Abstract
The expression of the MyoD gene homolog, nautilus (nau), in the Drosophila embryo defines a subset of mesodermal cells known as the muscle “pioneer” or “founder” cells. These cells are thought to establish the future muscle pattern in each hemisegment. Founders appear to recruit fusion-competent mesodermal cells to establish a particular muscle fiber type. In support of this concept every somatic muscle in the embryo is associated with one or more nautilus-positive cells. However, because of the lack of known (isolated) nautilus mutations, no direct test of the founder cell hypothesis has been possible. We now have utilized toxin ablation and genetic interference by double-stranded RNA (RNA interference or RNA-i) to determine both the role of the nautilus-expressing cells and the nautilus gene, respectively, in embryonic muscle formation. In the absence of nautilus-expressing cells muscle formation is severely disrupted or absent. A similar phenotype is observed with the elimination of the nautilus gene product by genetic interference upon injection of nautilus double-stranded RNA. These results define a crucial role for nautilus in embryonic muscle formation. The application of RNA interference to a variety of known Drosophila mutations as controls gave phenotypes essentially indistinguishable from the original mutation. RNA-i provides a powerful approach for the targeted disruption of a given genetic function in Drosophila.
Keywords: myogenesis, founder cells
The role of specific genes during development traditionally is determined by selecting or creating a mutation in the gene of interest followed by detailed analysis of the phenotype. Because there is no targeted homologous recombination in Drosophila to allow “knock-out” or gene-replacement strategies, mutations usually are obtained by a combination of genetic selection, P element insertion, and deficiency analysis after γ-irradiation. These methods are very labor-intensive and time-consuming and, on occasion, do not yield a clear answer.
We have been studying the role of the Drosophila MyoD homolog, nautilus, in the formation of embryonic muscle. Previous studies in Drosophila and grasshopper have suggested that particular sets of muscle precursor cells establish a muscle prepattern in each hemisegment and then recruit fusion-competent mesodermal cells to build the characteristic muscle groupings (1–3). This is known as the muscle “founder” or muscle “pioneer” theory of muscle formation. In Drosophila the nautilus-expressing cells are thought to be the muscle “founder cells.” Currently, no point mutations exist for the gene, although deficiencies have been characterized that are reported to eliminate nautilus (4–6). In the absence of a mutation in nautilus, two alternative strategies were employed to determine its role in myogenesis. First, we used the specificity of the nautilus promoter to target gal4-activated ricin toxin expression (7, 8) only in nautilus-positive cells (9), and, second, nautilus double-stranded RNA (dsRNA) was injected into embryos to genetically interfere with gene function, as reported previously in studies with Caenorhabditis elegans (10). Our goal was to determine the role of the “founder” cells in Drosophila embryo myogenesis and to investigate the direct role of nautilus in embryonic muscle formation.
Studies in C. elegans have demonstrated elegantly that the introduction of dsRNA corresponding to either a portion or the entire coding region of a particular gene into embryonic cells can interfere with the function of the endogenous gene to give a phenotype essentially equivalent to the known genetic mutation (10). Furthermore, injection into the adult resulted in genetic interference in the next generation of worms. This dsRNA interference (RNA-i) with genetic function was substantially more effective than the injection of either RNA strand alone. Surprisingly, the results strongly suggested that dsRNA was acting catalytically. The application of this method to Drosophila is demonstrated here in the analysis of nautilus function during muscle formation in the embryo. The general utility of RNA-i in the analysis of gene function during Drosophila development is demonstrated by the injection of dsRNAs representing a panel of genes expressed at various embryonic stages. The mutant phenotype was observed for essentially all genes tested.
MATERIALS AND METHODS
Preparation of dsRNA for Injection.
Sequences to be expressed as dsRNA were cloned into Bluescript KS(+), linearized with the appropriate restriction enzymes, and transcribed in vitro with the Ambion T3 and T7 Megascript kits following the manufacturer’s instructions. Transcripts were annealed in injection buffer (0.1 mM NaPO4, pH 7.8/5 mM KCl) after heating to 85°C and cooling to room temperature over a 1- to 24-hr period. All annealed transcripts were analyzed on agarose gels with DNA markers to confirm the size of the annealed RNA and quantitated as described previously (10). Injected RNA was not gel-purified because the efficacy was the same. We estimate that the injection of 0.1 nl of a 0.1- to 1.0-mg/ml solution of a 1-kb dsRNA corresponds to roughly 107 molecules. Sources for the DNA clones were as follows: nautilus (9), twist (11), daughterless (12, 13), engrailed (14), S59 (15), DMEF2 (16), β-galactosidase, and white (17).
Toxin Ablation System and Antisense Expression.
The targeted ablation strategy using controlled expression of wild-type ricin was developed by A. Brand (7, 8). Stocks of the UAS-ricin A gene flies were kindly provided by A. Brand. Briefly, to prevent transient expression of ricin toxin after injection, the white gene is inserted between the gal4UAS and the ricin A coding sequence and is flanked by flp recombinase target sites. Heat-shock flp recombinase is introduced by genetic cross. The UAS-ricin is balanced over CyO marked by en-lacZ at the wingless locus. The heat-shock flp line was injected with the gal4 P element driven by the 8.5-kb nautilus promoter that specifically expresses gal4 only in nautilus-positive cells at all stages of development, as described previously (9). To express nautilus antisense RNA in embryos only the coding region of nautilus was inserted in reverse orientation into the gal4 UAS plasmid and injected into embryos to produce the transgene stock (18). Four independent lines were established by standard procedures. Antisense expression was induced by crossing the gal4 UAS antisense nautilus line with the gal4 enhancer trap line 24B/twist gal4 that expresses high levels of gal4 from the twist promoter and the mesoderm-restricted enhancer trap line 24B (18, 19).
Injection of Embryos Through the Chorion.
Cages were set up using 2- to 4-day-old yw 67c flies. Agar–grape juice plates were alternated every hour to synchronize the egg collection for 1–2 days. The eggs were collected over a 30- to 60-min period for subsequent injection. The eggs were washed into a nylon mesh basket with tap water. Eggs were lined up anterior to posterior directly onto a glass slide. The eggs were injected in the posterior end slightly off-center using an Eppendorf transjector. After injection, slides were stored in a moist chamber to prevent drying out the embryos. The S59 lacZ line used for injections of S59 dsRNA was from M. Frasch (unpublished data).
General Methods.
Antibody stainings, β-galactosidase assays, and cuticle preparations were done following standard procedures and as described previously (9).
RESULTS
Requirement for nautilus-Expressing Cells in Drosophila Embryonic Muscle Formation.
We first investigated the specific ablation of nautilus-expressing cells by taking advantage of the gal4/UAS system to express wild-type ricin toxin in nautilus-positive embryonic cells (7, 8, 18, 19). We have shown previously that an 8.5-kb nautilus promoter fragment will drive lacZ expression specifically in the nautilus-positive embryonic cells to mimic the endogenous protein pattern (9). No ectopic expression was observed. This promoter fragment was used to express gal4 in the UAS ricin system. Activation of the wild-type ricin A chain is regulated by an flp cassette containing the white gene to block any toxin expression unless white is removed by heat-shock-activated flp recombinase. The ricin A fragment used in this system cannot cross cell membranes once it is released from ablated cells because it is missing the domain necessary to enter cells.
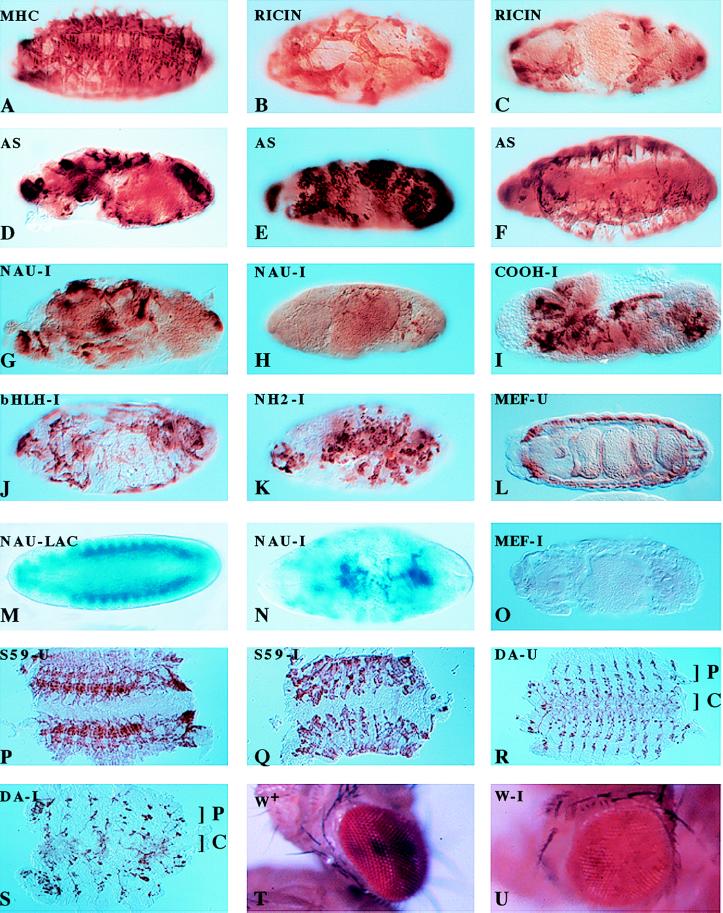
Flies expressing heat-shock FLPase that were also homozygous for a P element containing the nautilus promoter driving gal4 were crossed with flies carrying the UAS ricin cassette over a lacZ engrailed marker. Embryos were collected and heat-shocked 2 hr after egg laying to activate ricin expression in nautilus-positive cells. At various time points after heat shock embryos were fixed and analyzed for myosin and lacZ expression. Non-heat-shocked embryos were stained as controls for the normal muscle pattern by using myosin heavy chain antibody (9). Embryos not expressing ricin were marked by lacZ expression. The muscle pattern in all embryos expressing ricin was extremely disrupted (Fig. 1 B and C) whereas the control non-heat-shocked embryos had a normal embryonic muscle pattern (Fig. 1A). From the ricin toxin ablation experiments we conclude that the nautilus-expressing cells in the embryo are essential for muscle pattern formation, consistent with their role as muscle “founder” cells. The remaining mesodermal cells are unable to build and organize any muscle pattern in the absence of the nautilus-expressing cells. The variation in the ablation phenotype we observed is likely a result of the stage of muscle development at the time of ricin induction. Heat shock in larval instar stages also resulted in lethality (data not shown), implying that nautilus also plays a role in later Drosophila development, but we have not yet analyzed the pattern of muscle expression in these instances.
Figure 1.
Muscle formation and gene expression patterns in the Drosophila embryo as modulated by specific cell ablation, antisense expression, and RNA interference by the injection of dsRNA. Ablation of the nautilus-positive muscle founder cells by ricin toxin disrupts muscle formation. (A) Ricin not induced. (B and C) Ricin induced in nautilus-positive cells. Antisense expression of nautilus RNA disrupts muscle formation in three different UAS antisense (AS) nautilus lines (D– F). Injection of nautilus dsRNA blocks muscle formation (G and H) and does not depend on the bHLH domain for the disruption [dsRNA for the C terminus (I), dsRNA for the bHLH domain (J), and dsRNA for the amino terminus of nautilus (K)]. Injection of β-galactosidase dsRNA does not disrupt the muscle pattern but eliminates normal lacZ expression (shown in M) without affecting muscle pattern (similar to A), whereas injection of nautilus dsRNA into a nautilus lacZ line 14.1 disrupts the lacZ muscle pattern (compare M and N) and reduces lacZ expression. Injection of dsRNA for DMEF2 [uninjected (L) and injected (O)], S59 [uninjected (P) and injected (Q)], daughterless [uninjected (R) and injected (S); C, CNS; P, PNS], and white [uninjected (T) (w+) and injected (U)] results in the disruption of gene function for these genes. A–L and O–Q were stained with antimyosin; M and N were stained with 5-bromo-4-chloro-3-indolyl β-d-galactoside; R and S were stained with monoclonal 22C10 and horseradish peroxide for CNS and PNS. AS in the upper left-hand corner marks the nautilus antisense lines, -U indicates the uninjected phenotype for the designated gene, and -I indicates dsRNA injection for the indicated gene.
Disruption of the Drosophila Embryonic Muscle Pattern with the Expression of nautilus Antisense RNA in the Embryo.
“Knock-outs” and mutant screens in the mouse and C. elegans, respectively, have shown that MyoD-related proteins play a crucial role in myogenesis in these organisms (20, 21). To test whether the direct ablation of nautilus mRNA would result in a disrupted muscle phenotype in the Drosophila embryo, the gal4/UAS system was used to express nautilus antisense RNA throughout the mesoderm (18, 19). Females from the gal4 enhancer trap line 24B containing the twi-gal4 transgene were crossed with males from four independent lines homozygous for a gal4 UAS antisense nautilus transgene containing only the coding region in reverse orientation. The degree of disruption in the muscle pattern of the progeny flies depended partially on the particular antisense transgenic line used in the cross, as shown for three of these lines (Fig. 1 D–F). Previous studies with the overexpression of nautilus gave a phenotype that included the formation of some additional muscles and a disruption of the heart tube, presumably because of the formation of skeletal muscle cells in the heart tube itself (22). With the additional results from the antisense induction experiments we conclude that nautilus plays a major role in the formation of the muscle pattern and may be involved in the determination of the muscle “founder” cell lineage in the embryo, because the muscle phenotypes resulting from the ricin ablation of the nautilus-positive cells and the nautilus antisense expression were so similar.
Severe Disruption of the Muscle Pattern in the Embryo with the Injection of nautilus dsRNA.
Given the remarkable results obtained with the RNA interference studies in C. elegans (10), we decided to try the same approach in Drosophila, specifically addressing the question of nautilus function in embryonic muscle formation. To facilitate the rapid injection of 300–500 embryos for each dsRNA tested and the subsequent analysis of the mutant phenotypes, we developed methods to inject dsRNA directly through the chorion of the embryo. Our initial tests with full-length nautilus dsRNA gave a very severe pattern of muscle disruption (Fig. 1 G and H) and looked similar to the toxin ablation phenotype. Injection of either buffer alone or sodium azide gave no disruption of the muscle pattern (Table 1). To control for the specificity of the muscle pattern disruption, we injected nautilus dsRNA into a previously described transgenic line 14.1 in which the nautilus promoter drives lacZ expression in the founder cells and newly formed embryonic muscles (Fig. 1M) (9). Injection of nautilus dsRNA resulted in a severely disrupted blue muscle pattern (Fig. 1N) whereas injection of β-galactosidase dsRNA eliminated lacZ expression without any disruption in the muscle pattern (data not shown, but identical to Fig. 1A). Furthermore, the pattern of DMEF2 expression (23) essentially was normal in embryos injected with nautilus dsRNA (data not shown). Thus, the disruption of the muscle pattern observed depended on nautilus dsRNA.
Table 1.
Analysis of gene expression patterns using RNA interference
| Phenotype injected | RNA hybrid used | Known expression/mutant phenotype | Resulting phenotype | % of mutants |
|---|---|---|---|---|
| yw67c | Mook-injected buffer + dye | Wild-type MHC | Wild-type MHC | 0 |
| yw67c | Sodium azide | Unknown, should have a wild-type MHC | Wild-type MHC | 0 |
| yw67c | twist | Twisted embryos with marginal cuticular phenotype | Twisted embryos with marginal cuticular phenotype | 86 |
| yw67c | engrailed | Merged denticular bands, lawn of cuticle | Merged denticular bands, lawn of cuticle | 79 |
| yw67c | Mef2 | No muscle | No muscle | 72 |
| S59-β-gal | S59 | Muscles 18 and 25 absent | Muscles 18 and 25 absent | 75 |
| yw67c | daughterless | Most of the PNS absent; partly disrupted CNS | Most of the PNS absent; partly disrupted DNS | 85 |
| yw67c | nautilus | Mutant phenotype unknown | Disrupted muscle pattern; CNS and PNS normal | 76 |
| yw67c | 3′ Nautilus domain | Mutant phenotype unknown | Disrupted muscle pattern | 78 |
| yw67c | 5′ Nautilus domain | Mutant phenotype unknown | Disrupted muscle pattern | 76 |
| yw67c | bHLH domain | Mutant phenotype unknown | Disrupted muscle pattern | 74 |
| nau-β-gal | nautilus | Nautilus β-gal expression pattern | Disrupted β-gal muscle pattern | 80 |
| nau-β-gal | β-gal | Nautilus β-gal expression pattern | β-gal expression absent; wild-type MHC pattern | 80 |
MHC, major histocompatibility complex; β-gal, β-galactosidase.
It was formally possible that dsRNA representing the highly conserved basic helix–loop–helix (bHLH) domain in nautilus was acting nonspecifically with mRNA for another bHLH protein to disrupt the muscle pattern. This idea was used to explain the additional mutant phenotype observed with the injection of dsRNA encoding a conserved domain from the unc54C gene in C. elegans RNA-i studies (10). To address this possibility, we prepared dsRNAs representing the amino terminus (amino acids 1–147), the bHLH domain (amino acids 148–214), and the carboxyl terminus (amino acids 215-end) of nautilus for injection. All three domains produced a severe disruption of the muscle pattern (C terminus, bHLH domain, and amino terminus, Fig. 1 I–K, respectively), ruling out the nonspecific interaction of the nautilus bHLH dsRNA as a reason for the interference phenotype initially observed with full-length nautilus dsRNA.
RNA Interference and the Study of Gene Function in Drosophila.
The results from the injection of nautilus dsRNA pointed to a more general approach for the analysis of gene function during Drosophila development and suggested that the RNA interference method essentially would mimic a gene “knock-out” in the injected generation of Drosophila embryos. To test this idea we obtained a variety of cDNA clones representing a maternal gene expressed in the embryo (daughterless), additional genes involved in myogenesis (S59, DMEF2), homeobox genes (engrailed and S59), a gene important for gastrulation (twist), and a gene expressed in the adult eye (white). This panel of genes covers most stages of Drosophila development.
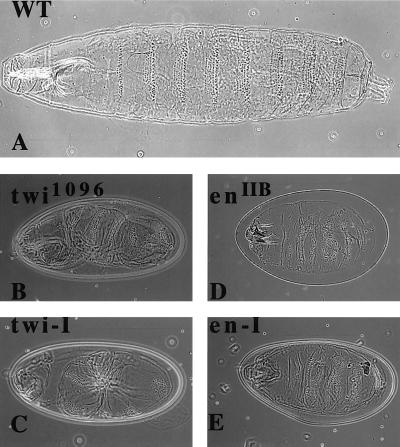
We initially tested twist because the mutant has a clear phenotype that is easy to score compared with the wild-type larva (Fig. 2A). The injection of twist dsRNA (the complete coding region) into embryos produced a twisted larval phenotype that was indistinguishable from the original twist mutation (Fig. 2B, tw 1096 mutant compared with the injected embryo in Fig. 2C). Similarly, injection of the first 1,200 bp of engrailed dsRNA produced the compressed dentical belt pattern characteristic of an engrailed null mutant (enIIB86) (Fig. 2D, en mutant compared with the injected embryo in Fig. 2E). Daughterless mRNA is both maternally loaded and expressed zygotically, and the mutant phenotype produces very characteristic disruptions in the central nervous system (CNS) and peripheral nervous system (PNS) (12, 13). It has been shown previously that mex3, a maternally loaded RNA in C. elegans, can be ablated by dsRNA injection into the gonads (10). We injected daughterless dsRNA (complete coding region) and looked for the characteristic neuronal phenotypes by using the mAb MAB 22C10 (12). The CNS as well as the PNS were disrupted to varying degrees in the injected embryos (Fig. 1S) compared with the uninjected embryos (Fig. 1R). The severity of the phenotype consistently showed a CNS disruption with a variable PNS pattern, possibly reflecting the fact that the CNS is formed before the PNS. This result suggested maternally loaded as well as zygotically expressed RNA can be affected by RNA-i in Drosophila. The homeobox gene S59 marks a subset of muscle founder cells for 5 of 29 muscles in each hemisegment of the embryo corresponding to muscles 5, 18, 25, 26, and 27 (15). Embryos with an S59 lacZ transgene marking muscles 18 and 25 (from M. Frasch) were injected with S59 dsRNA (complete coding region). The S59-specific lacZ antibody-staining pattern was abolished (data not shown). The total muscle pattern for embryos injected with S59 dsRNA (Fig. 1Q, injected embryos; Fig. 1P, uninjected embryos), although disrupted, still shows the presence of poorly organized muscle groups in each hemisegment. This is unlike the almost complete absence of muscle observed with the injection of nautilus dsRNA (Fig. 1 G and H). DMEF2, a member of the MADS domain transcription factor family, is essential for muscle formation in Drosophila (23). The DMEF2 −/− embryo has no muscle and is missing the characteristic gut constrictions found in the uninjected embryo (Fig. 1L). Injection of DMEF2 dsRNA (complete coding region) resulted in embryos lacking any detectable muscle and an absence of gut morphology (Fig. 1O).
Figure 2.
Cuticular patterns in early larvae induced by injection of twist and engrailed dsRNAs. (A) Wild-type cuticular pattern for early larva. (B) Twist phenotype of the known twist mutation twi1096. (C) Embryos injected with twist dsRNA show the same phenotype as in B. (D) Fused cuticular band phenotype seen for enIIB86 null mutants. (E) Embryos injected with the engrailed dsRNA show the same en null phenotype as in D.
Because particular RNA interference phenotypes were transferable to the next generation of C. elegans (10), we were particularly interested to see whether genes expressed in the adult eye could be affected by the injection of dsRNA into the embryo. We chose to look at the white gene, even though it is expressed throughout embryogenesis, and asked if any aspect of the white-eyed mutant phenotype could be observed after the injection of white dsRNA (the first 500 bp from the P element minigene) into wild-type embryos with red eyes. Phenotypes indicating interference with white gene function were observed in response to the RNA interference with white dsRNA (Fig. 1T, injected embryo; Fig. 1U, uninjected embryo), although the frequency of the mutant phenotype was extremely low (<3%) compared with the level of typical mutant phenotypes scored in the embryos injected with dsRNA (>75%). Similar to the results reported in C. elegans, very few molecules of white dsRNA appear to be required to obtain some evidence of interference with white gene function in the adult eye because we are injecting only on the order of 107 molecules. This last result supports the idea that RNA interference is acting catalytically because the transition from embryo to adult fly would substantially dilute the injected dsRNA. A summary of the phenotypes observed is given in Table 1.
DISCUSSION
Nautilus Is Required to Form Embryonic Muscle.
Unlike vertebrate myogenesis, where all skeletal muscle precursor cells express members of the MyoD family of regulatory proteins (21, 24), Drosophila muscle development involves two distinct cell types (1, 2, 6). One set of cells, denoted as the founder cells, is marked by the expression of the MyoD homolog, nautilus, and is thought to establish the muscle pattern in each hemisegment of the embryo. These cells then fuse with a second, uncommitted mesodermal cell population to build the muscle fiber pattern. Support for this idea comes from initial studies in the grasshopper where morphologically distinct mononucleated mesodermal cells, termed muscle “pioneers,” eliminate muscle formation when laser-ablated in the embryo (3). It is not known, however, whether the grasshopper muscle “pioneer” cells express MyoD because the homolog has not been isolated. Morphological examination of myogenesis in Drosophila has suggested a similar process is occurring, but it has not been directly demonstrated (1, 2).
The toxin ablation strategy (7, 8) has allowed us to eliminate specifically the nautilus-expressing cells and determine their role in myogenesis in the Drosophila embryo. The founder cell hypothesis predicts that loss of these cells should eliminate the muscle pattern and the formation of most muscle by the uncommitted mesodermal cells, and this is what was observed. This toxin strategy, developed to study the role of glial cells in axon tract formation, allowed the ablation of a selected population of glial cells in the Drosophila embryonic nervous system in a cell-autonomous manner and depended on only the targeted expression of gal4 through the appropriate promoter (8). Use of the nautilus promoter fragment to drive gal4 expression specifically and only in the nautilus-expressing mesodermal cells provided the same specificity in the ablation of these cells in the embryo (9).
Deficiencies have been reported that are thought to remove the nautilus gene with little effect on muscle pattern formation or viability (5, 6). This was interpreted to mean nautilus is not required for the formation of muscle precursors, but rather plays a role in their differentiation into mature fibers. Furthermore, nautilus was suggested to function in a subset of muscle precursors to implement their specific differentiation. Although the induction of antisense nautilus RNA with the gal4/UAS system produced a disruption of the muscle pattern, somewhat dependent on the antisense transgene line used, genetic interference through the injection of nautilus dsRNA completely eliminated the normal formation of the muscle groups. The myosin staining pattern was very similar to the one observed with the most severe ricin toxin ablation of the nautilus-expressing cells. These combined results suggest that nautilus expression defines the founder cell population and plays an essential role in myogenesis in the Drosophila embryo. The reasons for the discrepancy between our results and those published for the deficiencies reported to eliminate nautilus are not clear (6). However, the possible maternal contribution of nautilus was not ruled out completely. The cDNA probe used to determine the absence of the nautilus-coding region in the deficiency corresponded to the C terminus of nautilus and did not represent either the bHLH or amino-terminal portions of the protein. Therefore, it is theoretically possible that the deficiency still expresses a partially functional, truncated nautilus protein that could account for the minor nonlethal phenotype observed.
Genetic Interference by the Injection of dsRNA in Drosophila.
The severe muscle disruption phenotype observed with the injection of nautilus dsRNA representing the entire coding region gave us concern about the specificity of the interference. Because the bHLH domain is common to many regulatory proteins in Drosophila, including twist and daughterless, we injected dsRNA representing domains in nautilus outside the bHLH region. dsRNAs encoding regions of nautilus that do not contain the bHLH domain gave phenotypes essentially indistinguishable from that observed with the injection of the dsRNA for the entire coding region. This clearly demonstrated that the RNA interference phenotype observed for nautilus was specific and not a result of any homologous domain effect. This was supported further by the injection of dsRNA for β-galactosidase into the nautilus promoter-lacZ transgenic line 14.1 that expresses LacZ only in nautilus-positive cells that can be chased into most of the embryonic muscle groups: lacZ expression was eliminated with no effect on the muscle pattern. Likewise, early DMEF2 expression is not affected in embryos injected with nautilus dsRNA or induced for ricin expression (data not shown).
It was imperative that we established the specificity of the RNA interference approach in Drosophila. To do this we prepared a selection of dsRNAs for injection representing genes expressed maternally and zygotically in the embryo and, much later, in the adult eye. In practically every instance the phenotype observed from the injection of dsRNA was comparable to the original mutant, demonstrating the specificity of the procedure. This analysis underscores the utility of this approach. The function of any gene or combination of genes in Drosophila development, particularly in the embryo, can be disrupted simply by injecting dsRNAs representing all or a portion of the coding region of the gene or genes in question. The shortest piece of dsRNA we injected was roughly 200 bp, representing the bHLH region of nautilus. Shorter dsRNA fragments may work as well. RNA interference analysis of several genes in Drosophila as well as nautilus, combined with the fact that nautilus is expressed in roughly 30 muscle precursor cells in each hemisegment that are eventually incorporated into every somatic muscle of the embryo (9), allows us to conclude that nautilus does play a crucial role in defining the muscle founder cells and that nautilus gene function is essential for normal myogenesis in Drosophila.
There are several possible mechanisms for RNA interference involving catalytic degradation, interference with chromatin structure and/or transcription, or interaction between RNA and the genome. Whatever the mechanism, it does not involve promoter or intronic sequences. As mentioned previously, the fact that the endogenous mex3 transcript can be degraded in the C. elegans embryo suggests the mechanism operates posttranscriptionally, so an enzymatic process is most likely involved. However, the exact mechanism underlying RNA interference is not understood at the present time.
Acknowledgments
We thank Ward Odenwald for help with the filleted embryo preparations.
ABBREVIATIONS
- dsRNA
double-stranded RNA
- bHLH
basic helix–loop–helix
- CNS
central nervous system
- PNS
peripheral nervous system
Note Added in Proof
While my paper was in press, similar results with dsRNA-mediated genetic interference in Drosophila also were reported by Kennerdell and Carthew.
References
- 1.Rushton E, Drysdale R, Abmayr S M, Michelson A M, Bate M. Development. 1995;121:1979–1988. doi: 10.1242/dev.121.7.1979. [DOI] [PubMed] [Google Scholar]
- 2.Bate M. Development. 1990;110:791–804. doi: 10.1242/dev.110.3.791. [DOI] [PubMed] [Google Scholar]
- 3.Ho R K, Ball E E, Goodman C S. Nature (London) 1983;301:66–69. doi: 10.1038/301066a0. [DOI] [PubMed] [Google Scholar]
- 4.Keller C A, Grill M A, Abmayr S M. Dev Biol. 1998;202:153–156. doi: 10.1006/dbio.1998.9009. [DOI] [PubMed] [Google Scholar]
- 5.Abmayr S M, Erickson M S, Bour B A. Trends Genet. 1995;11:153–159. doi: 10.1016/s0168-9525(00)89030-7. [DOI] [PubMed] [Google Scholar]
- 6.Abmayr S M, Keller C A. Curr Top Dev Biol. 1998;38:35–80. doi: 10.1016/s0070-2153(08)60244-6. [DOI] [PubMed] [Google Scholar]
- 7.Hidalgo A, Urban J, Brand A H. Development. 1995;121:3703–3712. doi: 10.1242/dev.121.11.3703. [DOI] [PubMed] [Google Scholar]
- 8.Hidalgo A, Brand A H. Development. 1997;124:3253–3262. doi: 10.1242/dev.124.17.3253. [DOI] [PubMed] [Google Scholar]
- 9.Paterson B M, Walldorf U, Eldridge J, Dubendorfer A, Frasch M, Gehring W J. Proc Natl Acad Sci USA. 1991;88:3782–3786. doi: 10.1073/pnas.88.9.3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fire A, Xu S, Montgomery M K, Kostas S A, Driver S E, Mello C C. Nature (London) 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 11.Thisse B, Stoetzel C, Gorostiza-Thisse C, Perrin-Schmitt F. EMBO J. 1988;7:2175–2183. doi: 10.1002/j.1460-2075.1988.tb03056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caudy M, Grell E H, Dambly-Chaudiere C, Ghysen A, Jan L Y, Jan Y N. Genes Dev. 1988;2:843–852. doi: 10.1101/gad.2.7.843. [DOI] [PubMed] [Google Scholar]
- 13.Cronmiller C, Schedl P, Cline T W. Genes Dev. 1988;2:1666–1676. doi: 10.1101/gad.2.12a.1666. [DOI] [PubMed] [Google Scholar]
- 14.Kassis J A, Desplan C, Wright D K, O’Farrell P H. Mol Cell Biol. 1989;9:4304–4311. doi: 10.1128/mcb.9.10.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dohrmann C, Azpiazu N, Frasch M. Genes Dev. 1990;4:2098–2111. doi: 10.1101/gad.4.12a.2098. [DOI] [PubMed] [Google Scholar]
- 16.Lilly B, Galewsky S, Firulli A B, Schulz R A, Olson E N. Proc Natl Acad Sci USA. 1994;91:5662–5666. doi: 10.1073/pnas.91.12.5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pirrotta V. Biotechnology. 1988;10:437–456. doi: 10.1016/b978-0-409-90042-2.50028-3. [DOI] [PubMed] [Google Scholar]
- 18.Brand A H, Perrimon N. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 19.Buff E, Carmena A, Gisselbrecht S, Jimenez F, Michelson A M. Development. 1998;125:2075–2086. doi: 10.1242/dev.125.11.2075. [DOI] [PubMed] [Google Scholar]
- 20.Chen L, Krause M, Sepanski M, Fire A. Development. 1994;120:1631–1641. doi: 10.1242/dev.120.6.1631. [DOI] [PubMed] [Google Scholar]
- 21.Rudnicki M A, Jaenisch R. BioEssays. 1995;17:203–209. doi: 10.1002/bies.950170306. [DOI] [PubMed] [Google Scholar]
- 22.Keller C A, Erickson M S, Abmayr S M. Dev Biol. 1997;181:197–212. doi: 10.1006/dbio.1996.8434. [DOI] [PubMed] [Google Scholar]
- 23.Lilly B, Zhao B, Ranganayakulu G, Paterson B M, Schulz R A, Olson E N. Science. 1995;267:688–693. doi: 10.1126/science.7839146. [DOI] [PubMed] [Google Scholar]
- 24.Weintraub H. Cell. 1993;75:1241–1244. doi: 10.1016/0092-8674(93)90610-3. [DOI] [PubMed] [Google Scholar]
- 25.Kennerdell J R, Carthew R W. Cell. 1998;95:1017– 1026. doi: 10.1016/s0092-8674(00)81725-0. [DOI] [PubMed] [Google Scholar]