W. Young. 1983. Contribution of the symbiotic plasmid to the competitiveness of Rhizobium leguminosarum. J. Gen. Microbiol. 129:2973-2977. [Google Scholar]
- 6.Bromfield, E. S. P., L. R. Barran, and R. Wheatcroft. 1995. Relative genetic structure of a population of Rhizobium meliloti isolated directly from soil and from nodules of alfalfa (Medicago sativa) and sweet clover (Melilotus alba). Mol. Ecol. 4:183-188. [Google Scholar]
- 7.Bromfield, E. S. P., I. B. Sinha, and M. S. Wolynetz. 1989. Is frequency of occurrence of indigenous Rhizobium meliloti in nodules of field grown plants related to intrinsic competitiveness? Soil Biol. Biochem. 21:607-609. [Google Scholar]
- 8.Demezas, D. H., and P. J. Bottomley. 1986. Autoecology in rhizospheres and nodulating behavior of indigenous Rhizobium trifolii. Appl. Environ. Microbiol. 52:1014-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denarié, J., F. Debelle, and J. C. Prome. 1996. Rhizobium lipo-chitooligosaccharide nodulation factors: signaling molecules mediating recognition and morphogenesis. Annu. Rev. Biochem. 65:503-535. [DOI] [PubMed] [Google Scholar]
- 10.Dowling, D. N., and W. J. Broughton. 1986. Competition for nodulation of legumes. Annu. Rev. Microbiol. 40:131-157. [DOI] [PubMed] [Google Scholar]
- 11.Engvild, K. C., E. S. Jensen, and L. Skot. 1990. Parallel variation in isoenzyme and nitrogen fixation markers in a Rhizobium population. Plant Soil 128:283-286. [Google Scholar]
- 12.Evans, J., A. Gregory, N. Dobrowolski, S. G. Morris, G. E. O'Connor, and C. Wallace. 1996. Nodulation of field-grown Pisum sativum and Vicia faba: competitiveness of inoculant strains of Rhizobium leguminosarum bv. viciae determined by an indirect, competitive ELISA method. Soil Biol. Biochem. 28:247-255. [Google Scholar]
- 13.Excoffier, L., P. E. Smouse, and J. M. Quattro. 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: applications to human mitochondrial DNA restriction data. Genetics 131:479-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geniaux, E., G. Laguerre, and N. Amarger. 1993. Comparison of geographically distant populations of Rhizobium isolated from root nodules of Phaseolus vulgaris. Mol. Ecol. 2:295-302. [Google Scholar]
- 15.Handley, B. A., A. J. Hedges, and J. E. Beringer. 1998. Importance of host plants for detecting the population diversity of Rhizobium leguminosarum biovar viciae in soil. Soil Biol. Biochem. 30:241-249. [Google Scholar]
- 16.Hartmann, A., J. J. Giraud, and G. Catroux. 1998. Genotypic diversity of Sinorhizobium (formerly Rhizobium) meliloti strains isolated directly from a soil and from nodules of alfalfa (Medicago sativa) grown in the same soil. FEMS Microbiol. Ecol. 25:107-116. [Google Scholar]
- 17.Heinrich, K., D. M. Gordon, M. H. Ryder, and P. J. Murphy. 1999. A rhizopine strain of Sinorhizobium meliloti remains at a competitive nodulation advantage after an extended period in the soil. Soil Biol. Biochem. 31:1063-1065. [Google Scholar]
- 18.Heinrich, K., M. H. Ryder, and P. J. Murphy. 2001. Early production of rhizopine in nodules induced by Sinorhizobium meliloti strain L5-30. Can. J. Microbiol. 47:165-171. [DOI] [PubMed] [Google Scholar]
- 19.Hill, M. O. 1973. Diversity and evenness: a unifying notation and its consequences. Ecology 54:427-432. [Google Scholar]
- 20.Hirsch, P. R. 1979. Plasmid determined bacteriocin production by Rhizobium leguminosarum. J. Gen. Microbiol. 113:219-228. [Google Scholar]
- 21.Hirsch, P. R., M. Van Montagu, A. W. B. Johnston, N. J. Brewin, and J. Schell. 1980. Physical identification of bacteriocinogenic, nodulation and other plasmids in strains of Rhizobium leguminosarum. J. Gen. Microbiol. 120:403-412. [Google Scholar]
- 22.Hynes, M. F., and M. P. O'Connell. 1990. Host plant effect on competition among strains of Rhizobium leguminosarum. Can. J. Microbiol. 36:864-869. [Google Scholar]
- 23.Laguerre, G., E., Geniaux, S. I. Mazurier, R. Rodriguez Casartelli, and N. Amarger. 1993. Conformity and diversity among field isolates of Rhizobium leguminosarum bv. viciae, bv. trifolii, and bv. phaseoli revealed by DNA hybridization using chromosome and plasmid probes. Can. J. Microbiol. 39:412-419. [Google Scholar]
- 24.Laguerre, G., P. Mavingui, M. R. Allard, M. P. Charnay, P. Louvrier, S. I. Mazurier, L. Rigottier-Gois, and N. Amarger. 1996. Typing of rhizobia by PCR DNA fingerprinting and PCR-restriction fragment length polymorphism analysis of chromosomal and symbiotic gene regions: application to Rhizobium leguminosarum and its different biovars. Appl. Environ. Microbiol. 62:2029-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laguerre, G., S. I. Mazurier, and N. Amarger. 1992. Plasmid profiles and restriction fragment length polymorphism of Rhizobium leguminosarum bv. viciae in field populations. FEMS Microbiol. Ecol. 101:17-26. [Google Scholar]
- 26.Laguerre, G., S. M. Nour, V. Macheret, J. Sanjuan, P. Drouin, and N. Amarger. 2001. Classification of rhizobia based on nodC and nifH gene analysis reveals a close phylogenetic relationship among Phaseolus vulgaris symbionts. Microbiology 147:981-993. [DOI] [PubMed] [Google Scholar]
- 27.Leung, K., K. Yap, N. Dashti, and P. J. Bottomley. 1994. Serological and ecological characteristics of a nodule-dominant serotype from an indigenous soil population of Rhizobium leguminosarum bv. trifolii. Appl. Environ. Microbiol. 60:408-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leung, K., F. N. Wanjage, and P. J. Bottomley. 1994. Symbiotic characteristics of Rhizobium leguminosarum bv. trifolii isolates which represent major and minor nodule-occupying chromosomal types of field-grown subclover (Trifolium subterraneum L.). Appl. Environ. Microbiol. 60:427-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Louvrier, P., G. Laguerre, and N. Amarger. 1995. Semiselective medium for isolation of Rhizobium leguminosarum from soils. Soil Biol. Biochem. 27:919-924. [Google Scholar]
- 30.Louvrier, P., G. Laguerre, and N. Amarger. 1996. Distribution of symbiotic genotypes in Rhizobium leguminosarum biovar viciae populations isolated directly from soils. Appl. Environ. Microbiol. 62:4202-4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meade, J., P. Higgins, and F. O'Gara. 1985. Studies on the inoculation and competitiveness of a Rhizobium leguminosarum strain in soils containing indigenous rhizobia. Appl. Environ. Microbiol. 49:899-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moawad, H. A., W. R. Ellis, and E. L. Schmidt. 1984. Rhizosphere response as a factor in competition among three serogroups of indigenous Rhizobium japonicum for nodulation of field-grown soybeans. Appl. Environ. Microbiol. 47:607-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moënne-Loccoz, Y., and R. W. Weaver. 1995. Plasmids influence growth of rhizobia in the rhizosphere of clover. Soil Biol. Biochem. 27:1001-1004. [Google Scholar]
- 34.Murphy, P. J., W. Wexler, W. Grzemski, J. P. Rao, and D. Gordon. 1995. Rhizopines—their role in symbiosis and competition. Soil Biol. Biochem. 27:525-529. [Google Scholar]
- 35.Oresnik, I. J., L. A. Pacarynuk, S. A. P. O'Brien, C. K. Yost, and M. F. Hynes. 1998. Plasmid-encoded catabolic genes in Rhizobium leguminosarum bv. trifolii: evidence for a plant-inducible rhamnose locus involved in competition for nodulation. Mol. Plant Microbe Interact. 11:1175-1185. [Google Scholar]
- 36.Oresnik, I. J., S. Twelker, and M. F. Hynes. 1999. Cloning and characterization of a Rhizobium leguminosarum gene encoding a bacteriocin with similarities to RTX toxins. Appl. Environ. Microbiol. 65:2833-2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robert, F. M., and E. L. Schmidt. 1985. Response of three indigenous serogroups of Rhizobium japonicum to the rhizosphere of pre-emergent seedlings of soybean. Soil Biol. Biochem. 17:579-580. [Google Scholar]
- 38.Schofield, P. R., A. H. Gibson, W. F. Dudman, and J. M. Watson. 1987. Evidence for genetic exchange and recombination of Rhizobium symbiotic plasmids in a soil population. Appl. Environ. Microbiol. 53:2942-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Streeker, J. G. 1994. Failure of inoculant rhizobia to overcome the dominance of indigenous strains for nodulation. Can. J. Microbiol. 40:513-522. [Google Scholar]
- 40.Triplett, E. W., and M. J. Sadowsky. 1992. Genetics of competition for nodulation of legumes. Annu. Rev. Microbiol. 46:399-428. [DOI] [PubMed] [Google Scholar]
- 41.van Berkum, P., D. Beyene, F. T. Vera, and H. H. Keyser. 1995. Variability among Rhizobium strains originating from nodules of Vicia faba. Appl. Environ. Microbiol. 61:2649-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Egeraat, A. W. S. M. 1975. Exudation of ninhydrin-positive compounds by pea-seedling roots: a study of the sites of exudation and of the composition of the exudates. Plant Soil 42:15-36. [Google Scholar]
- 43.Velasquez, E., P. F. Mateos, N. Velasco, F. Santos, P. A. Burgos, P. Villadas, N. Toro, and E. Martinez-Molina. 1999. Symbiotic characteristics and selection of autochthonous strains of Sinorhizobium meliloti populations in different soils. Soil Biol. Biochem. 31:1039-1047. [Google Scholar]
- 44.Venter, A. P., S. Twelker, I. J. Oresnik, and M. F. Hynes. 2001. Analysis of the genetic region encoding a novel rhizobiocin from Rhizobium leguminosarum bv. viciae strain 306. Can. J. Microbiol. 47:495-502. [DOI] [PubMed] [Google Scholar]
- 45.Vincent, J. M. 1970. A manual for the practical study of root-nodule bacteria. IBP handbook no. 15. Blackwell Scientific Publications Ltd., Oxford, United Kingdom.
- 46.Vlassak, K. M., and J. Vanderleyden. 1997. Factors influencing nodule occupancy by inoculant rhizobia. Crit. Rev. Plant Sci. 16:163-229. [Google Scholar]
- 47.Wexler, M., D. Gordon, and P. J. Murphy. 1995. The distribution of inositol rhizopine genes in Rhizobium populations. Soil Biol. Biochem. 27:531-537. [Google Scholar]
- 48.Wilson, R. A., B. A. Handley, and J. E. Beringer. 1998. Bacteriocin production and resistance in a field population of Rhizobium leguminosarum biovar viciae. Soil Biol. Biochem. 30:413-417. [Google Scholar]
- 49.Yost, C. K., P. Rochepeau, and M. F. Hynes. 1998. Rhizobium leguminosarum contains a group of genes that appear to code for methyl-accepting chemotaxis proteins. Microbiology 144:1945-1956. [DOI] [PubMed] [Google Scholar]
- 50.Young, J. P. W., and M. Wexler. 1988. Sym plasmid and chromosomal genotypes are correlated in field populations of Rhizobium leguminosarum. J. Gen. Microbiol. 134:2731-2739. [Google Scholar]
- 51.Zézé, A., L. A. Mutch, and J. P. W. Young. 2001. Direct amplification of nodD from community DNA reveals the genetic diversity of Rhizobium leguminosarum in soil. Environ. Microbiol. 3:363-370. [DOI] [PubMed] [Google Scholar]
- 52.Zhang, X.-X., B. Kosier, and U. B. Priefer. 2001. Genetic diversity of indigenous Rhizobium leguminosarum bv. viciae isolates nodulating two different host plants during soil restoration with alfalfa. Mol. Ecol. 10:2297-2305. [DOI] [PubMed] [Google Scholar]