Abstract
Seasonal shifts in bacterioplankton community composition in Toolik Lake, a tundra lake on the North Slope of Alaska, were related to shifts in the source (terrestrial versus phytoplankton) and lability of dissolved organic matter (DOM). A shift in community composition, measured by denaturing gradient gel electrophoresis (DGGE) of 16S rRNA genes, occurred at 4°C in near-surface waters beneath seasonal ice and snow cover in spring. This shift was associated with an annual peak in bacterial productivity ([14C]leucine incorporation) driven by the large influx of labile terrestrial DOM associated with snow meltwater. A second shift occurred after the flux of terrestrial DOM had ended in early summer as ice left the lake and as the phytoplankton community developed. Bacterioplankton communities were composed of persistent populations present throughout the year and transient populations that appeared and disappeared. Most of the transient populations could be divided into those that were advected into the lake with terrestrial DOM in spring and those that grew up from low concentrations during the development of the phytoplankton community in early summer. Sequencing of DNA in DGGE bands demonstrated that most bands represented single ribotypes and that matching bands from different samples represented identical ribotypes. Bacteria were identified as members of globally distributed freshwater phylogenetic clusters within the α- and β-Proteobacteria, the Cytophaga-Flavobacteria-Bacteroides group, and the Actinobacteria.
Lake water contains allochthonous organic matter derived from terrestrial plants and soils and autochthonous organic matter produced by phytoplankton, aquatic plants, and benthic algae (45). Surveys of bacterial diversity in lakes have identified many populations common to freshwater systems worldwide (13, 53), but it is not known how these populations interact with and use different types of organic matter. Nor is it known whether different bacterial populations are responsible for decomposing the different types of organic matter that may be present simultaneously in the environment. In the Arctic, there is a clear seasonal separation between the major fluxes of allochthonous and autochthonous organic matter to lakes; terrestrial organic matter enters lakes during the spring runoff, and autochthonous organic matter is produced mainly in the summer and fall (50). We used this temporal separation to study seasonal shifts in the composition of lake bacterial communities and to determine how these shifts relate to changes in the supply of different types of organic matter.
Seasonal shifts in bacterioplankton community composition were investigated in Toolik Lake, an ultraoligotrophic Arctic lake on the North Slope of Alaska. Special attention was paid to the first month of the spring season when major changes occur in the supply of labile organic matter and the growth of bacterioplankton. In May, melting snow carries a large amount of organic matter and nutrients off the catchment and into the lake. The ice on Toolik Lake is covered by snow at that time, which restricts light penetration and thus primary production (7, 34, 50). In the days following this input of material, bacterial production beneath the ice cover increases by an order of magnitude and reaches an annual peak, presumably supported by this terrestrial organic matter (33). The supply of terrestrial organic matter diminishes with the end of the spring snowmelt. Over the following days to weeks, solar insolation increases and the snow on top of the lake ice melts and allows solar radiation to penetrate the ice and water column. Phytoplankton production reaches its annual peak as the ice leaves the lake (31) using winter-accumulated dissolved inorganic nutrients as well as nutrients washed in with the spring snowmelt. It is thought that bacterioplankton production during the ice-free season is maintained on a combination of phytoplankton exudates and terrestrial organic matter (33).
Evidence that bacteria in Toolik Lake use both terrestrial organic matter and phytoplankton-produced organic matter comes from comparing the rates of primary and secondary production. Annual primary production in Toolik Lake is very low (∼12 g of carbon m−2 year−1), classifying the lake as ultraoligotrophic. Bacterial production, however, is relatively high and has been estimated to be as much as 3 to 8 g of carbon m−2 year−1 or 66% of primary production (33). A comparative study of this ratio of bacterial to primary production in planktonic ecosystems determined that bacterial production is typically much lower, averaging only 20% of primary production (4). The relatively high rate of bacterial production in Toolik Lake could be the result of remarkably high bacterial growth efficiency, but it is more likely the result of bacterial growth on a combination of phytoplankton organic matter produced in the lake and terrestrial organic matter input from the catchment.
Biogeographical studies using DNA sequencing of 16S rRNA clone libraries have provided a picture of the bacterial diversity of Toolik Lake (1) and other planktonic systems (5, 10, 12, 46, 54). Comparison of these 16S rRNA gene sequences across different systems has identified many globally distributed phylogenetic clusters of bacteria (11, 13, 53). However, the small numbers of samples that can be processed by this method have limited the resolution of the microbial biogeography. Larger numbers of samples can be analyzed by community fingerprinting methods, such as denaturing gradient gel electrophoresis (DGGE) (32), terminal restriction fragment length polymorphism (TRFLP) (28), and automated ribosomal intergenic spacer analysis (ARISA) (9). All of these methods provide relatively rapid comparisons of bacterial community composition, but DGGE has the added advantage of allowing the identification of specific organisms represented in the fingerprint by excising and sequencing DNA from bands in the gel.
When applied to mixed laboratory cultures and field-based mesocosms, community fingerprinting methods indicate that the composition of planktonic bacterial communities shift readily with changes in environmental conditions, including grazing pressure and viral lysis (42, 48, 49), nutrient concentration (39), and organic matter composition (29). Shifts in organic matter supply as subtle as a change in phytoplankton species are enough to cause a species-level shift in bacterial community composition (47). These studies provide a sense of the breadth of environmental conditions that may influence the composition of natural bacterial communities. However, it is not always clear which environmental factors cause shifts in species composition.
We hypothesized that shifts in the species composition of the planktonic bacterial community would accompany major seasonal changes in the source and quality of dissolved organic matter (DOM) in Toolik Lake. We discovered that these shifts resulted from both the changes in the relative abundance of autochthonous bacteria as well as advection of allochthonous bacteria via the inlet stream during the spring thaw. Here we present results from two field seasons at Toolik Lake. Research in 1996 connects seasonal shifts in bacterial production to the quality of terrestrially derived DOM. Research in 2000 expanded on this work, linking changes in bacterial production and bacterial community composition to seasonal shifts in terrestrial organic matter influx and phytoplankton production in Toolik Lake and its primary inlet stream.
MATERIALS AND METHODS
Sampling sites.
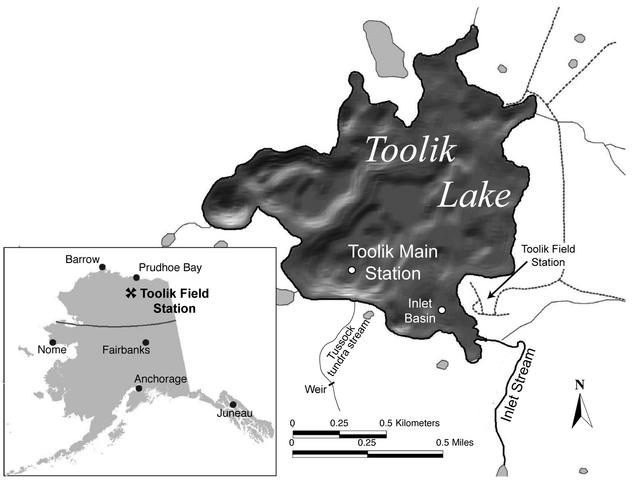
Toolik Lake (1.5 km2) is a deep (maximum depth, 25 m; mean depth, 7 m) kettle lake located on the North Slope of Alaska (68°38′00′′N, 149°36′15′′W), which has been the subject of continuous research since 1975 (17). The lake usually becomes ice free and thermally stratifies in late June and remains that way through September. Ice cover, which may exceed 1.4 m, forms in early October (33). Nearly all lake samples were collected at a main station in the southern basin (Fig. 1). Samples collected on 19 June 2000 were collected from the inlet basin because of thin ice.
FIG. 1.
Toolik Lake shown with shaded depth contours.
The catchment of Toolik Lake (65,000 ha) has vegetation dominated by tussock and upland heath tundra. Soils in the catchment have a maximum annual thaw depth of about 0.5 m and are underlain with permafrost (33, 50). The main inlet stream to Toolik Lake, entering from the southeast, drains 75% of this catchment and lies at the base of a chain of 12 smaller lakes (26). Stream flow usually begins in late May and quickly reaches its peak flow rate as snow on the catchment begins to melt (18).
On the south shore of the lake, a small primary stream drains a 1.5-ha catchment composed mainly of tussock tundra (Fig. 1). A weir channels water from the primary stream for flow measurements and sampling.
Measurements.
Prokaryotic cell concentration was counted directly with 4′,6′-diamidino-2-phenylindole (DAPI) (19, 35). Bacterial production was determined by measuring the incorporation of 14C-labeled l-leucine (30 nM final concentration) into the cold trichloroacetic acid (TCA)-insoluble fraction of macromolecules in two subsamples incubated for 2 to 4 h at in situ temperatures in the dark. TCA-precipitated macromolecules were collected on 0.2-μm-pore-size nitrocellulose filters (Millipore), washed twice with ice-cold 5% TCA, made transparent with 1 ml of methyl-Cellusolve, flooded with 6 ml of Scintisafe scintillation cocktail, and counted in a Packard Tri-Carb 2100 scintillation counter. Isotope dilution experiments confirmed the use of 30 nM leucine in the experiments (25). Bacterial production was expressed as the picomolar concentration of leucine incorporated per hour (25).
Water samples for dissolved organic carbon concentration were filtered in the field through Whatman GF/F filters and acidified to pH 3. Samples were kept in the dark at 4°C until analyzed on a Shimadzu TOC 5000 with platinum-catalyzed high-temperature combustion to CO2 and infrared detection.
Chlorophyll concentration was determined in samples collected from depths of 0, 1, 3, 5, 12, and 16 m at the main sampling station of Toolik Lake. Water was filtered through 47-mm-diameter Whatman GF/C filters. The filters were extracted in 10 ml of buffered 90% acetone (1 mg of MgCO3 liter−1) in the dark at room temperature for 24 h. Chlorophyll was measured with a Turner Designs 10-Au-005-CE fluorometer configured with a chlorophyll optical kit.
Bioassays.
During spring and summer of 1996, water samples were collected on 14 separate days from the Toolik inlet stream and on 7 separate days from the tussock tundra weir, starting on the day each stream began to flow. Duplicate 500-ml samples were filtered through 0.2-μm-pore-diameter polycarbonate filters to remove bacteria and were inoculated with 25 ml of filtered (0.6-μm pore diameter) water from the Toolik inlet or from 10-m depth in Toolik Lake collected on the same day. Samples were incubated at approximate in situ temperature (5°C for 14 May to 1 July, 10°C for 20 July to 21 August) for 12 days, and were subsampled daily for bacterial production, cell number, and dissolved oxygen concentration (DOC). Bacterial production usually attained its maximum rate of increase 4 to 6 days into the incubation and a plateau after 10 to 11 days.
Community composition. (i) Sample collection.
During spring and summer of 2000, water samples were collected from Toolik Lake and from the Toolik inlet stream (just upstream of the mouth) and stored at 4°C for up to 2 h. Plankton from 300 to 1,000 ml of water was collected on 0.2-μm-pore-diameter Sterivex-GP filter capsules (Millipore). After filtration, all liquid was forced out of the Sterivex filter, approximately 2 ml of DNA extraction buffer (DEB; 0.1 M Tris-HCl [pH 8], 0.1 M Na EDTA [pH 8], 0.1 M Na2H2PO4 [pH 8], 1.5 M NaCl, 5% hexadecyltrimethyl-ammonium bromide [CTAB]) was added through the in-port of the filter housing with a needle, and the in-port and out-port were capped. Samples were initially stored at −20°C for 1 to 3 months and subsequently were stored at −80°C until processed.
(ii) DNA extraction.
Fifty microliters of proteinase K (1%) was added to thawed samples, and samples were refrozen at −80°C and thawed at 34°C three times. After the final thaw, samples were incubated at 34°C for 30 min. One hundred microliters of sodium dodecyl sulfate (SDS; 20%; filter sterilized) was then added, and samples were incubated at 65°C for 2 h. The extraction buffer was then drawn out and replaced with 2 ml of DEB and 100 μl of 20% SDS. Filters were incubated at 65°C for 30 min, and buffer was drawn out and combined with the buffer from the first extraction. DNA was washed twice with buffered phenol-chloroform-isoamyl alcohol (pH 8.0), precipitated with 0.6 part isopropyl alcohol at room temperature overnight, resuspended in sterile water, and stored at −80°C (adapted from reference 52).
(iii) DGGE.
DGGE procedures were performed according to the method described by Muyzer et al. (32). PCR amplification (1× PCR buffer [Promega], 8 μM deoxynucleoside triphosphates [dNTPs], 1 μM primers, 2 U of Taq polymerase [Promega]) used primer 357f(g+c) (5′-CGCCCGCCGCGCCCCGCGCCCGGCCCGCCGCCCCCGCCCCCCTACGGGAGGCAGCAG-3′) which contains a GC clamp and is specific for most bacteria, and universal primer 519r (5′-ACCGCGGCTGCTGGCAC-3′), under the following conditions: initial denaturation for 5 min at 94°C; followed by cycles of 1 min at 94°C, 1 min at 65 to 55°C (reducing the temperature by 0.5 per cycle for 20 cycles plus further cycles at 55°C), and 1 min at 72°C; followed by 5 min at 72°C. Steps involving a temperature reduction were done at 0.3°C/s. In order to minimize heteroduplex formation during the plateau phase of PCR (22), we used a relatively high concentration of primers and optimized PCR cycle number for each sample so that PCR was halted while the product concentration was approximately one-quarter to one-half of the maximum concentration (between 20 and 30 cycles). The amount of template varied with the sample and was selected to optimize PCR amplification. In general, the entire volume of each 50-μl reaction mixture was used to load the DGGE gel.
Acrylamide (8%) gels were prepared with 30% acrylamide-bisacrylamide (37.5:1; Bio-Rad), and 0.5× TAE buffer (1× TAE is 40 mM Tris [pH 8.0], 20 mM acetic acid, 1 mM EDTA). Sixteen-centimeter-long, 1-mm-thick gels contained a linear gradient of denaturants (urea and formamide). A gradient of 30 to 50% was used for comparison of DGGE banding patterns. Electrophoresis was run in a Bio-Rad D-code system for 16 to 18 h at 70 V.
Magnified sections of DGGE gels were photographed with a ChemImager 4000 imaging system (Alpha Innotech), and complete images of each gel were reconstructed with Photoshop (Adobe). This provided a superior image of the gel in which each band spanned 5 to 10 pixels vertically. Bands were identified and marked in Adobe Illustrator. The position of the bands on the gel were determined based on the vertical position of the bands in a reference ladder (see below) run in four or five lanes across each gel. The relative vertical positions of the individual bands in each sample were confirmed by running the sample set on several gels and varying the order of the samples. Bands in each sample were scored as present or absent at each position. A pairwise distance matrix (Dice) was calculated from this binary data set and was analyzed with the Multidimensional Scaling module of the Statistica software package (StatSoft). The graphical representation of these analyses plots the DGGE banding patterns from each sample, such that samples containing many of the same bands are plotted close to each other. The points were then connected with lines to show the seasonal progression of bacterial community composition. The distance matrix was also analyzed with unweighted pair group mean average (UPGMA) cluster analysis and presented as a dendrogram.
(iv) DGGE band identification.
Four representative samples collected at a depth of 3 m (19 May, 19 June, 9 July, and 28 August) were amplified with DGGE primers and run on a DGGE gel as described above. Twelve bands per sample were selected for identification. Sterile pipette tips were stabbed into each band and swirled in PCR mix containing non-GC-clamp primers (G. Muyzer, personal communication). Amplification proceeded as described above, and PCR products were inserted into TOPO-TA cloning vectors (Invitrogen) and used to transform TOP10 chemically competent cells (Invitrogen) following the manufacturer's instructions. Inserts from four to eight clones per band were amplified with DGGE primers and run on DGGE gels along with the natural samples. Clones containing potential matches to bands in the original sample were run on DGGE gels a second time in lanes adjacent to the natural samples in order to confirm the band position match. Clones that exactly matched bands from the natural samples were sequenced. In addition, some clones that did not match the original bands in the natural sample, but rather aligned with bands nearby, were also sequenced.
(v) Clone library construction.
Clone libraries of nearly full-length 16S rRNA genes were constructed from two samples collected at a depth of 3 m at Toolik Main Station (13 May and 28 August). Each sample was amplified with PCR in 4 to 8 separate 100-μl reactions with bacterium-specific primer 8f (5′-AGAGTTTGATCCTGGCTCAG-3′) and universal primer 1492r (5′-GGTTACCTTGTTACGACTT-3′). PCR amplification began with a 1-min denaturation at 94°C followed by cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 2 min. The final cycle was extended for 5 min at 72°C. The number of PCR cycles used for each sample was chosen so that the reactions were stopped while the product concentration was still increasing (20 to 25 cycles). Products of PCR amplification were combined, concentrated, and purified with Qiaquick PCR purification columns (Qiagen) according to the manufacturer's instructions.
PCR products were A-tailed by combining purified PCR products with A-tailing buffer (2.5 mM MgCl, 0.8 mM dNTPs, 2.5 U of Taq DNA polymerase [Promega], 1× PCR buffer [Promega]) and incubation at 70°C for 15 min. PCR products were then cloned as described above.
(vi) DNA sequencing.
DNA sequences of DGGE band clones and environmental clone libraries were determined with a Beckman CEQ-2000 automated sequencer or an ABI 3700 automated sequencer according to the manufacturers' instructions. DGGE band clone sequences were determined for both complementary strands with plasmid-specific primers. Clone libraries were screened by sequencing the first 500 to 700 bp in one direction and comparing the sequences to those of clones from DGGE bands. All sequences submitted to GenBank were determined for both complementary strands. The primers used for sequencing were 8f (5′-ATRGTTTGATCCTGGCTCAG-3′), 357f (5′-CCTACGGGRGGCAGCAG-3′), 515f (5′-GTGCCAGCMGCCGCGGTAA-3′), 907f (5′-AAACTCAAAGGAATTGACGGG-3′), 519r (5′-ATTACCGCGGCTGCTGG-3′), 907r (5′-CCGTCAATTCCTTTRAGTTT-3′), and 1492r (5′-CGGCTACCTTGTTACGACTT-3′).
Phylogenetic analyses were accomplished with the program PAUP 4.0b10 for Macintosh (44). Substitution models for estimating distance matrices were chosen by using likelihood ratio tests (LRT) calculated with the program Modeltest, version 3.06 (36). Distance matrices were estimated by using these models under maximum-likelihood criteria. Minimum-evolution trees were determined with three iterations of tree and parameter estimations by tree bisection-reconnection branch swapping, with the last iteration including 100 random addition replicates. Bootstrapping of the data used both distance and parsimony estimations with the same models and parameters as heuristic searches on 100 replicates, with one random-addition replicate per bootstrap replicate, under minimum-evolution criteria. In most cases, branch swapping during bootstrap analyses was limited to 106 rearrangements.
Nucleotide sequence accession number.
The DNA sequences are available in the GenBank database under accession no. AF534425 to AF534464.
RESULTS
The Toolik inlet stream is typically frozen with no detectable flow from October to April. It usually begins to flow in mid to late May and, supplied with snow meltwater, flows at a very high rate for 2 to 4 weeks. Most of this water flows under the ice cover on the lake. Once ice leaves the lake, the stream has a very low summer flow rate except during storm events in the catchment. In 1996, the inlet stream began to flow at a slow rate on 13 May, reached its peak flow rate (9.8 m3 s−1) on 24 May (Fig. 2), and remained high until 12 June (a total of 19 days). During the entire period of high stream flow, Toolik Lake remained ice covered, becoming ice free after 19 June. The flow rate of the inlet stream then decreased rapidly and remained very low for the rest of the summer. In 2000, the inlet stream began to flow on 2 June, and the lake became ice free on 24 June.
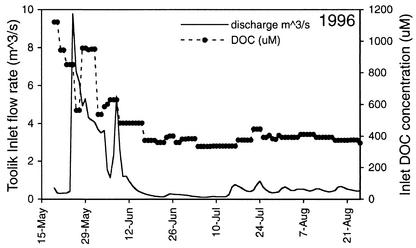
FIG. 2.
Discharge rate (solid line) and DOC concentration (dashed line with data points) of water in the primary inlet stream to Toolik Lake.
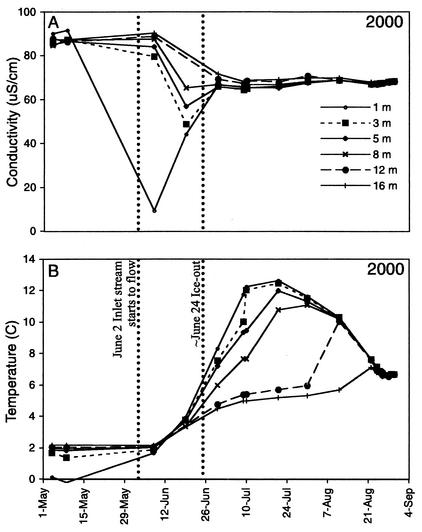
The temperature and conductivity of lake water in 2000 followed a typical seasonal progression. Inlet stream flow and melting ice produced a layer of low-conductivity water just beneath the ice in spring (Fig. 3A). After ice left the lake, thermal stratification developed in the upper 8 to 12 m (Fig. 3B). This stratification broke down in August as the lake cooled and mixed, and the surface of the lake began to freeze in September.
FIG. 3.
Conductivity (A) and temperature (B) in Toolik Lake in 2000.
The DOC concentration in the Toolik inlet stream in 1996 was highest in the spring when the flow rate of the stream was high (Fig. 2). The flux of DOC via the primary inlet stream reached a peak on 25 May, 12 days after the inlet stream began to flow and 3 days after daily average air temperature rose above 0°C. The flux of DOC remained at a high level, driven by rainfall and melting snow and ice until 11 June. After 11 June, the flux of DOC via the inlet stream remained very low for the rest of the season.
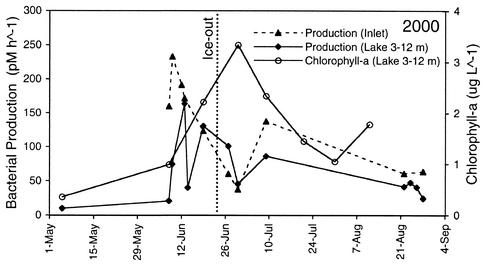
The chlorophyll concentration in the lake typically begins to increase below the ice as snow melts and reaches a peak after ice leaves the lake (33). In 2000, the chlorophyll concentration slowly increased after the start of stream flow and reached a peak after ice left the lake 28 days later (Fig. 4). The maximum chlorophyll concentration, measured at a depth of 3 m on 30 June, was 5.5 μg liter−1.
FIG. 4.
Bacterial production rate in the primary inlet stream (solid triangles), depth-averaged bacterial production rate (solid diamonds), and chlorophyll a concentration (solid circles, dashed line) in Toolik Lake in 2000.
Bacterial production and cell concentration.
Bacterial production increased rapidly in the Toolik inlet stream and in Toolik Lake during the first 10 days following the start of inlet stream flow in 2000 (Fig. 4). Bacterial production reached a peak in the inlet stream 7 days after flow began, followed by a peak in the lake 3 days later.
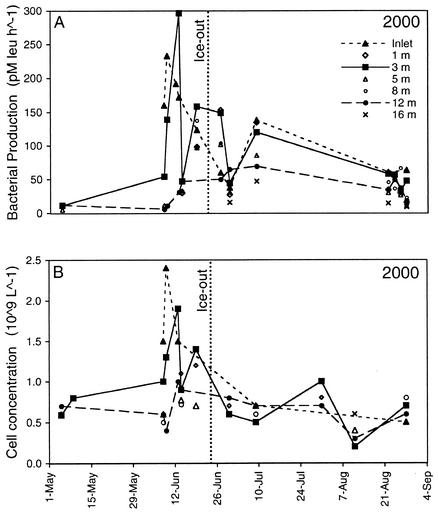
Bacterial production and cell concentration varied with depth in Toolik Lake during the first 10 days after the start of stream flow (Fig. 5). Bacterial production in near-surface waters (3 m below the bottom of the ice) at Toolik Main Station increased to nearly 30 times the winter production rate. Bacterial production at 12 to 16 m remained low, increasing only slightly to 2.5 times the winter rate after 10 days of stream flow.
FIG. 5.
Bacterial production rate (A) and prokaryotic cell concentration (B) in Toolik Lake and in the primary inlet stream in 2000.
Rates of bacterial production and cell concentration varied erratically at a depth of 3 m in 2000. The day after the highest measured rate, bacterial production at 3 m dropped to the same rate as at 12 m. Then, 3 days later in the sample collected in the inlet basin, bacterial production was at an intermediate level. The cell concentration followed a similar pattern. It is likely that the samples collected on 14 June were collected below the layer influenced by inlet stream water. Other measurements and DNA samples were not collected on June 14.
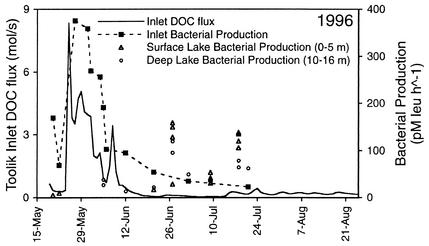
The bacterial production rate in the Toolik inlet stream in 1996 increased with the flux of DOC and reached a peak on 27 May, 2 days after the highest rate of DOC flux to Toolik Lake (Fig. 6). Bacterial production in Toolik Lake was only measured in the summer after stream flow had diminished, except for a few measurements in deep water samples made during the spring.
FIG. 6.
DOC flux via the primary inlet stream (solid line) and bacterial production rate in the inlet stream (solid squares), in Toolik Lake surface waters (open triangles), and in Toolik Lake deep waters (open circles) in 1996.
Bioassays.
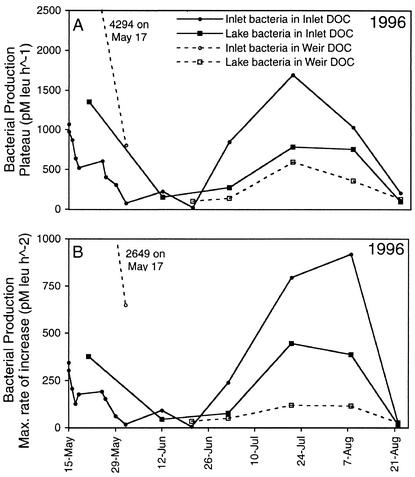
Two measurements from the bioassay incubations were used to estimate the relative quantity of available DOM in Toolik Lake and the Toolik inlet stream. The first was the rate of bacterial production at the upper growth plateau (picomoles of leucine incorporated per liter per hour). The second was the maximum slope between two successive measurements of bacterial production (picomoles of leucine incorporated per liter per hour squared).
Bacterial production in bioassay incubations was low for a few days, went through a period of rapid increase, and reached a plateau after 10 to 11 days. The quantity of labile DOM in the inlet stream decreased steadily from the first day of stream flow and reached a minimum on 1 June, 7 days after peak DOC flux. The quantity of labile DOM followed a similar pattern in the tussock tundra stream weir, with the first sample containing a great deal of labile organic matter (Fig. 7).
FIG. 7.
Quantity of labile DOC measured as the plateau in bacterial production rate (A) and the maximum rate of increase in bacterial production rate (B) during the bioassay incubations in 1996 for Toolik inlet stream DOC incubated with Toolik inlet bacteria (solid circles) and Toolik Lake bacteria (solid squares) and for tussock tundra weir DOC incubated with Toolik inlet bacteria (open circles) and with Toolik Lake bacteria (open squares). Note that measurements from the tussock tundra stream weir on 17 May 1996 were extremely high and therefore were off the scale of these graphs.
After the inlet stream flow diminished in early June, the quantity of labile DOM in the inlet stream remained low, but it rose again after ice left the lake in late June. However, by this time, DOC flux via the inlet was very low and likely had little influence on lake bacterial production.
DGGE.
A total of 30 samples were analyzed and compared by using DGGE banding patterns. We found that identical DGGE banding patterns were produced in separate PCRs of the same sample and were also produced for samples collected in duplicate. One difficulty we faced when comparing banding patterns across many samples on a DGGE gel was that the relative vertical position of some bands could not always be confirmed. This uncertainty was resolved by running multiple gels and loading samples in different orders so that the relative positions of bands could be compared side by side. This information was taken into account when matching bands in the DGGE gel.
Bacterial community composition was compared in all samples collected from Toolik inlet and from depths of 3 and 12 m at Toolik Lake Main Station as well as the sample collected on 19 June at a depth of 3 m in the inlet basin. DGGE bands were identified at 96 different positions in the gel, 12 of which appeared in all samples and 3 of which appeared in only one sample. Each sample contained between 38 and 55 identifiable bands in the analyzed range.
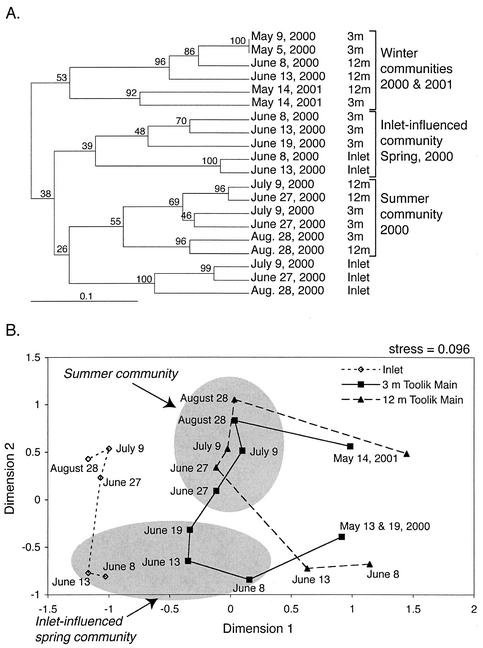
UPGMA cluster analysis (Fig. 8A) of these banding patterns showed a dramatic shift in bacterial community composition below the ice at 3 m between 19 May and 8 June, which appeared to be caused by the input of organisms from the inlet stream on 8 and 13 June. This shift did not occur at depth of 12 m, indicating that the inlet stream plume was restricted to the upper water column, consistent with the physical and chemical variables we measured. Another major shift in bacterial community composition was detected between 19 and 27 June, but this time, the shift was detected at both the 3- and 12-m depths. In the time between the first and second shifts, the inlet stream flow decreased to a very low level, ice on the surface of the lake melted, the water column became thermally stratified, and the chlorophyll concentration reached its seasonal maximum. Then, after the second shift, the bacterial community composition in the lake appeared to change more gradually as the season progressed to 28 August and through the winter to May of 2001.
FIG. 8.
UPGMA cluster analysis (with bootstrap values, 1,000 replications) (A) and multidimensional scaling analysis (B) (with stress value) of Dice distance matrix calculated from DGGE banding patterns. Brackets in UPGMA analysis and lines and gray circles on multidimensional scaling diagram were added to highlight the seasonal shifts in bacterial community composition as represented by DGGE banding patterns.
Multidimensional scaling analysis (Fig. 8B) highlights the seasonal succession of the bacterial community shifts. At a depth of 3 m on 8 June, the winter community began a gradual shift in the direction of the inlet stream community. On 27 June, after ice left the lake, the community shifted again to a relatively constant summer community that persisted from 27 June to 28 August. At a depth of 12 m, the winter community persisted until 13 June and then shifted directly from the winter community to the summer community. Another community shift occurred at both depths sometime between 28 August and 14 May of the following year.
DGGE analysis of samples collected at a range of depths demonstrated that bacterial community composition was fairly constant with depth after ice left the lake, despite thermal stratification. DGGE patterns were nearly identical in samples collected at 3, 5, 8, 12, and 16 m on 27 June, 9 July, and 28 August (data not shown).
DGGE band DNA sequencing. (i) Testing assumptions.
Sequencing revealed a great deal about the nature of DGGE banding patterns. Our first question was whether individual bands represented more than one organism. This was investigated by sequencing the same band from different samples and also by screening numerous clones prepared with the DNA extracted from each band (Fig. 9). In general, these clones contained at least one insert that matched the original band in the natural sample when run on a DGGE gel. However, many of the clones did not match up with the original band from the natural sample. Some were slightly offset, and others matched perfectly with other bands from the natural sample. Therefore it is essential when sequencing the DNA from DGGE bands to run each piece of DNA on a DGGE gel alongside the original natural sample to confirm that it matches the original band.
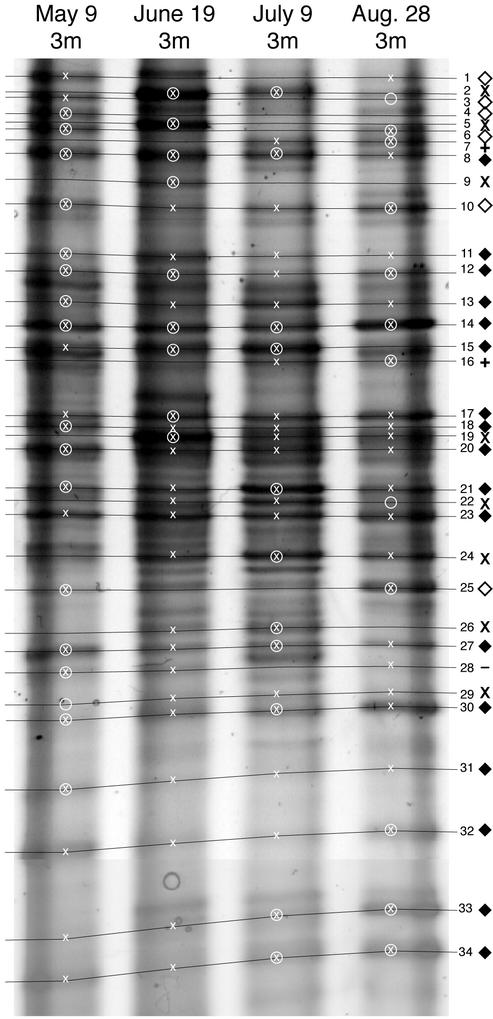
FIG. 9.
DGGE patterns of the four samples used for DGGE band sequencing. Samples were collected at a depth of 3 m in Toolik Lake. Lines connect bands at the same position in the gel. White X's indicate the presence of a band in a sample. White circles indicate the samples from which the bands were sequenced. Symbols to the right of the lines categorize the sequenced DGGE bands as persistent bands (solid diamonds), bands that appear below the ice in the spring (black X's), bands that disappear below the ice in the spring (open diamonds), bands that appear only after ice leaves the lake (+), and bands that do not fall into these categories (−).
In most cases, each DGGE band identified in this study represented one unique DNA sequence (Table 1). However, 5 of the 39 DGGE bands contained two unique sequences, and one band contained five unique sequences. In all cases, at least two of the unique sequences were drawn from the same natural sample. For these six bands, we cannot determine whether the pairs or groups of organisms are present in all samples, or, alternatively, whether one organism is present in one sample and another is present in another sample. Two of these six bands represent pairs of closely related bacteria belonging to the FukuN47 cluster of the Cytophaga-Flavobacteria-Bacteroides phylum. The rest represented phylogenetically distant organisms.
TABLE 1.
| Band no. | Phylum | Cluster | DGGE clone sequence | Nearly full-length sequence | Accession no. | Closest match | Source | Accession no. | % Similarity |
|---|---|---|---|---|---|---|---|---|---|
| Persistent bands | |||||||||
| 14 | Alpha | LD12 | TLMdgge14 | TLM01 | AF534425 | LD12 | Lake Loosdrecht | Z99997 | 100 |
| 21 | Beta | GKS98 | TLMdgge21 | TLM02 | AF534426 | GKS98 | Lake Gossenklle | AJ224990 | 100 |
| 27 | Beta | P. necessariusb | TLMdgge27 | AF534460 | CR-FL22 | Columbia River | AF141404 | 100 | |
| 33 | Beta | P. necessarius | TLMdgge33 | TLM03 | AF534427 | FukuS35 | Lake Fuchskuhle | AJ290013 | 100 |
| 13 | CFBg | FukuN47 | TLMdgge13 | AF534449 | SC-1-91 | Agricultural soil | AJ252666 | 97 | |
| 15 | CFB | Unknown | TLMdgge15 | AF534450 | ESR-14 | Lake Esrum | AF268298 | 99 | |
| 32 | CFB | Unknown | TLMdgge32 | AF534464 | Sta4-22 | Lake IJssel | AJ416239 | 96 | |
| 30 | Actinobacteria | ACK-M1 | TLMdgge30 | TLM06 | AF534430 | FukuN30 | Lake Fuchskuhle | AJ289996 | 100 |
| 31 | Actinobacteria | Unknown | TLMdgge31 | AF534463 | CR-FL30 | Columbia River | AF141411 | 99 | |
| 34 | Actinobacteria | Sta2-30 | TLMdgge34 | TLM07 | AF534431 | Sta2-30 | Lake IJssel | AJ416212 | 100 |
| 11 | Chloroplast | Chlorellaceae | TLMdgge11 | AF534447 | Plastidc | Freshwater | X65689 | 99 | |
| 17 | Chloroplast | Ochromonadaceae | TLMdgge17 | (TLM14)f | AF534453 | CL0-93 | Crater Lake | AF316708 | 98 |
| 18 | Chloroplast | Cryptomonadaceae | TLMdgge18 | AF534454 | LCK38 | Lake Cadagno | AF107328 | 99 | |
| 20 | Chloroplast | Cryptomonadaceae | TLMdgge20 | TLM13 | AF534437 | LCK38 | Lake Cadagno | AF107328 | 99 |
| 8a | CFB | FukuN47 | TLMdgge08a | AF534444 | SY6-50 | Lake Soyang | AF296201 | 98 | |
| 8b | CFB | FukuN47 | TLMdgge08b | AF534445 | CL0-11 | Crater Lake | AF316795 | 99 | |
| 12a | CFB | FukuN47 | TLMdgge12a | TLM09 | AF534433 | Clone 09 | Rimov Reservoir | AF361195 | 98 |
| 12b | CFB | FukuN47 | TLMdgge12b | AF534448 | FukuN47 | Lake Fuchskuhle | AJ290005 | 97 | |
| 23 | 5 different taxa | ||||||||
| Bands that appear below ice | |||||||||
| 22 | Alpha | Rickettsia | TLMdgge22 | AF534456 | AY-52 | Marine | AJ298360 | 97 | |
| 2 | Beta | BAL47 | TLMdgge02 | AF534439 | PRD01a007B | Parker River | AF289161 | 97 | |
| 26 | Beta | Methyloplilus | TLMdgge26 | AF534459 | GOBB3-CL275 | N. Baltic Sea | AF448464 | 99 | |
| 5 | CFB | Flavobacterium | TLMdgge05 | AF534440 | F. psychrophiliumd | Freshwater | AY034478 | 97 | |
| 24 | CFB | Flavobacterium | TLMdgge24 | (TLM12)f | AF534457 | ESR-18 | Lake Esrum | AF268302 | 100 |
| 9 | CFB | FukuN47 | TLMdgge09 | AF534446 | SY4-2 | Lake Soyang | AF107525 | 99 | |
| 29 | CFB | FukuN47 | TLMdgge29 | AF534462 | SC-1-91 | Agricultural soil | AJ252666 | 97 | |
| 19 | Chloroplast | Ochromonadaceae | TLMdgge19 | AF534455 | CL0-93 | Crater Lake | AF316708 | 99 | |
| Bands that disappear below ice | |||||||||
| 3 | Beta | BAL47 | TLMdgge03 | TLM04 | AF534428 | CR-FL9 | Columbia River | AF141392 | 99 |
| 10 | Beta | BAL47 | TLMdgge10 | TLM05 | AF534429 | CL120-116 | Crater Lake | AF316738 | 100 |
| 6 | CFB | Sphingobacterium | TLMdgge06 | AF534441 | Urk0-13 | Lake IJssel | AJ416172 | 98 | |
| 1 | CFB | Unknown | TLMdgge01 | TLM10 | AF534434 | Sta4-22 | Lake IJssel | AJ416239 | 94 |
| 4 | CFB | Unknown | TLMdgge04 | TLM11 | AF534435 | Unknown | |||
| 25a | Beta | P. necessarius | TLMdgge25a | TLM08 | AF534432 | CR-FL23 | Columbia River | AF141405 | 99 |
| 25b | Gamma | TLMdgge25b | AF534458 | Sta0-34 | Lake IJssel | AJ416166 | 97 | ||
| Bands that appear after ice out | |||||||||
| 7a | CFB | Flavobacterium | TLMdgge07a | AF534442 | CRE-PA32 | Columbia estuary | AF141515 | 98 | |
| 7b | CFB | LD2 | TLMdgge07b | AF534443 | LD2 | Lake Loosdrecht | AJ007871 | 98 | |
| 16a | Gamma | Legionella | TLMdgge16a | AF534451 | CL120-7 | Crater Lake | AF316800 | 99 | |
| 16b | Alpha | Unknown | TLMdgge16b | AF534452 | Endosymbionte | Freshwater | AF069963 | 94 | |
| No category | |||||||||
| 28 | Beta | P. necessarius | TLMdgge28 | AF534461 | CR-FL22 | Columbia River | AF141404 | 98 |
Included are major phylum, phylogenetic cluster clone name and accession number, closest match using BLAST analysis, environmental source and accession number of closest match, and percent similarity of DNA sequence in the DGGE region. Bands are grouped by categories indicating detection during the season. Nearly-full-length sequences drawn from environmental clone libraries exactly match DGGE band sequences and share the same accession number. Nearly-full-length sequences in parentheses are very close matches to DGGE band sequences and have separate accession numbers.
Polynucleobacter necessarius (15).
Chlorella sorokiniana.
Flavobacterium psychrophilum.
Endosymbiont of Acanthamoeba.
CFB, Cytophaga-Flavobacterium-Bacteroides group.
(ii) Community shifts before ice out.
Tracing the seasonal progression of individual DGGE bands and identifying the organisms they represent allow a more detailed look at these shifts in bacterial community composition. Most of the DGGE bands could be categorized as those that appeared or disappeared below the ice, those that appeared or disappeared after ice left the lake, and those that were present in all lake samples.
It is clear that bacteria from the inlet stream contributed to the initial shift in bacterial community composition at a depth of 3 m in the lake. A total of 24 new bands appeared under the ice at 3 m on 8, 13, or 19 June. Eighteen of these bands were also found in the inlet stream samples from 8 or 13 June. On 27 June, after ice left the lake, only eight of these bands could be found at 12 m, indicating that these organisms were restricted to the upper water column. The persistence of these 18 inlet bands in the lake was variable, but only 6 were still detectable on 28 August, and 4 remained until May of the following year.
Sequencing identified eight of the bands that appeared below the ice in the spring: seven bacteria and one chloroplast. Four were cytophaga-like organisms belonging to the cosmopolitan freshwater cluster FukuN47 (TLMdgge09 and TLMdgge29) and to the phylogenetically diverse genus Flavobacterium (TLMdgge05 and TLMdgge24). Two were β-Proteobacteria belonging to the freshwater clusters Rhodoferax sp. strain BAL47 (TLMdgge02) and Methylophilus methylotrophus (TLMdgge26). One was a strain of α-Proteobacteria (TLMdgge22), but it was not closely related to any other organism, except for one sequence from the ocean (Table 1 and Fig. 10).
FIG. 10.
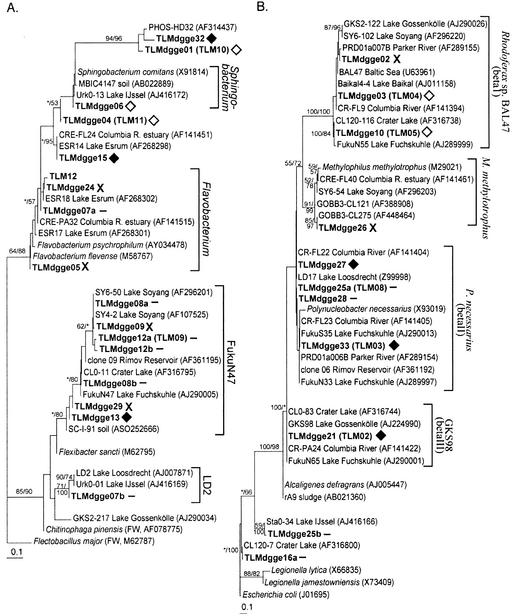
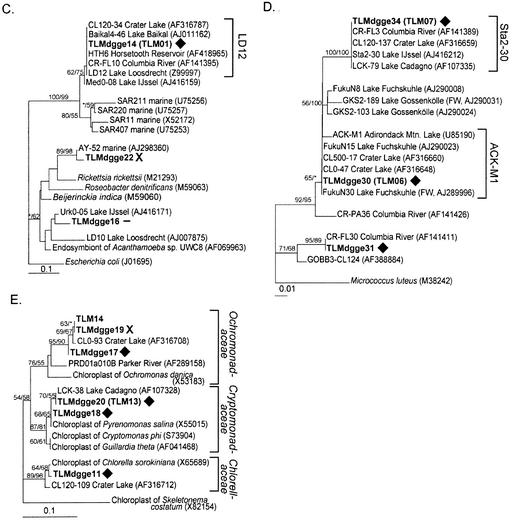
Minimum evolution trees showing the phylogenetic positions of organisms within the Cytophaga-Flavobacterium-Bacteroides group (A), β- and γ-Proteobacteria (B), α-Proteobacteria (C), Actinobacteria (D), and chloroplasts (E). Sequences from this study are in boldface type. Symbols following the sequences indicate whether the DGGE band was persistent (solid diamonds), disappeared below the ice in the spring (open diamonds), appeared below the ice in the spring (X's), or could not be categorized either because it was sequenced from a band containing more than one organism or because it did not fit into one of the previous categories (minus sign). Clusters are named after cultivated organisms or after the name of the longest available 16S rRNA gene sequence from an environmental clone. Parenthetical cluster names are from Glöckner et al. (13).
There were a total of 49 bands in the inlet stream on 8 and 13 June. As mentioned previously, 18 of these bands appeared at a depth of 3 m below the ice. Eight bands did not appear below the ice in the lake. The remaining 23 were already present in the lake when the inlet stream started to flow, and of these, 17 were persistent bands present in all lake samples collected in 2000.
Of the 43 winter bands present in the lake on 5 and 9 May 2000, 18 disappeared during early June at a depth of 3 m. At a depth of 12 m, however, all 18 of these bands persisted until 13 June or later, further indicating that the initial shift in bacterial community composition was restricted to the upper water column. Twelve of these bands reappeared at a depth of 3 m, although two bands did not reappear until May of the following year.
Five bands that disappeared below the ice in spring were identified through sequencing. Three were cytophaga-like organisms, two of which were related to the Sphingobacterium sp. (TLMdgge06) and to activated sludge clone sequence PHOS-HD32 (TLMdgge01). The other two clones were β-Proteobacteria belonging to the cosmopolitan freshwater cluster Rhodoferax sp. strain BAL47 (TLMdgge03, TLMdgge10). These β-Proteobacteria organisms were replaced with another, closely related member of the β-Proteobacteria (TLMdgge02) that washed into the lake via the inlet stream (Table 1 and Fig. 10)
(iii) After ice out.
After ice left the lake, 14 new bands appeared in summer samples collected at 3 m (27 June, 9 July, and 28 August). Eleven of these bands also appeared in 12-m samples. Only five of these bands were still detectable in May of 2001, suggesting that they represent organisms that are only active in the summer months. Two bands from this set were identified, but both represented more than one organism (Table 1), so the persistence of these organisms throughout the summer cannot be determined. However, none of these organisms was present before ice left the lake. Band 7 represented two cytophaga-like organisms belonging to the cosmopolitan freshwater cluster LD2 (TLMdgge07a) and to the genus Flavobacterium (TLMdgge07b). Band 16 represented a member of the γ-Proteobacteria that was nearly identical to an organism found in Crater Lake, Oreg. (TLMdgge16a), and a member of α-Proteobacteria distantly related to Rickettsia (Table 1 and Fig. 10).
(iv) Persistent bands.
The DGGE banding patterns provide evidence of a persistent bacterial community in the lake. Twenty bands were present in every lake sample collected in 2000, 18 of which were still present in May of the following year. Nineteen of these bands also appeared in some of the inlet samples, suggesting that these organisms may also be common in the smaller lakes farther upstream. Four of the five chloroplast bands identified were among these persistent bands. Seven bands represented members of cosmopolitan freshwater clusters of the α-Proteobacteria, β-Proteobacteria, Actinobacteria, and cytophaga-like organisms. TLMdgge14 was 100% identical to sequences from different freshwater systems and belonged to the LD12 cluster, otherwise known as the freshwater SAR11 cluster. Eight of the 10 persistent bacteria identified in this study (not including persistent bands representing more than one organism or chloroplasts) are 99 to 100% similar to organisms found in other freshwater plankton (Table 1 and Fig. 10).
(v) Winter bands.
Samples collected in May 2000 and May 2001 shared many bands. Of the 43 bands in samples from 5 and 9 May of 2000, a total of 33 appeared in one of the two samples collected on 14 May 2001. Eighteen of these bands were persistent lake bacteria, and 15 disappeared at some point during the summer and then reappeared.
DISCUSSION
The 2 to 4 weeks following the start of snowmelt and the initiation of stream flow is a time of rapid change for the planktonic communities existing below the ice in ultraoligotrophic Toolik Lake and potentially in other deep lakes of the Arctic tundra. Melting snow carries a large fraction of the total annual inputs of organic carbon, nitrogen, and phosphorus into Toolik Lake (50), stimulating the highest rates of bacterial production measured throughout the year. The peak in bacterial production that results from this stimulation occurs when the lake temperature is about 4°C, even though the lake temperature can be as high as 17°C in mid-summer (33). It is clear from the present study that this stimulation is due, in part, to the relatively high lability of this organic matter. At the same time as this stimulation of bacterial production, rapid shifts occur in the composition of planktonic bacterial communities. However, analysis of community composition demonstrated that the bacterial populations that benefit from this organic matter are not necessarily native to the lake, but rather are composed of bacteria that wash into the lake with the organic matter via the inlet stream.
After the flux of terrestrial organic matter slows and after ice leaves the lake, the planktonic food web of Toolik Lake takes on the characteristics of a typical open water oligotrophic system. A summer phytoplankton community develops, producing fresh organic matter that either complements or replaces terrestrial organic matter as the principle source of food for the bacterial community (33). The composition of the planktonic bacterial community changes with the development of this phytoplankton community and then remains relatively stable for the rest of the summer.
DOC inputs to Arctic lakes.
During the early spring most of the water passing into Toolik Lake via the inlet stream is surface runoff from snow meltwater, so changes in the quantity of labile DOM in the inlet stream are directly related to changes in the chemistry of snow meltwater during the spring thaw. Melting snow itself contains a relatively low concentration of organic matter (7). However, as the snow begins to melt, it becomes saturated with water, and this water leaches DOM from surface soil and dead plant material (7, 34). Research from the Arctic and Antarctic has demonstrated that the quantity and quality of this DOM are enhanced by the effect of seasonal freezing and thawing on the leaching process. Frozen dead plant leaves in the sub-Antarctic were shown to release up to 80% of soluble leaf carbohydrates when thawed (21). Moreover, freezing and thawing of Arctic soils were shown to increase bioactive DOM due to cell lysis of the microbial biomass (41). These dissolved materials are transported by meltwater flow over the surface of the frozen tundra soils and are washed into streams and lakes.
The subsequent decrease in the quantity of labile DOM in the inlet stream and the tussock tundra stream was probably caused by two factors. First, DOM becomes more dilute as the rate of leaching slows down and as more of the snowpack melts. Second, the thaw depth of the tundra increases over time and the flowpath of meltwater increasingly passes through soils where DOM can be removed or altered by soil microbes before reaching streams and lakes (30, 43).
As the rate of snowmelt and stream flow decreases, water from the catchment is altered as it spends more time in the chain of small lakes upstream of Toolik Lake (26). Phytoplankton production in these lakes is probably responsible for much of the labile organic matter detected in the inlet during the summer. Inlet flow and DOM supply, however, were much lower than during the spring and so probably had little impact on total bacterial production in Toolik Lake during low-flow conditions in summer.
In contrast with the DOM in the Toolik inlet stream, the quantity of labile DOM in the primary stream passing through the tussock tundra weir remains, for the most part, relatively low during the summer months. The important difference between these two streams is that the tussock tundra stream flows directly out of the tundra of this small catchment and does not pass through a lake. Therefore, it provides a better example of seasonal changes in the quality of terrestrially derived DOM. During the summer months the water in this stream is a combination of rainwater and tundra soil water. Before emerging in the stream, this water is in contact with the root zone of the tundra plants where it could acquire DOM (23). It appears, however, that this DOM contains much less labile substrate than snowmelt-associated DOM and somewhat less labile substrate than the DOM produced by phytoplankton in the small lakes upstream of Toolik Lake. It is likely that an active soil microbial community uses up much of the labile DOM before it emerges in the stream water.
Shifts in bacterial community composition.
This study demonstrates two primary mechanisms for shifts in bacterioplankton community composition: (i) succession resulting from in situ changes in new cell production among different populations of bacteria and (ii) introduction by advection of allochthonous populations of bacteria from inflowing streams. The shifts involved in the first mechanism are almost certainly complex phenomena driven by numerous environmental factors. Among those tested in laboratory experiments are phytoplankton species composition, protist grazer abundance, viral lysis, and selective grazing (14, 47-49). Results from our field study suggest that changes in the biochemical composition of DOM can also cause shifts in bacterial community composition. These environmental factors may influence different populations of bacteria such that rare organisms increase their net cell production and abundant organisms decrease their net cell production.
Based on the results of this study, the second mechanism, advection, must also be considered when drawing conclusions about shifts in community composition. The advection of allochthonous organisms, entrained in a particular water mass, is expected in aquatic systems like lakes, estuaries, and oceanic upwelling zones where different water masses mix. A bacterial cell budget of Lake Ortrasket, Sweden, determined that 29% of the new cells in some layers of the lake were imported by river flow (2). Lindstrom found a weak correlation between DGGE banding patterns and changes in the amount of water entering a boreal forest lake, suggesting that lake communities were influenced by imported bacterial populations (27). Crump et al. demonstrated the mixing of marine and riverine bacterial populations in the Columbia River estuary (5). Schauer et al. detected a shift in bacterial community composition along the Catalan coast and suggested that it could be caused by the intrusion of offshore slope waters via a submarine canyon (40). By sampling the source of advecting bacterial communities (i.e., the inlet stream), we were able to identify allochthonous populations in Toolik Lake and demonstrate that these populations were responsible for a shift in bacterial community composition.
This allochthonous bacterial community probably began to develop in water-saturated snow at the base of the thawing snowpack. Eventually, bacteria were added to the water from soils, vegetation, streams, and lakes as it flowed down through the catchment. Identifying the original source of each population in this community is not yet possible; however, one recent study suggested that bacteria living in lakes and rivers can be distinguished from bacteria living in soils by 16S rRNA gene sequences (53). The authors of this study identified 34 phylogenetic clusters of bacteria common to freshwater planktonic systems. In Toolik Lake, five of the seven allochthonous populations identified through DNA sequencing were members of these freshwater clusters, but only one population was closely related to an environmental clone from soil. So it appears the allochthonous bacterial community was composed primarily of freshwater bacterioplankton populations that developed in the spring with the influx of terrestrial organic matter.
The fate of this allochthonous community once it enters Toolik Lake depends, to some extent, on the fate of the stream water that carries it. As this water flows into Toolik Lake in the spring, it forms a thick layer below the ice and appears to entrain some lake water and lake bacteria based on DGGE banding patterns. Bacterial production in this layer was on the same scale as bacterial production in the inlet stream, suggesting that allochthonous bacterial populations continued to be active during the spring. However, most of these populations disappeared from the lake sometime during the summer. It is possible that these organisms were outcompeted by organisms growing on phytoplankton-produced organic matter in the summer, but it is also possible that many of these organisms were advected out of the lake at the end of the spring with the layer of water formed by the inlet stream. Inlet stream water flowing below the ice accounts for approximately 40% of annual inflow to Toolik Lake (50) and generally raises the lake level to its maximum annual height (33). A 1979 study monitored the flow of this stream water under the ice by measuring the distribution of rhodamine dye injected into the inlet stream (18). This study showed that the 2- to 5-m-thick layer formed by stream water crossed the lake quickly and appeared at the outlet stream at the north end of the lake in only 5 days. However, the actual quantity of stream and surface water leaving the lake has not been measured, so the importance of this loss term for the allochthonous community is unknown.
Once phytoplankton production increased and ice left the lake, the principal mechanism for shifts in bacterial community composition became succession. The shift in community composition during this period occurred rapidly at first when phytoplankton production was at its peak, as indicated by the shift at 12 m between 19 and 27 June (Fig. 8B). However, after this shift, changes in community composition occurred much more slowly for the rest of the summer. A few studies have linked shifts in bacterial community composition to phytoplankton growth, but most have been centered on the role of phytoplankton blooms. Mesocosm studies (3, 37) and studies of natural systems (8, 20, 51) have characterized numerous shifts in bacterial community composition during the progression of phytoplankton blooms. We detected a shift in bacterial community composition with the development of the phytoplankton community early in the summer, but unlike results from a number of these studies, that bacterial community remained fairly constant for the rest of the summer. This difference may be due to a difference in the scale of phytoplankton production. Phytoplankton growth in Toolik Lake is limited by low nutrient levels and so does not undergo the same dynamics as blooms typical of mesotrophic or eutrophic systems. Therefore, we did not see shifts in bacterial community composition associated with the various stages of growth and decline in a phytoplankton bloom.
The identity of populations responsible for bacterial production in Toolik Lake cannot be determined with certainty because of the potential influence of grazing and other top-down controls. Heterotrophic nanoflagellates (HNF) are the most important grazers of bacteria in Toolik Lake, and their grazing rate can be very high (16, 38). Bacterivory by HNF should increase the rate of succession in bacterial communities by cropping slow or nongrowing populations. But this is complicated by the phenomenon of selective grazing. HNF graze more efficiently on relatively large bacterial cells (24) and may graze preferentially on actively growing cells (6). Therefore, some active populations may be grazed too heavily to become abundant, and some inactive populations may escape grazing and remain abundant. However, changes in community composition over time can provide some information about the activity of individual populations. Populations that appeared during community succession, such as those that appeared in Toolik Lake during the summer, were probably active, because their numbers increased to a point that they became detectable. Likewise, populations that disappeared during community succession either became less active or encountered some new form of mortality that decreased their relative abundance (e.g., grazing and viral lysis). The same logic cannot be applied to the populations that appeared below the ice in the spring, because many were advected into the lake via the inlet stream. The activity of persistent bacterial populations also cannot be linked to their appearance in the water column, but the continued presence of these populations throughout the year may indicate that their growth rate is high enough to counter losses due to grazing and other forms of mortality.
Persistent bacterial populations.
Overall, shifts in bacterial community composition in Toolik Lake and other ultraoligotrophic systems can be expected to occur slowly due to the slow growth rate of the microorganisms. And indeed, the community shifts described in our research were not rapid or radical. A large fraction of the bacterial community in the lake, nearly half the populations identified in each sample, was made up of persistent populations present in all other samples from this lake during the year. Most of these persistent populations were also found in the inlet stream, indicating that they were present in the smaller lakes upstream.
The fact that these persistent populations appeared unaffected by shifting sources of labile organic matter suggests that either they are generalists capable of growing on both terrestrially and phytoplankton-produced organic matter or they gain their energy from other sources, such as the large pool of recalcitrant DOM present in the lake at all times. Many of the persistent populations are nearly identical to organisms found in a wide variety of freshwater environments. For example, the organism represented by band 14 is a member of the LD12 phylogenetic cluster (53), which is the freshwater cluster of the widely distributed (and principally marine) SAR11 α-Proteobacteria group. This organism has been found in both ultraoligotrophic Toolik Lake (1) and highly eutrophic Lake Loosdrecht (53). Also, the DNA sequences from three persistent bands were related to the Actinobacteria and were 99 to 100% identical to organisms found in many other freshwater systems. The global distribution of these populations and the high degree of relatedness within their phylogenetic clusters suggest that they are extremely adaptable to many different freshwater systems with different supplies of nutrients and labile organic matter.
Conclusions.
Planktonic bacterial communities in Toolik Lake, Alaska, are composed of persistent populations and transient populations. Most transient populations can be divided into those that are advected by snow meltwater into the upper water column of the lake below the ice in spring and those that grow up from low concentrations in response to the development of the phytoplankton community in early summer. Advected bacteria are probably responsible for high levels of bacterial production below the ice in early spring, consuming labile organic matter carried off the tundra and into the lake by snow meltwater. A different community of bacteria is responsible for bacterial production after ice leaves the lake, likely consuming organic matter released by phytoplankton. Most of the bacteria identified by sequencing the DNA from DGGE bands were related to organisms found in other freshwater planktonic systems worldwide, and many were members of globally distributed phylogenetic clusters of freshwater bacteria.
Acknowledgments
This work was supported by National Science Foundation LTER grant no. 9810222.
Primary production data for 2000 were supplied by M. A. Evans. We thank M. Sogin and the Bay Paul Center for Comparative Molecular Biology for laboratory facilities and expertise; A. Bollmann, E. van Hannen, and G. Zwart for help with DGGE; and K. Judd, G. Kipphut, K. Riseng, and M. Costa for assistance in the field.
REFERENCES
- 1.Bahr, M., J. E. Hobbie, and M. L. Sogin. 1996. Bacterial diversity in an Arctic lake: a freshwater SAR11 cluster. Aquat. Microb. Ecol. 11:271-277. [Google Scholar]
- 2.Bergström A.-K., and M. Jansson. 2000. Bacterioplankton production in humic Lake Örträsket in relation to input of bacterial cells and input of allochthonous organic carbon. Microb. Ecol. 39:101-115. [DOI] [PubMed] [Google Scholar]
- 3.Castberg, T., A. Larsen, R. A. Sandaa, C. P. D. Brussaard, J. K. Egge, M. Heldal, R. Thyrhaug, E. J. van Hannen, and G. Bratbak. 2001. Microbial population dynamics and diversity during a bloom of marine coccolithophorid Emiliania huxleyi (Haptophyta). Mar. Ecol. Prog. Ser. 221:39-46. [Google Scholar]
- 4.Cole, J. J., S. Findlay, and M. L. Pace. 1988. Bacterial production in fresh and saltwater ecosystems: a cross-system overview. Mar. Ecol. Prog. Ser. 43:1-10. [Google Scholar]
- 5.Crump, B. C., E. V. Armbrust, and J. A. Baross. 1999. Phylogenetic analysis of particle-attached and free-living bacterial communities in the Columbia River, its estuary, and the adjacent coastal ocean. Appl. Environ. Microbiol. 65:3192-3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.del Giorgio, P. A., J. M. Gasol, D. Vaqué, P. Mura, S. Agustí, and C. M. Duarte. 1996. Bacterioplankton community structure: protists control net production and the proportion of active bacteria in a coastal marine community. Limnol. Oceanogr. 41:1169-1179. [Google Scholar]
- 7.Everett, K. R., D. L. Kane, and L. D. Hinzman. 1996. Surface water chemistry and hydrology of a small Arctic drainage basin, p. 185-201. In J. F. Reynolds and J. D. Tenhunen (ed.), Landscape function and disturbance in Arctic tundra. Springer-Verlag, New York, N.Y.
- 8.Fandino, L. B., L. Riemann, G. F. Steward, R. A. Long, and F. Azam. 2001. Variations in bacterial community structure during a dinoflagellate bloom analyzed by DGGE and 16S rDNA sequencing. Aquat. Microb. Ecol. 23:119-130. [Google Scholar]
- 9.Fisher, M. M., and E. W. Triplett. 1999. Automated approach for ribosomal intergenic spacer analysis of microbial diversity and its application to freshwater bacterial communities. Appl. Environ. Microbiol. 65:4630-4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuhrman, J. A., K. McCallum, and A. A. Davis. 1993. Phylogenetic diversity of subsurface microbial communities from the Atlantic and Pacific Oceans. Appl. Environ. Microbiol. 59:1294-1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giovannoni, S. J., and M. Rappe. 2000. Evolution, diversity, and molecular ecology of marine prokaryotes, p. 47-84. In D. L. Kirchman (ed.), Microbial ecology of the oceans. Wiley-Liss, New York, N.Y.
- 12.Giovannoni, S. J., T. B. Britschgi, C. L. Moyer, and K. G. Field. 1990. Genetic diversity in Sargasso Sea bacterioplankton. Nature 345:60-62. [DOI] [PubMed] [Google Scholar]
- 13.Glöckner, F. O., E. Zaichikov, N. Belkova, L. Denissova, J. Pernthaler, A. Pernthaler, and R. Amann. 2000. Comparative 16S rRNA analysis of lake bacterioplankton reveals globally distributed phylogenetic clusters including an abundant group of actinobacteria. Appl. Environ. Microbiol. 66:5053-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hahn, M. W., and M. G. Höfle. 1999. Flagellate predation on a bacterial model community: interplay of size-selective grazing, specific bacterial cell size, and bacterial community composition. Appl. Environ. Microbiol. 65:4863-4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heckmann, K., and H. J. Schmidt. 1987. Polynucleobacter necessarius gen. nov., sp. nov., an obligately endosymbiotic bacterium living in the cytoplasm of Euplotes aediculatus. Int. J. Syst. Bacteriol. 37:456-457. [Google Scholar]
- 16.Hobbie, J. E., M. Bahr, and P. A. Rublee. 1999. Controls on microbial food webs in oligotrophic arctic lakes. Arch. Hydrobiol. Spec. Issues Advanc. Limnol. 54:61-76. [Google Scholar]
- 17.Hobbie, J. E. 1997. History of limnology in Alaska: expeditions and major projects, p. 45-60. In A. M. Milner and M. W. Oswood (ed.), Freshwaters of Alaska. Springer-Verlag, New York, N.Y.
- 18.Hobbie, J. E., T. L. Corliss, and B. J. Peterson. 1983. Seasonal patterns of bacterial abundance in an Arctic lake. Arct. Alp. Res. 15:253-259. [Google Scholar]
- 19.Hobbie, J. E., R. J. Daley, and S. Jasper. 1977. Use of Nuclepore filters for counting bacteria by fluorescence microscopy. Appl. Environ. Microbiol. 33:1225-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Höfle, M. G., H. Haas, and K. Dominik. 1999. Seasonal dynamics of bacterioplankton community structure in a eutrophic lake as determined by 5S rRNA analysis. Appl. Environ. Microbiol. 65:3164-3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hurst, J. L., G. J. F. Pugh, and D. W. H. Walton. 1985. The effects of freeze-thaw cycles and leaching on the loss of soluble carbohydrates from leaf material of two subantarctic plants. Polar Biol. 4:27-31. [Google Scholar]
- 22.Jensen, M. A., and N. Straus. 1993. Effect of PCR conditions on the formation of heteroduplex and single-stranded-DNA products in the amplification of bacterial ribosomal DNA spacer regions. PCR Methods Appl. 3:186-194. [DOI] [PubMed] [Google Scholar]
- 23.Judd, K. E., and G. W. Kling. 2002. Production and export of dissolved carbon in arctic tundra mesocosms: the roles of vegetation and water flow. Biogeochemistry 60:213-234. [Google Scholar]
- 24.Jürgens, K., and H. Güde. 1994. The potential importance of grazing-resistant bacteria in planktonic systems. Mar. Ecol. Prog. Ser. 112:169-188. [Google Scholar]
- 25.Kirchman, D. L. 1993. Leucine incorporation as a measure of biomass production by heterotrophic bacteria, p. 513-517. In P. F. Kemp, B. F. Sherr, E. B. Sherr, and J. J. Cole (ed.), Handbook of methods in aquatic microbial ecology. Lewis Publishers, Boca Raton, Fla.
- 26.Kling, G. W., G. W. Kipphut, M. M. Miller, and W. J. O'Brien. 2000. Integration of lakes and streams in a landscape perspective: the importance of material processing in spatial patterns and temporal coherence. Freshwater Biol. 43:477-497. [Google Scholar]
- 27.Lindstrom, E. S. 1998. Bacterioplankton community composition in a boreal forest lake. FEMS Microbiol. Ecol. 27:163-174. [Google Scholar]
- 28.Liu, W.-T., T. L. Marsh, H. Cheng, and L. J. Forney. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Massana, R., C. Pedros-Alio, E. O. Casamayor, and J. M. Gasol. 2001. Changes in marine bacterioplankton phylogenetic composition during incubations designed to measure biogeochemically significant parameters. Limnol. Oceanogr. 46:1181-1188. [Google Scholar]
- 30.Michaelson, G. J., C. L. Ping, G. W. Kling, and J. E. Hobbie. 1998. The character and bioactivity of dissolved organic matter at thaw and in the spring runoff waters of the Arctic tundra north slope, Alaska. J. Geophys. Res. 103:28939-28946. [Google Scholar]
- 31.Miller, M. C., G. R. Hater, P. Spatt, P. Westlake, and P. Yeakel. 1986. Primary production and its control in Toolik Lake, Alaska. Arch. Hydrobiol. 74:97-134. [Google Scholar]
- 32.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Brien, W. J., M. Bahr, A. E. Hershey, J. E. Hobbie, G. W. Kipphut, G. W. Kling, H. Kling, M. McDonald, M. C. Miller, P. Rublee, and J. R. Vestal. 1997. The limnology of Toolik Lake, p. 61-106. In A. M. Milner and M. W. Oswood (ed.), Freshwaters of Alaska. Springer-Verlag, New York, N.Y.
- 34.Oswood, M. W., G. Irons III, and D. M. Schell. 1996. Dynamics of dissolved and particulate carbon in an Arctic stream, p. 275-289. In J. F. Reynolds and J. D. Tenhunen (ed.), Landscape function and disturbance in Arctic tundra. Springer-Verlag, New York, N.Y.
- 35.Porter, K. G., and Y. S. Feig. 1980. The use of DAPI for identifying and counting aquatic microflora. Limnol. Oceanogr. 25:943-948. [Google Scholar]
- 36.Posada, D., and K. A. Crandall. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 37.Riemann, L., G. F. Steward, and F. Azam. 2000. Dynamics of bacterial community composition and activity during a mesocosm diatom bloom. Appl. Environ. Microbiol. 66:578-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rublee, P. A. 1992. Community structure and bottom-up regulation of heterotrophic microplankton in Arctic LTER lakes. Hydrobiology 240:133-141. [Google Scholar]
- 39.Schafer, H., L. Bernard, C. Courties, P. Lebaron, P. Servais, R. Pukall, E. Stackebrandt, M. Troussellier, T. Guindulain, J. Vives-Rego, and G. Muyzer. 2001. Microbial community dynamics in Mediterranean nutrient-enriched seawater mesocosms: changes in the genetic diversity of bacterial populations. FEMS Microbiol. Ecol. 34:243-253. [DOI] [PubMed] [Google Scholar]
- 40.Schauer, M., R. Massana, and C. Pedros-Alio. 2000. Spatial differences in bacterioplankton composition along the Catalan coast (NW Mediterranean) assessed by molecular fingerprinting. FEMS Microbiol. Ecol. 33:51-59. [DOI] [PubMed] [Google Scholar]
- 41.Schimel, J. P., and J. S. Clein. 1996. Microbial response to freeze-thaw cycles in tundra and taiga soils. Soil Biol. Biochem. 28:1016-1066. [Google Scholar]
- 42.Sîmek, K., P. Kojecka, J. Nedoma, P. Hartman, J. Vrba, and J. R. Dolan. 1999. Shifts in bacterial community composition associated with different microzooplankton size fractions in a eutrophic reservoir. Limnol. Oceanogr. 44:1634-1664. [Google Scholar]
- 43.Skogland, T., S. Lomeland, and J. Goksoyr. 1988. Respiratory burst after freezing and thawing of soil: experiments with soil bacteria. Soil Biol. Biochem. 20:851-856. [Google Scholar]
- 44.Swofford, D. L. 1999. PAUP* Phylogenetic Analysis Using Parsimony (*and Other Methods), version 4. Sinauer Associates, Sunderland, Mass.
- 45.Thurman, E. M. 1985. Organic geochemistry of natural waters. Martinus Nijhoff/Dr. W. Junk, Publishers, Dordrecht, The Netherlands.
- 46.Urbach, E., K. L. Vergin, A. Morse, G. L. Larson, and S. J. Giovannoni. 2001. Unusual bacterioplankton community structure in ultra-oligotrophic Crater Lake. Limnol. Oceanogr. 46:557-572. [Google Scholar]
- 47.Van Hannen, E. J., W. Mooji, M. P. van Agterveld, H. J. Gons, and H. J. Laanbroek. 1999. Detritus-dependent development of the microbial community in an experimental system: qualitative analysis by denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 65:2478-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Hannen, E. J., M. Veninga, J. Bloem, H. J. Gons, and H. J. Laanbroek. 1999. Genetic changes in the bacterial community structure associated with protistan grazers. Arch. Hydrobiol. 145:25-38. [Google Scholar]
- 49.Van Hannen, E. J., G. Zwart, M. P. van Agterveld, H. J. Gons, J. Ebert, and H. J. Laanbroek. 1999. Changes in the bacterial and eukaryotic community structure after mass lysis of filamentous cyanobacteria associated with viruses. Appl. Environ. Microbiol. 65:795-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Whalen, S. C., and J. C. Cornwell. 1985. Nitrogen, phosphorous, and organic carbon cycling in an Arctic lake. Can. J. Fish. Aquat. Sci. 42:797-808. [Google Scholar]
- 51.Yager, P. L., T. L. Connelly, B. Mortazavi, K. E. Wommack, N. Band, J. E. Bauer, S. Opsahl, and J. T. Hollibaugh. 2001. Dynamic bacterial and viral response to an algal bloom at subzero temperatures. Limnol. Oceanogr. 46:790-801. [Google Scholar]
- 52.Zhou, J., M. A. Bruns, and J. M. Tiedje. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:316-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zwart, G., B. C. Crump, M. P. Kamst-van Agterveld, F. Hagen, and S. K. Han. 2002. Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of freshwater lakes and rivers. Aquat. Microb. Ecol. 28:141-155. [Google Scholar]
- 54.Zwart, G., W. D. Hiorns, B. A. Methe, M. P. van Agterveld, R. Huismans, S. C. Nold, J. P. Zehr, and H. J. Laanbroek. 1998. Nearly identical 16S rRNA sequences recovered from lakes in North America and Europe indicate the existence of clades of globally distributed freshwater bacteria. Syst. Appl. Microbiol. 21:546-556. [DOI] [PubMed] [Google Scholar]