Abstract
The effect of signal compounds and of different incubation conditions on the culturability (i.e., the fraction of all cells capable of growth) of natural bacterioplankton from the eutrophic lake Zwischenahner Meer was investigated over a period of 20 months. Numbers of growing cells were determined by the most-probable-number technique in liquid media containing low concentrations (10 μM) of the signal compounds N-(oxohexanoyl)-dl-homoserine lactone, N-(butyryl)-dl-homoserine lactone, cyclic AMP (cAMP), or ATP. cAMP was the most effective signal compound, leading to significantly increased cultivation efficiencies of up to 10% of the total bacterial counts. Microautoradiography with [2,8-3H]cAMP, combined with fluorescence in situ hybridization, demonstrated that cAMP was taken up by 18% of all cells. The bacterial cAMP uptake systems had a very low Km value of ≤1 nM. Analysis of the cultured bacteria by 16S rRNA gene fingerprinting showed that different bacterial phylotypes were recovered in the presence and in the absence of cAMP. Consequently, the addition of cAMP caused a stimulation of otherwise nonculturable bacteria. Phylogenetically different bacteria were also recovered at different temperatures and oxygen partial pressures. Throughout the study period, mainly members of the β-subclass of the Proteobacteria were cultivated. In addition, some members of the Actinomycetales were enriched. Quantification by culture-independent fluorescence in situ hybridization demonstrated that β-Proteobacteria and Actinomycetales also dominated the natural bacterioplankton assemblage. Sequence comparison revealed that two members of the Actinomycetales which reached high numbers in the natural bacterioplankton assemblage could actually be enriched by our cultivation approach.
Culture-independent studies continue to reveal novel 16S rRNA sequence types. One-third of the ∼40 bacterial divisions recognized so far consist entirely of not-yet-cultured bacteria which have been described solely by their 16S rRNA sequences (19, 26). Others, such as the Acidobacteria or green nonsulfur bacteria, comprise only very few cultured representatives but numerous not-yet-cultured bacteria (9, 17, 21, 26, 37). Other microbial communities consist mainly of well-known divisions such as the β-Proteobacteria, which dominate the freshwater bacterioplankton (22). Still, the sequences obtained in almost all cases do not correspond to cultured members but rather represent novel lineages (23, 25). Since these novel bacterial divisions and lineages dominate in many environments (9, 17), existing enrichment and isolation strategies need to be refined and novel cultivation approaches need to be developed in order to fully understand the functional role of bacteria in their natural environment.
We define culturability as the fraction of all cells capable of growth in artificial laboratory media. One important prerequisite for increased culturability are low nutrient concentrations, which improve the recovery of bacteria from natural samples (1, 14, 15, 20), possibly because this prevents starved cells from substrate-accelerated death (16). Among other factors (8), cell-to-cell communication is essential for the growth of some bacteria in artificial media (8, 13, 24, 38). Acyl homoserine lactones (acyl-HSLs), especially N-(oxohexanoyl)-dl-HSL (OHHL) and N-(butyryl)-dl-HSL (BHL), represent the best-understood signal compounds and are used by many gram-negative bacteria capable of quorum sensing (D. Kirke, The quorum sensing site [http://www.nottingham.ac.uk/quorum/table.htm], 2001). Gram-positive bacteria have been shown to communicate by using peptides (32). Recently, cyclic AMP (cAMP), which in gram-negative bacteria is involved in regulation of a large number of genes (11, 47), was shown to significantly enhance the cultivation success of bacterioplankton from the Baltic Sea (13). It is unknown whether cAMP has a similar effect on freshwater planktonic bacteria and whether it affects all members of the bacterioplankton communities equally during their seasonal succession. Another compound of potential significance for the cultivation of bacteria is ATP, which occurs in concentrations of up to 1 nM in seawater (6) and freshwater (44) and is probably derived from phytoplankton. Dissolved ATP is utilized by natural bacterioplankton at high turnover rates (44), but its effect on the culturability of planktonic bacteria is not known.
Because of the high diversity of natural bacterial communities, it is unlikely that one set of incubation conditions will permit the growth of the majority of bacteria. The cultivation efficiency can be increased by using different growth media in parallel (7). However, the effects of different incubation conditions, such as oxygen partial pressure or temperature, on cultivation success have not been investigated in a systematic manner so far.
In the present study, the effects of acyl-HSLs, cAMP, ATP, and different incubation conditions on the culturability of freshwater bacterioplankton were assessed in a systematic manner. Freshwater bacterioplankton was chosen because such communities may contain around 550 species (29) and therefore are much less diverse and easier to investigate than soil microbial communities, in which bacterial diversity is 2 orders of magnitude higher (46, 50).
MATERIALS AND METHODS
Sampling site and sample processing.
The shallow eutrophic lake Zwischenahner Meer in northern Germany (53°12′N, 8°0′E) was chosen for our investigations. The lake has a surface area of 520 ha, a mean depth of 3.3 m, and a maximum depth of 5.0 m and is characterized by a simple hydrodynamic structure, since the entire water column is mixed frequently (29). The sampling site was located at the head of a pier extending 20 m from the east shore. Twenty-five water samples were collected from a depth of 0.3 m at regular intervals over a time period of 20 months (between September 1999 and April 2001).
Lake water samples were prefiltered through a 20-μm mesh plankton net and then immediately processed by inoculating microtiter plates for cultivation approaches and by fixation of a subsample for total cell counts, using glutaraldehyde at a final concentration of 2% (vol/vol). For extraction of genomic DNA, 250-ml water samples were concentrated on 0.2-μm-pore-size polycarbonate filters (Millipore, Bedford, Mass.). Water temperature and conductivity were determined with an LF95 conductivity meter equipped with a TetraCon 96 electrode (WTW, Weilheim, Germany).
Bacterial abundance.
Fixed water samples were filtered onto 0.1-μm-pore-size polycarbonate filters (Nuclepore Track-Etch membrane; Whatman, Springfield Mill, United Kingdom). Total bacterial cell counts were determined by epifluorescence microscopy after staining with 4′,6-diamidino-2-phenylindole (DAPI) as described earlier (13).
Effect of signal molecules on cultivation success.
The effect of different signal compounds on the cultivation of heterotrophic bacterioplankton was studied in most-probable-number (MPN) series. Synthetic freshwater (10) served as the basal medium and was supplemented with a fatty acid mixture containing formate, acetate, and propionate (200 μM each); an amino acid mixture containing all 20 amino acids (200 μM each); Tween 80 (0.001%, vol/vol); glucose; and the sodium salts of pyruvate, citrate, succinate, and 2-oxoglutarate (200 μM each) (30).
cAMP (Sigma Chemical Co., St. Louis, Mo.), N-(ketocaproyl)-dl-HSL (synonymous with OHHL) (Sigma), BHL (Fluka Chemie AG, Buchs, Switzerland), or ATP (Sigma) was added to the basal medium to a final concentration of 10 μM. Mixtures for control experiments received AMP (Sigma) instead of cAMP or ATP, capronic acid sodium salt (synonymous with hexanoic acid) (Fluka) in combination with (S)-α-amino-γ-butyrolactone hydrochloride (synonymous with HSL) (Aldrich, Sheboygan, Wis.) instead of OHHL, and butyric acid sodium salt (Fluka) in combination with HSL instead of BHL.
The MPN series were set up in 10-fold serial dilutions and in seven parallels in 96-well polystyrene microtiter plates (Corning Inc., New York, N.Y.). Each well contained a volume of 200 μl. Incubations for experiments with cAMP as signal compound were carried out under different partial pressures of oxygen and temperatures: (i) 21% O2 and 20°C, (ii) 3% O2 and 20°C, (iii) 0% O2 and 20°C, (iv) 21% O2 and 25°C, (v) 21% O2 and 15°C, and (vi) 21% O2 and 4°C. By contrast, incubations for experiments with ATP, BHL, and OHHL were all carried out at 21% O2 and 20°C. Thus, a maximum of nine different cultivation experiments could be performed in parallel. After 6 weeks, growth was monitored visually by turbidity and the most probable cell numbers were calculated by using a computer program (31). The statistical significance of differences in MPNs between samples were calculated as described by Jones (30).
Subsamples from the highest positive dilutions were plated onto solidified (1.2% agar) complex medium YPG, which contains yeast extract (0.075%, wt/vol), peptone (0.15%, wt/vol), and glucose (0.075%, wt/vol) (10). This method of subcultivation repeatedly proved to be successful, indicating that the bacteria enriched in liquid media are also capable of growing on solid complex media.
DNA extraction.
To remove contaminating humic acids, which were abundant in water from Zwischenahner Meer (10), DNA of bacterial cells collected on the polycarbonate filters was extracted with Genomic tips 20/G from Qiagen (Hilden, Germany), by using a modification of the protocol of the manufacturer. Filters were cut in small pieces and treated as specified by the manufacturer, but all volumes of the necessary lysis and washing buffers were increased by a factor of 1.5. The DNA eluted from the genomic tips was concentrated by using Centricon YM-100 centrifugal filter devices (Millipore). DNA from cultured bacteria in the highest positive dilutions of each MPN series was liberated by the freeze-thaw technique as described recently (13).
16S rRNA gene fingerprints.
Bacterial 16S rRNA genes from natural the bacterial community and cultured bacteria were amplified by using primers GC341f and 907r (39). As the template, 50 ng of extracted DNA or 1 μl of the freeze-thawed DNA preparations was used. Amplification was performed by step-down PCR (42), which includes 10 cycles at an annealing temperature of 62°C, followed by 20 cycles at 57°C. Each cycle started with a melting step at 96°C and ended with an extension step at 72°C.
PCR products were separated by denaturing gradient gel electrophoresis (DGGE) in 6% (wt/vol) polyacrylamide gels containing a linear gradient of 30 to 70% denaturant as described earlier (42). After the gels were stained with ethidium bromide, DNA bands of interest were excised with a sterile scalpel, and the DNA was eluted overnight at 4°C in 20 μl of sterile double-distilled water. One microliter of the eluate was reamplified, and the DNA was purified and quantified by dye binding with PicoGreen (42).
Sequencing and phylogenetic analysis.
Sequencing of the 16S rRNA gene fragments from DGGE bands was performed with the ABI Prism BigDye terminator cycle sequencing ready-reaction kit (Perkin-Elmer Applied Biosystems GmbH, Weiterstadt, Germany) and the ABI Prism 310 genetic analyzer (Applied Biosystems, Foster City, Calif.) as specified by the manufacturer. The 16S rRNA gene sequences obtained were added to the ribosomal DNA sequence database of the Technical University of Munich (spring 2001 release), using the program package ARB (http://www.mikro.biologie.tu-muenchen.de). Sequences of closest relatives to the sequences of this study (according to a BLAST search in the nucleotide database of the National Center for Biotechnology Information) were also imported into the ARB database. The Fast Aligner version 1.03 tool was used for automatic sequence alignment, which was checked and corrected manually according to secondary-structure information. Sequences longer than 1,400 bp were used for constructing a tree by maximum likelihood, with a 50% filter. Shorter sequences were then added by the parsimony tool, again with a 50% filter.
Fluorescence in situ hybridization (FISH).
Water samples from 4 and 22 April 2002 were fixed in 50% (vol/vol) ethanol and stored at 4°C. Teflon-coated glass slides (Omnilab, Bremen, Germany) treated with gelatin (0.1%) and KCr(SO4)2 (0.01%) were used for hybridization. Four 20-μl portions of sample were applied to each well and allowed to dry at 60°C. Dehydration and hybridization were carried out at 46°C as previously described (12). Twenty-five nanograms of the probes EUB338, NON338, ALF968, BET42a, BET42a competitor, GAM42a, GAM42a competitor, and CF319a (22) was used for hybridization at a formamide concentration of 35% (22). For detection of gram-positive bacteria, probe HGC69a (45) was employed. Dried samples on the glass slides were pretreated with a lysozyme solution (10 mg ml−1 in phosphate-buffered saline [PBS] buffer [130 mM NaCl, 30 mM Na phosphate, pH 7.2]) for 20 min at room temperature. Subsequently, the slides were rinsed with distilled water and then dehydrated and hybridized (with 20% formamide).
After hybridization, all of the slides were washed for 20 min at 48°C in buffer containing appropriate NaCl concentrations for the formamide concentrations applied (48) and then rinsed with distilled water. After drying, samples were stained with DAPI (12). For each sample, 40 randomly chosen microscopic fields containing an average of 80 to 100 cells were enumerated. All probe-specific cell counts are presented as the percentage of the cells visualized by DAPI. Cell counts were corrected by subtracting the counts obtained with the negative control NON338.
cAMP uptake experiments.
Uptake of cAMP was assessed with water samples from 22 April 2002. The time course of cAMP uptake was studied by adding 2 μCi of [2,8-3H]cAMP (Moravek Biochemicals, Brea, Calif.) to 7-ml samples of lake water (corresponding to a final cAMP concentration of 16.8 nM). Samples were incubated at 15°C. At regular time intervals, biological activity in samples was stopped by addition of ethanol to a final concentration of 50% (vol/vol). To determine whether cAMP is taken up by a specific transport system rather than by simple diffusion, competitive uptake experiments with unlabeled cAMP were conducted. In these experiments, 7-ml samples supplemented with unlabeled cAMP at various concentrations were preincubated for 45 min at 15°C. Control samples received formaldehyde at a final concentration of 4% (vol/vol). Subsequently, 2 μCi of [2,8-3H]cAMP was added and incubation was carried out for 2 h, after which uptake was stopped by addition of ethanol. A volume of 2.8 ml of ethanol-fixed cells was then withdrawn and centrifuged for 15 min at 15,000 × g. Cell pellets were washed in 500 μl of PBS buffer, centrifuged again, and then resuspended in 500 μl of PBS. This solution was added to 1 ml of scintillation fluid (Zinsser Analytik, Frankfurt, Germany), and the radioactivity was counted by liquid scintillation spectrometry in a Rackbeta 1209 liquid scintillation counter (Perkin-Elmer Wallac, Freiburg, Germany).
In order to determine the percentage of natural bacteria capable of taking up cAMP, we applied microautoradiography in combination with FISH (34, 41). Cells of the natural bacterioplankton were first incubated in the presence of radiolabeled cAMP and then fixed and washed as described above. Subsequently, in situ hybridization was performed with probe EUB338 (see above), using coverslips instead of glass slides. After hybridization, the dried coverslips were covered with the photographic emulsion (type NTB2; Kodak, Rochester, N.Y.) as described previously (40). Coverslips were stored for 14 days at 4°C for emulsion exposure and subsequently developed. The coverslips were then placed upside down (i.e., with cells facing down) on a glass slide coated with the antifading solution DABCO (13). By this technique even small radiolabeled cells can be detected, because silver grains are found below the cells rather than covering them.
Nucleotide sequence accession numbers.
The GenBank accession numbers of the sequences determined in this study are AF488636 to AF488674.
RESULTS
Effect of signal compounds, oxygen, and temperature on culturability.
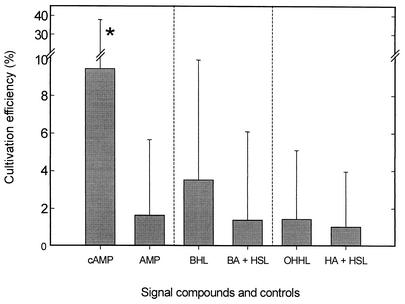
During the study period, maximum cultivation efficiencies for heterotrophic bacteria ranged between 0.25 and 10.4% of the total cell numbers. Values of around 10% were reached on several occasions and during different seasons (Table 1). On 14 sampling dates, maximum cultivation efficiencies were attained in growth media supplemented with cAMP. In six of these cases, the increase in culturability upon addition of cAMP was statistically significant. ATP was the only other compound which stimulated the growth of bacteria in three additional cases (Table 1). In contrast, no significant effect on bacterial growth was observed in media containing BHL or OHHL (Table 1; Fig. 1). Of the different incubation conditions tested, an oxygen partial pressure of 21% most frequently resulted in maximum cultivation efficiencies, whereas anoxic conditions were always less favorable. Optimum temperatures for cultivation ranged between 15 and 25°C. Incubation at a temperature of 4°C never yielded significantly enhanced MPNs (Table 1).
TABLE 1.
Cultivation efficiencies and conditions for maximum culturability of planktonic bacteria from Zwischenahner Meera
| Date (day.mo.yr) | Conditions for maximum culturability | Maximum culturability (% of total counts)b | Control culturability (% of total counts) | Level of significance (P) |
|---|---|---|---|---|
| 27.9.1999 | cAMP, 21% O2, 20°C | 1.38 (4.98/0.20) | 0.23 (0.72/0.02) | 0.01 |
| 21.10.1999 | cAMP, 21% O2, 20°C | 9.44 (37.8/1.52) | 1.64 (5.65/0.40) | 0.01 |
| 23.11.1999 | cAMP, 21% O2, 15°C | 0.75 (2.72/0.11) | 1.14 (3.69/0.13) | —c |
| 14.1.2000 | AMP, 3% O2, 20°C | 0.40 (1.19/0.04) | 0.48 (1.67/0.05) | — |
| 10.2.2000 | cAMP, 3% O2, 20°C | 2.97 (8.76/0.26) | 2.14 (6.39/0.20) | — |
| 21.2.2000 | cAMP, 21% O2, 15°C | 0.29 (0.84/0.03) | 0.29 (0.84/0.03) | — |
| 30.3.2000 | cAMP, 3% O2, 20°C | 6.51 (17.0/0.58) | 1.19 (7.77/0.33) | 0.10 |
| 6.4.2000 | cAMP, 21% O2, 20°C | 0.25 (0.75/0.02) | 0.17 (0.54/0.02) | — |
| 4.5.2000 | cAMP, 21% O2, 20°C | 2.06 (6.22/0.19) | 1.98 (5.99/0.55) | — |
| 11.5.2000 | BHL, 21% O2, 15°C | 1.89 (6.02/0.20) | 0.89 (3.45/0.16) | — |
| 16.6.2000 | cAMP, 21% O2, 25°C | 10.4 (38.3/1.56) | 5.59 (22.2/0.62) | — |
| 23.6.2000 | ATP, 21% O2, 20°C | 1.74 (5.24/0.16) | 0.34 (1.64/0.09) | 0.02 |
| 27.7.2000 | AMP, 21% O2, 15°C | 5.43 (18.6/0.54) | 5.18 (16.7/0.50) | — |
| 11.8.2000 | BHL, 21% O2, 20°C | 1.84 (5.84/0.20) | 1.12 (4.05/0.16) | — |
| 15.8.2000 | ATP, 21% O2, 20°C | 3.16 (9.33/0.28) | 2.83 (8.28/0.25) | — |
| 24.8.2000 | cAMP, 21% O2, 15°C | 0.53 (2.03/0.10) | 0.10 (0.33/0.03) | 0.02 |
| 22.9.2000 | cAMP, 21% O2, 15°C | 0.85 (3.16/0.17) | 0.58 (2.37/0.08) | — |
| 19.10.2000 | AMP, 3% O2, 20°C | 0.32 (1.49/0.07) | 0.15 (0.46/0.01) | 0.20 |
| 21.11.2000 | ATP, 21% O2, 20°C | 6.69 (25.9/1.23) | 0.45 (1.79/0.05) | 0.001 |
| 23.11.2000 | cAMP, 3% O2, 20°C | 1.51 (4.80/0.16) | 0.67 (2.61/0.11) | 0.20 |
| 13.12.2000 | AMP, 21% O2, 25°C | 1.37 (4.94/0.20) | 0.83 (3.38/0.12) | — |
| 16.1.2001 | cAMP, 21% O2, 15°C | 1.18 (3.83/0.13) | 0.24 (0.71/0.02) | 0.05 |
| 21.2.2001 | cAMP, 3% O2, 20°C | 3.34 (9.87/0.30) | 1.58 (5.05/0.17) | — |
| 20.3.2001 | ATP, 21% O2, 15°C | 9.16 (36.0/1.61) | 3.03 (9.08/0.28) | 0.20 |
| 24.4.2001 | AMP, 21% O2, 20°C | 0.89 (2.85/0.10) | 1.21 (3.65/0.11) | — |
Not listed are MPN series which exhibited a significant increase in culturability compared to the controls but which yielded lower absolute culturability values.
Upper and lower 95% confidence levels given in parentheses; values in boldface indicate cultivation efficiencies of >5%.
—, not significant (P > 0.20).
FIG. 1.
Effect of signal compounds on the efficiency of cultivation of planktonic bacteria from Zwischenahner Meer (sampled on 21 October 1999). Cultivation efficiency was determined by the MPN technique and is given as the percentage of total cell counts. Error bars indicate 95% confidence intervals. A significant increase in cultivation success compared to the control (P < 0.01) is indicated by an asterisk. Control compounds were AMP, butyric acid (BA), hexanoic acid (HA), and HSL.
Effect of cultivation conditions on the phylotypes of bacteria recovered by the MPN approach.
Changes in the cultivation efficiencies per se cannot provide information on the mechanism of stimulation by signal compounds. An increase of MPN values in the presence of cAMP or ATP may be caused by a selective stimulation of otherwise nongrowing bacteria. Alternatively, a larger fraction of the cells of the same bacterial population may start growing upon addition of a signal compound. Hence, the bacteria stimulated in the presence of cAMP or ATP were further characterized by 16S rRNA gene fingerprinting.
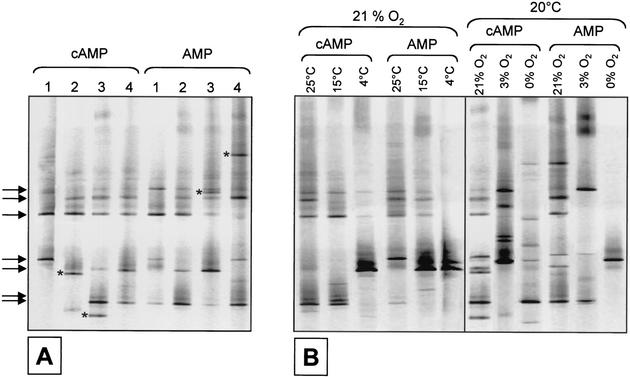
It was suspected that, even under identical incubation conditions, the taxonomic composition of growing bacteria may vary. In order to be able to account for such statistical fluctuations, we first analyzed samples in which cAMP or ATP did not exhibit a stimulating effect (Fig. 2A). In four identical MPN series inoculated in parallel, seven of the analyzed 16S rRNA gene fingerprints were detected in the majority of samples; a few DNA bands were present in only one of the MPN series (Fig. 2A). Also, similar fingerprints were obtained irrespective of the addition of cAMP or AMP (Fig. 2A). By contrast, different bacterial phylotypes were cultured in the highest positive dilutions when MPN series were incubated at different temperatures (Fig. 2B, left panel). Incubation at different oxygen partial pressures clearly caused the most pronounced changes in the composition of cultured bacteria (Fig. 2B, right panel).
FIG. 2.
Analysis of 16S rRNA gene fingerprints from the highest positive dilutions of MPN series. As an example, results obtained with a sample from 24 April 2001 are shown. On this date, cAMP did not affect the cultivation success. (A) Variability of fingerprints recovered in four different parallels of media containing cAMP or AMP. Arrows indicate fingerprints recovered from media containing cAMP as well as AMP. Asterisks indicate unique fingerprints. (B) Effect of incubation temperature (left panel) and oxygen partial pressure (right panel) on the phylotypes obtained in the highest positive dilutions of media supplemented with either cAMP or AMP.
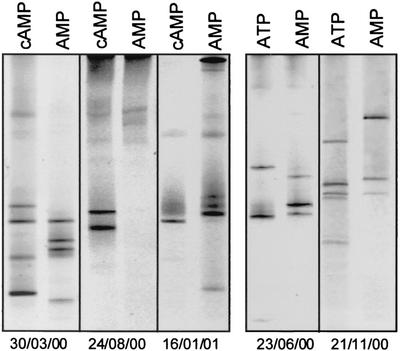
Subsequently, we investigated whether a significant increase in culturability could be attributed to a stimulation of otherwise nongrowing bacteria. Based on the observed increases in MPN values, five different experiments were selected (Table 1; Fig. 3). In these cases, most of the 16S rRNA gene fingerprints detected in the highest positive dilutions of media containing cAMP or ATP differed from those of the respective controls (Fig. 3). Analysis of other MPN series in which cAMP also led to significant increases but yielded lower absolute cultivation efficiencies (≤1%) showed similar results (data not shown).
FIG. 3.
Comparison of 16S rRNA gene fingerprints of bacteria growing in the presence of cAMP or ATP with those detected in control assay mixtures supplemented with AMP. Only MPN series in which bacterial growth was significantly stimulated by cAMP or ATP are shown (compare Table 1).
Diversity of the natural bacterial community and the cultured fraction.
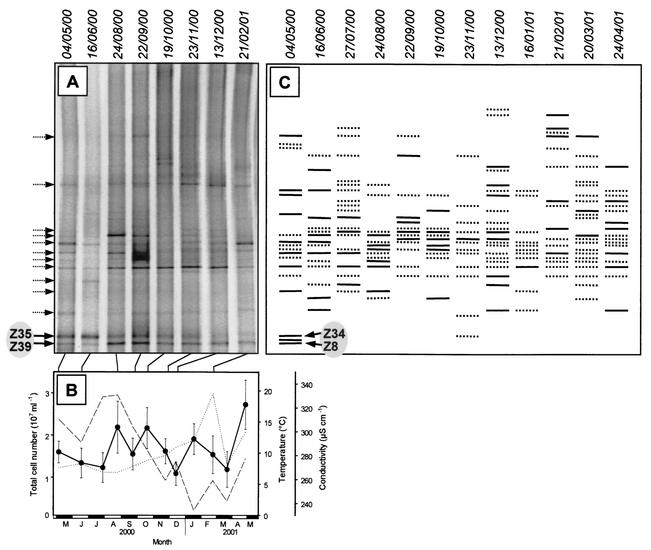
The ultimate goal of the present study was to cultivate those bacteria which dominate in natural habitats but so far have escaped all previous cultivation attempts. Single wells of the highest positive dilutions in most (i.e., 71%) of the cases yielded only one 16S rRNA gene fingerprint on DGGE gels and hence most likely harbored pure cultures. In order to more easily detect among our cultures those bacteria which were potential candidates for numerically dominant members of the natural bacterioplankton assemblage, the fingerprints generated from the highest positive dilutions of all MPN series were combined and the resulting patterns were compared to the fingerprints of the natural bacterioplankton community (Fig. 4A and C). Despite the considerable seasonal changes in environmental conditions (Fig. 4B), only minor fluctuations were observed in fingerprint patterns generated from the total bacterial community from Zwischenahner Meer (Fig. 4A). This stability of the bacterioplankton assemblage corresponds to the time course of total cell numbers, which also exhibited only slight seasonal variations of between 1 × 107 and 3 × 107 cells ml−1. In contrast to the entire bacterioplankton community, however, the diversity of the cultured fraction varied strongly over the study period (Fig. 4C). This observation indicates that the same incubation conditions stimulated the growth of different members of the bacterioplankton assemblage when applied at different sampling dates. Hence, the composition of the cultivated fraction appears to be controlled by the physiological state of individual bacteria and the specific incubation conditions chosen rather than the overall composition of the bacterioplankton community.
FIG. 4.
(A) Composition of bacterioplankton communities in Zwischenahner Meer as analyzed by 16S rRNA gene fingerprinting. (B) Total cell numbers (•), water temperature (- - -), and conductivity (. . .) at the same sampling dates as in panel A. (C) Schematic representation of cumulative 16S rRNA gene fingerprints obtained from separate enrichments on different dates, either in the presence of cAMP alone (solid lines) or with AMP or both cAMP and AMP (dotted lines), at all different incubation temperatures and oxygen partial pressures. Dotted arrows on the left of panel A indicate positions at which fingerprints of cultured strains correspond to those of the natural community but, when sequenced, nevertheless contained different sequences. The sequence obtained from band Z34 was identical to that from Z35, and the sequence from band Z8 was identical to that from Z39 (compare Fig. 5).
Among the 41 fingerprints of the most frequently cultured bacteria, 13 bands (i.e., 32%) were also present in the natural community (Fig. 4A). These corresponding DNA bands were therefore excised and sequenced. In cases, in which one of the 13 melting types was retrieved on multiple occasions, several bands with the same melting behavior were analyzed. Other frequently occurring melting types were also included in the sequence analyses, yielding a total of 34 sequences of cultured bacteria (Fig. 5).
FIG.5.
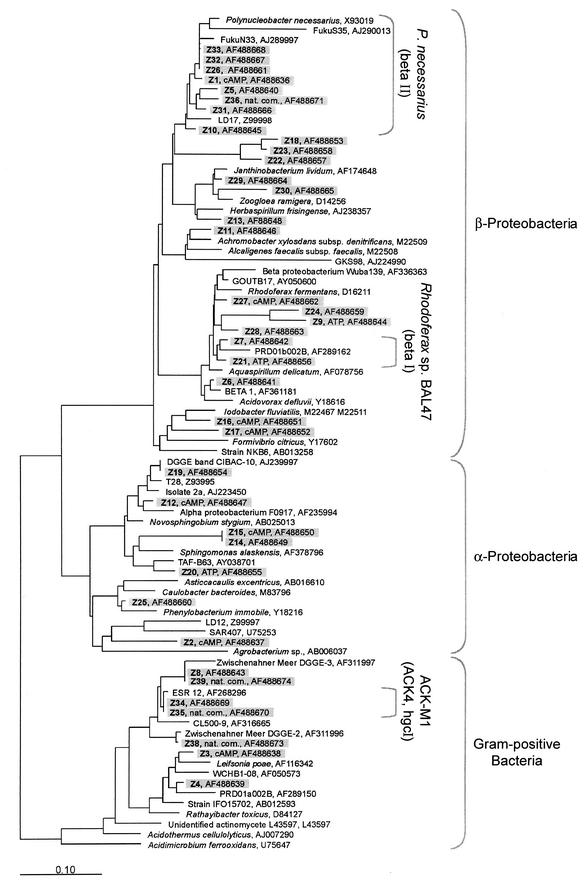
Phylogenetic tree calculated by maximum likelihood, comparing 16S rRNA gene sequences recovered from different MPN enrichments and from the natural community of bacteria from Zwischenahner Meer with those of their closest relatives. Sequences obtained in the present study are shaded. cAMP and ATP indicate sequences originating from highest positive dilutions of series in which cAMP or ATP resulted in significantly increased MPN values. Nat. com., natural bacterioplankton community from Zwischenahner Meer. Three of the freshwater clusters (Polynucleobacter necessarius, Rhodoferax sp. strain BAL47, and ACK-M1) suggested by Zwart et al. (52) are identified by brackets. The bar indicates 10% sequence divergence.
Sequence comparison revealed that DNA bands exhibiting an identical melting behavior in most cases contained different sequences, (Fig. 5; compare sequences Z29, Z30, and Z36, which originated from the same band position on DGGE gels). Accordingly, most of the sequences were recovered only once (29 of 34 sequenced bands), and only sequences Z33, Z32, and Z26, as well as Z15 and Z14, were identical (Fig. 5). Obviously, the 41 different 16S rRNA gene fingerprints represent a considerable underestimate of the actual diversity of the bacteria which can be recovered with our cultivation approach.
The phylogenetic analysis of all sequences demonstrated an affiliation with only three phylogenetic groups (Fig. 5). Most bacteria were identified as members of the β-subclass of the Proteobacteria, seven were affiliated with members of the α-subclass of the Proteobacteria, and only four sequences were related to the Actinomycetales. Since this relative composition of the cultured bacteria remained rather constant throughout the study period, we also determined the fraction of the different phylogenetic groups within the total bacterioplankton community at the end of the study by FISH (Table 2). Similar to their dominance among the cultured representatives, β-Proteobacteria also constituted a significant fraction of all detected eubacterial cells in the natural assemblage (9 to 13% of DAPI-stained cells). However, gram-positive bacteria with high GC contents were even more abundant (16 to 17% of all bacterial cells), which is in sharp contrast to the always low number of cultured representatives of this group. During in situ hybridization, the gram-positive bacteria were characterized by small (0.2 to 0.5 μm in diameter), dimly fluorescent cells which could be visualized only by hybridization after pretreatment with lysozyme (data not shown). Thus, two phylogenetic groups accounted for more than 60% of the cells hybridized with the Eubacteria-specific probe (Table 2). Members of the α- and γ-subclasses of the Proteobacteria, as well as members of the Cytophaga and Flavobacteria, formed only a small part of the natural bacterioplankton, together accounting for no more than 5.5% of the DAPI-stained cells.
TABLE 2.
Abundances of different phylogenetic groups in situ
| Date (day.mo.yr) | Total cell counts,a 107 ml−1 (mean ± SD) | % of total cells (mean ± SD) detected with probeb:
|
|||||
|---|---|---|---|---|---|---|---|
| EUB338 | ALF968 | BET42a | GAM42a | CF319a | HGC69a | ||
| 4.4.2002 | 1.17 ± 0.42 | 43.7 ± 10.5 | 1.4 ± 1.1 | 13.2 ± 7.4 | 0.7 ± 0.9 | 1.6 ± 1.7 | 15.9 ± 6.3 |
| 22.4.2002 | 1.96 ± 0.55 | 42.7 ± 8.2 | 1.1 ± 2.0 | 9.3 ± 5.0 | 0.2 ± 0.6 | 4.2 ± 2.7 | 17.4 ± 9.3 |
As determined by DAPI counting.
Numbers are corrected for unspecific binding of oligonucleotides (counts with probe NON338 were 0.59 to 2.01%).
Recovery of so-far uncultured and dominant members of the bacterioplankton community.
Based on the phylogenetic comparison of 16S rRNA gene sequences, the closest relatives of many of the cultured β-Proteobacteria are so far uncultured freshwater bacteria (sequences Z1, Z5, Z7, Z10, Z26, and Z31 to Z33 [Fig. 5]). The same is true for the two α-proteobacterial sequences Z19 and Z20. The partial sequence Z19 was actually identical to that of an environmental sequence dominating in the oxic epilimnion of a Spanish lake. Our culture may therefore harbor this so-far uncultured bacterium. Most of the remaining sequences are most closely related to those of typical aquatic bacteria.
Remarkably, the DGGE analysis of 16S rRNA gene fingerprints from bacterioplankton sampled in May 2000 suggested that two dominant fingerprints of the natural bacterial community in Zwischenahner Meer were also recovered in MPN media supplemented with cAMP (compare bands Z8, Z34, Z35, and Z39 in Fig. 4A and C). Subsequent sequencing revealed that the sequences of bands Z34 and Z35, as well as those of Z8 and Z39, were indeed identical. Closely related to the sequences Z34 and Z35 is the hitherto uncultured freshwater firmicute ESR 12 (Fig. 5). Similarly, sequences Z8 and Z39 are related to another uncultured gram-positive bacterium. These results strongly indicate that two dominant members of the natural bacterioplankton assemblage could be cultivated in our liquid media supplied with cAMP.
Characterization of cAMP transport by natural bacterioplankton.
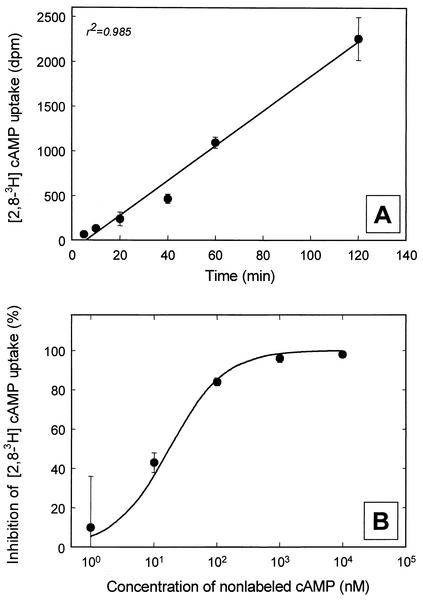
The addition of the signal compound cAMP repeatedly yielded increased numbers of viable cells and also led to the cultivation of two dominant members of the natural bacterioplankton community. At the end of the study period, it was therefore investigated whether a specific uptake mechanism for this compound exists in planktonic bacteria. [3H]cAMP uptake began within a few minutes after the start of the experiments and continued at a constant rate of 11 pM h−1 for at least 2 h (Fig. 6A) (linear correlation coefficient [r2] = 0.985). No unspecific adsorption of [3H]cAMP was observed in formalin-killed controls. A competitive uptake experiment using different concentrations of unlabeled cAMP demonstrated an increased inhibition of the [3H]cAMP uptake with increasing concentrations of unlabeled cAMP (Fig. 6B). These data suggest that cAMP enters bacterioplankton cells via a specific transporter.
FIG. 6.
Uptake of [3H]cAMP by natural freshwater bacterioplankton in water samples from 22 April 2002. (A) Time course of [3H]cAMP uptake. (B) Inhibition of [3H]cAMP uptake by different concentrations of unlabeled cAMP. The curve fit was calculated with a Km for cAMP uptake of 1 nM, as described in the text.
For further characterization of the uptake mechanism, the data were used to estimate the average half-saturation constant Km for cAMP transport into the bacterioplankton cells. Based on the formula for the transport of a labeled carbon substrate, added at a concentration A in the presence of different concentrations X of the corresponding unlabeled molecules (51), the inhibition (I) of [3H]cAMP transport should follow the equation
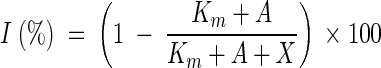 |
(1) |
The data obtained could be fitted by this equation. At the concentration A of 16.9 nM employed in our experiment, the best curve fits were obtained with Km values of ≤1 nM (Fig. 6B). This result indicates that cAMP transport in bacterioplankton cells from Zwischenahner Meer had a very high substrate affinity.
In parallel, the percentage of Bacteria capable of cAMP transport was determined by microautoradiography combined with FISH. In these experiments, 25% ± 10.1% of bacteria detected with the oligonucleotide probe EUB338 for Bacteria and 18% ± 7.3% of all planktonic bacteria were found to take up [3H]cAMP, as visualized by silver grains below and around the cells.
DISCUSSION
Generally, the efficiency of cultivation of planktonic bacteria from natural samples is very low; it typically ranges below 1% (2). In the present study, incubation conditions were varied in a systematic manner and the number and taxonomic composition of cultured bacteria were monitored. The goal was to identify conditions which (i) significantly increase the cultivation efficiency of freshwater bacteria and (ii) especially favor the growth of those bacteria which dominate the natural bacterioplankton assemblage but nevertheless have repeatedly escaped cultivation. The major focus of the work was the effect of signal compounds on culturability, since a previous study had demonstrated a stimulating effect of cAMP and the acyl-HSLs BHL and OHHL on the growth of marine bacterioplankton (13).
Effect of different incubation conditions and signal compounds on the cultivation of freshwater bacterioplankton.
Throughout the study period, media incubated at 4°C or under an anoxic atmosphere consistently yielded the lowest MPN values. Obviously, psychrophilic or anaerobic bacteria do not constitute a major fraction of the bacterioplankton in Zwischenahner Meer. The effect of a reduced oxygen partial pressure of 3% was tested since it may reduce the formation of toxic oxygen radicals and therefore prevent cellular damage caused by oxidative stress (28). However, reduced oxygen concentrations in most cases did not result in increased cultivation efficiency.
The low concentrations of carbon substrates as employed in the present study had an effect on the type of bacteria recovered. Members of the β-subclass of the Proteobacteria dominated among those cultured bacteria which were sequenced. Correspondingly, FISH also demonstrated a high abundance of this group in the natural community of Zwischenahner Meer. This observation is consistent with those from other freshwater lakes (22, 25). In a previous study, cultivation of bacterioplankton from Zwischenahner Meer in more-nutrient-rich media yielded only one isolate affiliated with the β-Proteobacteria, while α-Proteobacteria and members of the Cytophaga-Flavobacterium group dominated among the isolates (29). The frequent growth of β-Proteobacteria in our media is likely to be the result of the lower concentrations of carbon substrates employed in the present investigation.
cAMP was the most effective signal compound, leading to significantly increased MPNs on several occasions. It can be concluded that the stimulating effect of cAMP is not limited to marine bacterioplankton assemblages (13) but, during certain time periods, also occurs in freshwater environments. In the absence of cell-bound phosphodiesterases, no chemical hydrolysis or hydrolysis by dissolved enzymes of cAMP was observed over a 26-h period (3). Under natural conditions, dissolved cAMP is thus rather stable in the absence of microorganisms.
Interestingly, ATP also stimulated growth of the natural bacterioplankton in several cases. Dissolved ATP has been shown to occur in seawater in concentrations of 0.2 to 1.2 nM and is utilized rapidly by marine bacteria (6). ATP is known to be involved in the regulation of various cellular processes. It is, for instance, involved in DNA replication by activation of the DnaA protein (27). This mechanism might be involved in the growth stimulation of planktonic bacteria as observed in the present study. In contrast to marine bacterioplankton (13), the well-known bacterial signal compounds BHL and OHHL had no effect on cultivation of heterotrophic bacteria from Zwischenahner Meer, which is in agreement with results of cultivation experiments with water from Lake Constance (14).
Role of cAMP uptake in natural bacterioplankton.
The involvement of cAMP in the regulation of catabolic enzymes has been demonstrated for a wide range of bacteria (11). Moreover, other roles of cAMP, e.g., in DNA replication or the stationary-phase response in Escherichia coli, are known (27, 33, 49). In enterobacteria, cAMP is involved in the regulation of the majority of genes expressed under starvation conditions (47). The addition of extracellular cAMP was found to prevent substrate-accelerated death in starved laboratory cultures (16). Furthermore, the transcription factor σS (the gene product of rpoS) is involved in the transition to stationary phase. cAMP, in a complex with the cAMP receptor protein, acts as a negative regulator of the transcription of rpoS (36). Addition of extracellular cAMP has the same effect (33). It is therefore possible that the addition of extracellular cAMP hinders bacterial cells in entering the protective stationary phase but rather maintains them in a nutrient-scavenging state more favorable for cultivation. It has also been suggested that the ability of natural bacteria to acquire cAMP from the environment obviates the need to accumulate cAMP intracellularly in preparation for recovery from starvation (18). The cAMP present in seawater (3) in turn may originate from dividing planktonic bacteria and could serve as a signal to stimulate the growth of accompanying dormant cells (13).
Microautoradiography confirmed that a significant fraction of the bacteria in Zwischenahner Meer (18%) are capable of cAMP uptake. So far, uptake of cAMP has been shown in only a few studies of marine bacterioplankton. In these cases, only a significantly lower fraction of bacterial cells (2 to 7% of the bacterial community) (4) was capable of cAMP uptake.
In natural aquatic systems, cAMP reaches concentrations in the picomolar range (1 to 35 pM) (3). Hence, a prerequisite for the utilization of this compound is the expression of high-affinity uptake systems, as have been postulated for marine bacterioplankton cells based upon the uptake observed at concentrations of 50 pM (3, 4). cAMP is taken up by high-affinity transport systems having stringent structural requirements, allowing transport of cyclic nucleotides but not of structurally related noncyclic compounds such as AMP (5). Our data provide a first estimate of the affinity of cAMP uptake systems present in freshwater planktonic bacteria. The Km of ≤1 nM is consistent with the range of cAMP concentrations detected in marine planktonic habitats and similar to the Kms of marine bacteria (4).
Recovery of previously unculturable bacteria.
Actinomycetales, formerly considered typical soil bacteria, have been shown to be abundant and globally distributed in freshwater habitats (23). However, the high abundance of these bacteria may in fact be due to size-selective grazing behavior of protistan predators which results in a selective advantage of small bacteria (43). FISH confirmed that, similarly to other aquatic environments, the actinobacteria present in Zwischenahner Meer were represented by small cells (0.2 to 0.5 μm in diameter).
In contrast to gram-negative bacteria, their gram-positive counterparts do not seem to utilize cAMP as a signal compound (35). Clearly, detailed physiological and molecular studies of freshwater actinobacteria are necessary in order to elucidate the role of cAMP in the physiology of these bacteria and to ultimately identify the cellular mechanism underlying the phenomenon of their so-called nonculturability.
Conclusions.
By applying dilution series of liquid media with low nutrient concentrations, pure cultures of typical aquatic bacteria can be recovered from natural planktonic assemblages. In these media, the addition of the signal compound cAMP most effectively increases cultivation success. The bacterial phylotypes obtained depend on the physiological state of bacterial cells and the incubation conditions chosen rather than on the overall composition of the bacterioplankton community.
To our knowledge, the present study for the first time demonstrates the cultivation of planktonic actinobacteria (Z8 and Z34 [Fig. 4 and 5]) in fully synthetic laboratory media. The two actinobacteria most likely represent dominant indigenous bacteria. Our results show that increases in culturability should not be taken as the only criterion for the improvement of cultivation media. Instead, numerically significant but previously nonculturable bacteria can be recovered even under conditions where overall culturability remains comparably low.
Acknowledgments
We thank Bert Engelen and Natascha Selje for help with the software package ARB.
This work was funded by a grant from the Bundesministerium für Bildung, Wissenschaft, Forschung und Technologie (grant no. 0311949) to J. Overmann and H. Cypionka.
REFERENCES
- 1.Aagot, N., O. Nybroe, P. Nielsen, and K. Johnsen. 2001. An altered Pseudomonas diversity is recovered from soil by using nutrient-poor Pseudomonas-selective soil extract media. Appl. Environ. Microbiol. 67:5233-5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R. I. 1995. Fluorescently labelled, rRNA-targeted oligonucleotide probes in the study of microbial ecology. Mol. Ecol. 4:543-553. [Google Scholar]
- 3.Ammerman, J. W., and F. Azam. 1981. Dissolved cyclic adenosine monophosphate (cAMP) in the sea and uptake of cAMP by marine bacteria. Mar. Ecol. Prog. Ser. 5:85-89. [Google Scholar]
- 4.Ammerman, J. W., and F. Azam. 1982. Uptake of cyclic AMP by natural populations of marine bacteria. Appl. Environ. Microbiol. 43:869-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ammerman, J. W., and F. Azam. 1987. Characteristics of cyclic AMP transport by marine bacteria. Appl. Environ. Microbiol. 53:2963-2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Azam, F., and R. E. Hodson. 1977. Dissolved ATP in the sea and its utilization by marine bacteria. Nature 267:696-698. [DOI] [PubMed] [Google Scholar]
- 7.Balestra, G. M., and I. J. Misaghi. 1997. Increasing the efficiency of the plate counting method for estimating bacterial diversity. J. Microbiol. Methods 30:111-117. [Google Scholar]
- 8.Barer, M. R., and C. R. Harwood. 1999. Bacterial viability and culturability. Adv. Microb. Physiol. 41:93-137. [DOI] [PubMed] [Google Scholar]
- 9.Barns, S. M., S. L. Takala, and C. R. Kuske. 1999. Wide distribution and diversity of members of the bacterial kingdom Acidobacterium in the environment. Appl. Environ. Microbiol. 65:1731-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartscht, K., H. Cypionka, and J. Overmann. 1999. Evaluation of cell activity and of methods for the cultivation of bacteria from a natural lake community. FEMS Microbiol. Ecol. 28:249-259. [Google Scholar]
- 11.Botsford, J. L., and J. G. Harman. 1992. Cyclic AMP in prokaryotes. Microbiol. Rev. 56:100-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bruns, A., and L. Berthe-Corti. 1998. In situ detection of bacteria in continuous-flow cultures of seawater sediment suspensions with fluorescently labeled rRNA-directed oligonucleotide probes. Microbiology 144:2783-2790. [DOI] [PubMed] [Google Scholar]
- 13.Bruns, A., H. Cypionka, and J. Overmann. 2002. Cyclic AMP and acyl homoserine lactones increase the cultivation efficiency of heterotrophic bacteria from the central Baltic Sea. Appl. Environ. Microbiol. 68:3978-3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bussmann, I., B. Philipp, and B. Schink. 2001. Factors influencing the cultivability of lake water bacteria. J. Microbiol. Methods 47:41-50. [DOI] [PubMed] [Google Scholar]
- 15.Button, D. K., F. Schut, P. Quang, R. Martin, and B. R. Robertson. 1993. Viability and isolation of marine bacteria by dilution culture: theory, procedures, and initial results. Appl. Environ. Microbiol. 59:881-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Calcott, P. H., and J. R. Postgate. 1972. On substrate-accelerated death in Klebsiella aerogenes. J. Gen. Microbiol. 70:115-122. [DOI] [PubMed] [Google Scholar]
- 17.Coolen, M. L., H. Cypionka, A. M. Sass, H. Sass, and J. Overmann. 2002. Ongoing modification of Mediterranean pleistocene sapropels mediated by prokaryotes. Science 296:2407-2410. [DOI] [PubMed] [Google Scholar]
- 18.Deming, J. W., and J. A. Baross. 2000. Survival, dormancy, and nonculturable cells in extreme deep-sea environments, p. 147-197. In R. R. Colwell and D. J. Grimes (ed.), Nonculturable microorganisms in the environment. ASM Press, Washington, D.C.
- 19.Dojka, M. A., J. K. Harris, and N. R. Pace. 2000. Expanding the known diversity and environmental distribution of an uncultured phylogenetic division of bacteria. Appl. Environ. Microbiol. 66:1617-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eguchi, M., M. Ostrowski, F. Fegatella, J. Bowman, D. Nichols, T. Nishino, and R. Cavicchioli. 2001. Sphingomonas alaskensis strain AFO1, an abundant oligotrophic ultramicrobacterium from the North Pacific. Appl. Environ. Microbiol. 67:4945-4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gich, F., J. Garcia-Gil, and J. Overmann. 2001. Previously unknown and phylogenetically diverse members of the green nonsulfur bacteria are indigenous to freshwater lakes. Arch. Microbiol. 177:1-10. [DOI] [PubMed] [Google Scholar]
- 22.Glöckner, F. O., B. M. Fuchs, and R. Amann. 1999. Bacterioplankton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl. Environ. Microbiol. 65:3721-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glöckner, F. O., E. Zaichikov, N. Belkova, L. Denissova, J. Pernthaler, A. Pernthaler, and R. Amann. 2000. Comparative 16S rRNA analysis of lake bacterioplankton reveals globally distributed phylogenetic clusters including an abundant group of actinobacteria. Appl. Environ. Microbiol. 66:5053-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guan, L. L., H. I. Onuki, and K. Kamino. 2000. Bacterial growth stimulation with exogenous siderophore and synthetic N-acyl homoserine lactone autoinducers under iron-limited and low-nutrient conditions. Appl. Environ. Microbiol. 66:2797-2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hiorns, W., B. Methe, S. Nierzwicki-Bauer, and J. Zehr. 1997. Bacterial diversity in Adirondack mountain lakes as revealed by 16S rRNA gene sequences. Appl. Environ. Microbiol. 63:2957-2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hughes, P., A. Landoulsi, and M. Kohiyama. 1988. A novel role for cAMP in control of the activity of the E. coli chromosome replication initiator protein, DnaA. Cell 55:343-350. [DOI] [PubMed] [Google Scholar]
- 28.Jannasch, H. W., and R. I. Mateles. 1974. Experimental bacterial ecology studied in continuous culture. Adv. Microb. Physiol. 11:165-212. [Google Scholar]
- 29.Jaspers, E., K. Nauhaus, H. Cypionka, and J. Overmann. 2001. Multitude and temporal variability of ecological niches as indicated by the diversity of cultivated bacterioplankton FEMS Microbiol. Ecol. 36:153-164. [DOI] [PubMed] [Google Scholar]
- 30.Jones, J. G. 1979. A guide to methods for estimating microbial numbers and biomass in fresh water. Scientific publication no. 39. Freshwater Biological Association, Ambleside, United Kingdom.
- 31.Klee, A. J. 1993. A computer program for the determination of most probable number and its confidence limits. J. Microbiol. Methods 18:91-98. [Google Scholar]
- 32.Kleerebezem, M., L. E. N. Quadri, O. P. Kuipers, and W. M. de Vos. 1997. Quorum sensing by peptide pheromones and two component signal transduction systems in Gram-positive bacteria. Mol. Microbiol. 24:895-904. [DOI] [PubMed] [Google Scholar]
- 33.Lange, R., and R. Hengge-Aronis. 1991. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol. Microbiol. 5:49-59. [DOI] [PubMed] [Google Scholar]
- 34.Lee, N., P. H. Nielsen, K. H. Andreasen, S. Juretschko, J. L. Nielsen, K.-H. Schleifer, and M. Wagner. 1999. Combination of fluorescent in situ hybridization and microautoradiography: a new tool for structure-function analyses in microbial ecology. Appl. Environ. Microbiol. 65:1289-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lengeler, J. W., G. Drews, and H. G. Schlegel. 1999. Biology of prokaryotes. Georg Thieme Verlag, Stuttgart, Germany.
- 36.Loewen, P. C., B. Hu, J. Strutinsky, and R. Sparling. 1998. Regulation in the rpoS regulon of Escherichia coli. Can. J. Microbiol. 44:707-717. [DOI] [PubMed] [Google Scholar]
- 37.Ludwig, W., S. H. Bauer, M. Bauer, I. Held, G. Kirchhof, R. Schulze, I. Huber, S. Spring, A. Hartmann, and K. H. Schleifer. 1997. Detection and in situ identification of representatives of a widely distributed new bacterial phylum. FEMS Microbiol. Lett. 153:181-190. [DOI] [PubMed] [Google Scholar]
- 38.Mukamolova, G. V., A. S. Kaprelyants, D. I. Young, M. Young, and D. Kell. 1998. A bacterial cytokine. Proc. Natl. Acad. Sci. USA 95:8916-8921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muyzer, G., S. Hottenträger, A. Teske, and C. Waver. 1995. Denaturing gradient gel electrophoresis of PCR-amplified 16S rDNA—a new molecular approach to analyse the genetic diversity of mixed microbial communities, p. 3.4.4.1-3.4.4.22. In A. D. L. Akkermans, J. D. Van Elsas, and F. J. De Bruijn (ed.), Molecular microbial ecology manual. Kluwer, Dordrecht, The Netherlands.
- 40.Nübel, U., M. M. Bateson, V. Vandieken, A. Wieland, M. Kühl, and D. M. Ward. 2002. Microscopic examination of distribution and phenotypic properties of phylogenetically diverse Chloroflexaceae-related bacteria in hot spring microbial mats. Appl. Environ. Microbiol. 68:4593-4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ouverney, C. C., and J. A. Fuhrman. 1999. Combined microautoradiography-16S rRNA probe technique for determination of radioisotope uptake by specific microbial cell types in situ. Appl. Environ. Microbiol. 65:1746-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Overmann, J., M. J. L. Coolen, and C. Tuschak. 1999. Specific detection of different phylogenetic groups of chemocline bacteria based on PCR and denaturing gradient gel electrophoresis of 16S rRNA gene fragments. Arch. Microbiol. 172:83-94. [DOI] [PubMed] [Google Scholar]
- 43.Pernthaler, J., T. Posch, K. Simek, J. Vrba, A. Pernthaler, F. O. Glockner, U. Nübel, R. Psenner, and R. Amann. 2001. Predator-specific enrichment of actinobacteria from a cosmopolitan freshwater clade in mixed continuous culture. Appl. Environ. Microbiol. 67:2145-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Riemann, B. 1979. The occurrence and ecological importance of dissolved ATP in fresh water. Freshwater Biol. 9:481-490. [Google Scholar]
- 45.Roller, C., M. Wagner, R. Amann, W. Ludwig, and K.-H. Schleifer. 1995. In situ probing of gram-positive bacteria with high DNA G+C content using 23S rRNA-targeted oligonucleotides. Microbiology 141:1269.. [DOI] [PubMed] [Google Scholar]
- 46.Sandaa, R. A., V. Torsvik, O. Enger, F. L. Daae, T. Castberg, and D. Hahn. 1999. Analysis of bacterial communities in heavy metal-contaminated soils at different levels of resolution. FEMS Microbiol. Ecol. 30:237-251. [DOI] [PubMed] [Google Scholar]
- 47.Schultz, J. E., G. I. Latter, and A. Matin. 1988. Differential regulation by cyclic AMP of starvation protein synthesis in Escherichia coli. J. Bacteriol. 170:3903-3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Snaidr, J., R. Amann, I. Huber, W. Ludwig, and K. Schleifer. 1997. Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl. Environ. Microbiol. 63:2884-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taddei, F., I. Matic, and M. Radman. 1995. cAMP-dependent SOS induction and mutagenesis in resting bacterial populations. Proc. Natl. Acad. Sci. USA 92:11736-11740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Torsvik, V., L. Ovrebs, and T. F. Thingstad. 2002. Prokaryotic diversity—magnitude, dynamics, and controlling factors. Science 296:1064-1066. [DOI] [PubMed] [Google Scholar]
- 51.Wright, R. T., and J. E. Hobbie. 1966. Use of glucose and acetate by bacteria and algae in aquatic ecosystems. Ecology 47:447-464. [Google Scholar]
- 52.Zwart, G., B. C. Crump, M. P. Kamst-van Agterveld, F. Hagen, and S. K. Han. 2002. Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquat. Microb. Ecol. 28:141-155. [Google Scholar]