Abstract
Collective transcriptional analysis of heat shock response in the hyperthermophilic archaeon Pyrococcus furiosus was examined by using a targeted cDNA microarray in conjunction with Northern analyses. Differential gene expression suggests that P. furiosus relies on a cooperative strategy of rescue (thermosome [Hsp60], small heat shock protein [Hsp20], and two VAT-related chaperones), proteolysis (proteasome), and stabilization (compatible solute formation) to cope with polypeptide processing during thermal stress.
Information gleaned from genome sequence data indicates that heat shock response in hyperthermophilic archaea has several distinguishing features. For example, hyperthermophilic archaea lack Hsp70 (DnaK), Hsp40 (DnaJ), and GrpE, all of which are centrally important in the heat shock response of most known microorganisms (26). Also, the major chaperonin found thus far in thermophilic archaea, an Hsp60 homolog referred to as the thermosome, is more closely related to chaperonins associated with the eukaryotic cytosol (TriCC/CCT complex) than to the bacterial GroEL/ES system (19, 32). Energy-dependent proteolysis plays a major role during heat shock in bacteria in which genes encoding ATP-dependent proteases, such as lon, clp, and hfl, are linked to heat shock promoters (13). However, based on available genome sequence data, hyperthermophilic archaea lack the Clp and HflB (FtsH) family of proteins and have a different version of the Lon protease (43). Hyperthermophilic archaea, which typically have proteasomes, lack the eukaryotic ubiquitination pathway for selective protein degradation by the proteasome and, therefore, seem to modulate proteolysis at the protease level. Another interesting feature of hyperthermophilic archaeal heat shock response is the induced formation of unique compatible solutes that have been proposed to stabilize intracellular proteins against thermal denaturation (33). Whether compatible solutes reduce the need for protein turnover mechanisms is not known.
The relative contributions to the collective response of chaperones, chaperonins, proteases, and compatible solutes during heat shock in hyperthermophilic archaea have yet to be examined. Here, the heat shock response of the hyperthermophilic archaeon Pyrococcus furiosus (9) was investigated by using Northern analyses in conjunction with a targeted cDNA microarray, based on genes encoding the thermosome, molecular chaperones, proteases, glycoside hydrolases, and other relevant cellular functions expected to be affected during thermal stress.
Experimental approach and data analysis.
Relevant open reading frames (ORFs) were located from the P. furiosus genome at NCBI (http://www.ncbi.nlm.nih.gov/cgi-bin/Entrez/framik?db=genome&gi=228) and BLAST searches of prokaryotic genomes found at The Institute for Genomic Research (www.TIGR.org). SCANPROSITE (http://expasy.cbr.nrc.ca/tools/scnpsit1.html) and PFAM HMM (>http://pfam.wustl.edu/hmmsearch.shtml) search tools were used to verify the presence of putative catalytic domains. ORF fragments (generally 400 to 700 bp) were selected from 201 different genes (about 10% of the genome) and were PCR amplified and purified as reported previously (6) by using P. furiosus genomic DNA. Purified PCR products were quantified (100 ng/μl), randomized, and printed onto CMT-GAPS aminosilane-coated slides (Corning, Corning, N.Y.) with an arrayer (model 417; Affymetrix, Santa Clara, Calif.). A total of six replicates of each ORF were printed onto each slide. DNA was attached to the substrate by UV cross-linking in at 250 mJ and baking at 75°C for 2 h. Five separate first-strand cDNA reactions (per sample, per slide) were prepared from P. furiosus total RNA with Stratascript (Stratagene) and random hexamers (Invitrogen Life Technologies, Carlsbad, Calif.) by using indirect incorporation as previously described (15). The cDNA generated from each of the five reactions was pooled before hybridization to the slides; hybridizations and washes were performed as described previously (16), except that no poly(T) was added to the hybridization mixture. Slides were scanned and signal intensity data was extracted by using the Scanarray 4000 scanner and Quantarray software, respectively (GSI Lumonics, Billerica, Mass.). Local background intensity was subtracted from each spot signal, and spotting buffer (50% dimethyl sulfoxide [DMSO]) was used to subtract global background. The signal from the Cy-3 channel was normalized to the signal from the Cy-5 channel based on total signal intensity.
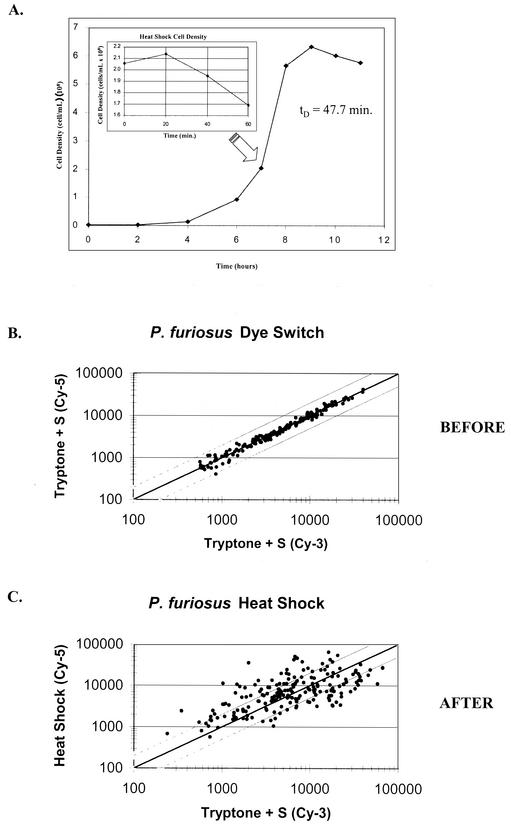
P. furiosus DSM 3638 was grown on a sea salts-based medium (SSM) with 0.5% (wt/vol) tryptone and 0.2% (wt/vol) yeast extract as carbon and energy sources as described previously (42) and monitored by cell enumeration as described elsewhere (6). RNA isolations were performed as described previously (6), except that RNA was further purified by using the RNAqeuous RNA isolation kit (Ambion) after the first acid phenol extraction and subsequent ethanol precipitation (42). Northern analysis was carried out as described previously (6), with 20 μg of total RNA loaded in each lane. Cells were grown until mid-exponential phase at 90°C on SSM and shifted to 105°C for 1 h; control cultures were allowed to continue growing at 90°C for the same period of time (Fig. 1A). The detection limit was determined by examining scatter plots of dye switch experiments. As shown in Fig. 1B, differential dye incorporation did not produce an effect greater than twofold for any ORF on the array and is not significant for signal intensity values greater than 2,000 units. Therefore, the results were not further data corrected for a dye effect. However, many genes were differentially expressed by twofold or more with statistical significance (Fig. 1C).
FIG. 1.
(A) Growth curve for P. furiosus grown on tryptone + S0 (1% wt/vol) in SSM at 90°C. Cells were subjected to a 60-min heat shock at 105°C during exponential growth. (B) Fluorescence signal intensities of reciprocally labeled cDNA from cells grown at 90°C. (C) Fluorescent signal intensities of heat shock versus unperturbed growth. The upper and lower diagonal lines indicate twofold differential expression.
Three separate hybridization experiments were performed with the same RNA sample, and the results were compared with data from an independent isolation and hybridization (Table 1). Negative controls (genes from mouse) were spotted onto the array to assess background noise. Ratios of microarray intensity data from the hybridization experiments were combined and converted to a log2 scale; ORFs containing a negative ratio value (generally indicating very low transcript abundance) were discarded from further analysis. Genes that showed corrected intensity ratios of approximately twofold induction or repression (log2Ri > 1 or log2 Ri < −1, where Ri is the intensity ratio) with a paired t test (between unperturbed and perturbed growth) significance level of P < 0.01 (as shown in Table 1) were considered differentially expressed.
TABLE 1.
Differential expression of selected ORFs
| Gene type and description | Locus | Expression, 1st replicate
|
Expression, 2nd replicate
|
||
|---|---|---|---|---|---|
| Log2 ratio intensity ± SD | Fold | Log2 ratio intensity ± SD | Fold | ||
| Chaperone-related genes | |||||
| Small heat shock protein (class I) | PF1883 | 2.95 ± 0.34 | >7.7 | 2.80 ± 0.12 | >6.9 |
| VAT homolog | PF1882 | 2.78 ± 0.18 | 6.9 | 1.89 ± 0.04 | 3.7 |
| VAT homolog | PF0963 | 2.21 ± 0.27 | 4.6 | 1.45 ± 0.05 | 2.7 |
| Thermosome, single subunit | PF1974 | 2.00 ± 0.25 | >4.0 | 2.07 ± 0.16 | >4.2 |
| Prefoldin homolog (beta subunit) | PF0380 | −0.76 ± 0.19 | −1.7 | −1.02 ± 0.03 | −2.0 |
| ATP-dependent proteases | |||||
| Proteasome, subunit beta (PsmB-1) | PF1404 | 1.03 ± 0.33 | 2.0 | 0.98 ± 0.05 | 2.0 |
| Proteasome, subunit beta (PsmB-2) | PF0159 | 0.89 ± 0.21 | 1.9 | 0.47 ± 0.05 | 1.4 |
| ATP-dependent Regulatory Subunit (PAN) | PF0115 | 0.25 ± 0.16 | 1.2 | −0.12 ± 0.05 | −1.1 |
| ATP-dependent LA (Lon)a | PF0467 | −1.53 ± 0.24 | −2.9 | −0.38 ± 0.14 | >−1.3 |
| Proteasome, subunit alpha (PsmA) | PF1571 | −1.96 ± 0.28 | −3.9 | −1.16 ± 0.03 | −2.2 |
| ATP-independent proteases and peptidases | |||||
| Subtilisin-like protease | PF0688 | 2.81 ± 0.31 | 7.0 | 4.04 ± 0.07 | 16.5 |
| ArgE/peptidase | PF1185 | 2.18 ± 0.22 | 4.5 | 1.89 ± 0.05 | 3.7 |
| HtpX heat shock protein | PF1597 | 2.16 ± 0.10 | 4.5 | 1.60 ± 0.04 | 3.0 |
| Similar to endo-1,4-beta-glucanase (ytoP) | PF1861 | 1.67 ± 0.23 | 3.2 | 1.21 ± 0.07 | 2.3 |
| D-aminopeptidase | PF1924 | 1.52 ± 0.18 | 2.9 | 1.96 ± 0.05 | 3.9 |
| Signal sequence peptidase I, SECII | PF0313 | 1.10 ± 0.23b | 2.1 | 1.43 ± 0.05 | 2.7 |
| Methionine aminopeptidase (MAP) (Pep M) | PF0541 | −1.00 ± 0.15 | −2.0 | 0.41 ± 0.03 | 1.3 |
| Putative proline dipeptidase | PF0747 | −1.18 ± 0.21 | −2.3 | 0.32 ± 0.02 | 1.3 |
| Heat shock protein X | PF1597 | −1.18 ± 0.31 | −2.3 | −0.58 ± 0.05 | −1.5 |
| Carboxypeptidase I | PF0456 | −1.27 ± 0.19 | −2.4 | −2.26 ± 0.03 | −4.8 |
| Pyrolysin | PF0287 | −2.29 ± 0.19 | >−4.9 | −1.53 ± 0.05 | −2.9 |
| Glycoside hydrolases | |||||
| Beta-glucosidase (CellA) | PF0073 | 4.18 ± 0.20 | 18.1 | 3.25 ± 0.09 | 9.5 |
| Putative methyltransferase (deacetylase) | PF0137 | 3.47 ± 0.28 | 11.1 | 1.83 ± 0.01 | 3.6 |
| Endo-beta-1,3-glucanase (Lam16) | PF0076 | 2.67 ± 1.13 | 6.4 | 1.93 ± 0.15 | 3.8 |
| Chitinase (Chi18A) | PF1234 | 2.14 ± 0.29 | 4.4 | 2.62 ± 0.11 | 6.2 |
| Beta-galactosidase precursor Gal35 (put.) | PF0363 | 1.72 ± 0.21 | 3.3 | 1.85 ± 0.03 | 3.6 |
| Alpha amylase (Amy57) | PF0272 | 1.64 ± 0.56 | 3.1 | −1.34 ± 0.07 | −2.5 |
| Beta-mannosidase (Man 1) | PF1208 | 1.46 ± 0.16 | 2.8 | 1.25 ± 0.03 | 2.4 |
| Put. alpha-dextrin endo-1,6-α-glucosidase | PF1108 | 1.34 ± 0.27 | 2.5 | 1.59 ± 0.05 | 3.0 |
| Chitinase (Chi18B) | PF1233 | 1.26 ± 0.67 | 2.4 | 2.87 ± 0.14 | 7.3 |
| Beta-glucosidase (CellB) | PF0442 | 1.17 ± 0.23 | 2.3 | 0.39 ± 0.05 | 1.3 |
| Other | |||||
| Trehalose/maltose binding protein (malE) | PF1739 | 2.69 ± 0.17 | 6.5 | 1.29 ± 0.06 | 2.4 |
| Spermidine synthase | PF0127 | 1.41 ± 0.28 | 2.7 | 1.99 ± 0.02 | 4.0 |
| Recombinase, radA | PF1926 | 1.39 ± 0.20 | >2.6 | 1.02 ± 0.04 | 2.0 |
| Putative sugar binding protein (malE-like) | PF1938 | 1.34 ± 0.19 | 2.5 | −1.48 ± 0.21 | −2.8 |
| Putative trehalose synthase | PF1742 | 1.29 ± 0.31 | 2.4 | 0.78 ± 0.08 | 1.7 |
| Recombinase, radB | PF0021 | 0.46 ± 0.32 | 1.4 | −0.24 ± 0.07 | −1.2 |
| Damage-inducible protein (dinF homolog) | PF1850 | 0.08 ± 0.23b | 1.1 | −0.46 ± 0.06 | −1.4 |
Archaeal Lon proteins are missing an ATP-binding domain (43).
P < 0.02 (P < 0.01 for all other reported observations).
For complete information on signal intensity and fold changes for all genes included on the array, see our website(http://www.che.ncsu.edu/extremophiles/publications/Pfu_heatshock.html ).
Differential expression of genes during heat shock.
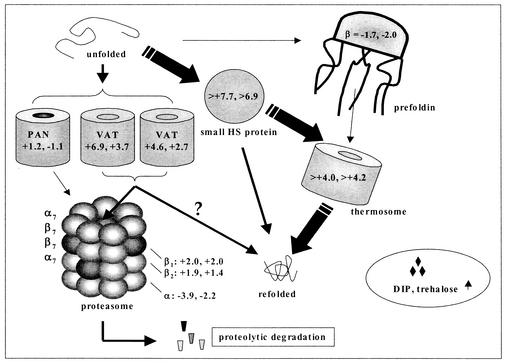
Thermal stress was apparent by the effects on growth (Fig. 1A) and the induction of known and putative stress genes present in P. furiosus (see Table 1). Therefore, the results given here are based on the combined effects of thermal stress and reduced cell growth that are collectively described by the term heat shock. The genes encoding the major Hsp60-like chaperonin (thermosome) in P. furiosus (19) and the Hsp20-like small heat shock protein (23) were strongly induced, as were two other molecular chaperones (VAT) belonging to the CDC48/p97 branch of the AAA+ family. VAT is thought to participate in both protein unfolding (for proteolysis) and refolding processes (12, 32). The P. furiosus genome encodes the α- and β-subunits of prefoldin, an ATP-independent chaperone found primarily in eukaryotes and archaea (25). The gene encoding the prefoldin β-subunit was down regulated upon heat shock, although the corresponding ORF was expressed at relatively high levels in both cases, which is consistent with reports suggesting that the genes encoding prefoldin are not induced by stress (27). The proposed protein folding cascade in P. furiosus, containing all known chaperone homologs in the organism, is shown in Fig. 2.
FIG. 2.
Protein folding in P. furiosus based on differential gene expression of ORFs. Fold values are presented for the heat shock experiment and the biological replicate. P. furiosus appears to utilize primarily Hsp20 and Hsp60 (and possibly VAT) as the major components of the refolding cascade while relying on the proteasome for energy-dependent proteolysis.
RadA and RadB in P. furiosus correspond functionally to RecA and Rad51 in bacteria and eukaryotes, respectively (20). The gene encoding RadA was elevated twofold during heat shock, while RadB was unaffected. Although it was previously proposed that radA was constitutively expressed in P. furiosus during a 1-h heat shock from 95 to 108°C (20), the results here indicate that a heat shock-inducible DNA repair system is present in this organism. It is not known whether P. furiosus has an adaptive DNA repair system akin to the bacterial SOS response, but here the gene encoding the E. coli DinF homolog in P. furiosus (4) was expressed at low levels under both conditions tested.
The proteasome appears be the only true ATP-dependent protease in P. furiosus (2). The only other ATP-dependent protease candidate encoded in the P. furiosus genome is an archaeal version of Lon that, unlike bacterial versions, lacks an ATP-binding domain sequence (10, 43). Here, the P. furiosus lon was not induced by heat shock, although this gene was expressed at relatively high levels under both stressed and unstressed conditions. Both proteasome β-subunits (β1 and β2) were induced somewhat upon heat shock (twofold or less), while the expression of the α-subunit decreased two- to fourfold. The reasons for the decrease in α-subunit gene expression of the P. furiosus proteasome upon heat shock is not known, although a similar observation has been made in mammalian cells (22). The conserved gene cluster containing the α-subunit and the exosome (20) was differentially expressed in a concerted manner (data not shown). This result is consistent with predictions based on comparative genomic analyses (21, 29) and provides experimental support for the possible functional or physical coupling between selective protein degradation and RNA processing in archaea. The gene encoding PAN, the ATPase component of the 26S proteasome, was unaffected by heat shock.
The role of the proteasome in heat-shocked hyperthermophilic archaea and other prokaryotes is not known but presumably involves polypeptide processing. The thermophilic archaeon Thermoplasma acidophilum cannot grow without proteasome activity under heat shock conditions (31), although the proteasome is not essential during heat shock in the actinomycetes (8). The up-regulation of the β1-subunit during heat shock is intriguing, since this was found to be absent from the native 20S proteasome purified from P. furiosus (2).
Several ATP-independent proteases were also affected by heat shock (Table 1). Notably, the gene encoding pyrolysin, a membrane-associated protease with an endo-acting and subtilisin-like catalytic domain (41), was strongly repressed, while a subtilisin-like protease (18) was strongly induced. Five peptidase-encoding genes were induced, including the gene encoding HtpX, which has been implicated elsewhere in surface protein expression related to changes in adhesiveness, cellular morphology, and levels of surface-active antigens (38).
Hyperthermophiles accumulate compatible solutes during exposure to thermal stress (33). Levels of di-myo-inositol phosphate (DIP), the only reported temperature-dependent compatible solute in P. furiosus, were reported to increase as much as 20-fold when subjected to a temperature shift from 95 to 101°C (28). In Pyrococcus woesei, DIP is presumed to be synthesized from glucose-6-phosphate by two enzymes: l-myo-inositol 1-phosphate synthase and a putative DIP-synthetase (34). Here, the gene encoding the former (PF1616) was strongly induced by thermal stress; the gene encoding the latter has yet to be identified. Another known compatible solute, trehalose, has been shown to stabilize proteins of various origins against thermal stress in vitro (17) but has not been reported to accumulate in P. furiosus under conditions of salt or thermal stress. Trehalose metabolism appears to be induced by heat shock in yeast (40) and Salmonella enterica (5). The closely related hyperthermophilic archaeon Thermococcus litoralis takes up trehalose through a high-affinity maltose/trehalose transport system (44). The genes encoding the putative trehalose synthase and MalE sugar binding protein were induced under heat shock (see Table 1), suggesting that P. furiosus, like T. litoralis, might take up trehalose available from yeast extract in its medium during stressed conditions or might synthesize this compound (24).
Expression levels of glycoside hydrolase genes were, in general, very low during growth at 90°C on peptide-based medium (data not shown). However, there was a significant induction of these genes upon heat shock (see Table 1). The cellular motivation for expressing genes related to carbohydrate acquisition may relate to the increased demand in ATP during thermal stress, which could be met by increased glycolysis (30). However, the gene encoding glyceraldehyde-3-phosphate ferredoxin oxidoreductase (37) was down-regulated significantly upon heat shock. It is also possible that futile cycles may be operational during heat shock response whereby the synthesis and catabolism of trehalose, glycerol, and glycogen help to control the energy balance of the cell, as observed in the stress response in Saccharomyces cerevisiae (1). Here, glycogen synthase and trehalose synthase gene expression levels (data not shown) were high during heat shock, which could trigger heightened glycoside hydrolase gene levels. Given the known capacity for P. furiosus and other hyperthermophiles to produce saccharide-based compatible solutes when subjected to stress (33), the acquisition of carbohydrates mediated by heightened levels of glycoside hydrolases for this purpose also needs to be considered. As such, the presence of small amounts of glycosides from yeast extract in the medium could have stimulated glycoside hydrolase gene expression under thermal stress.
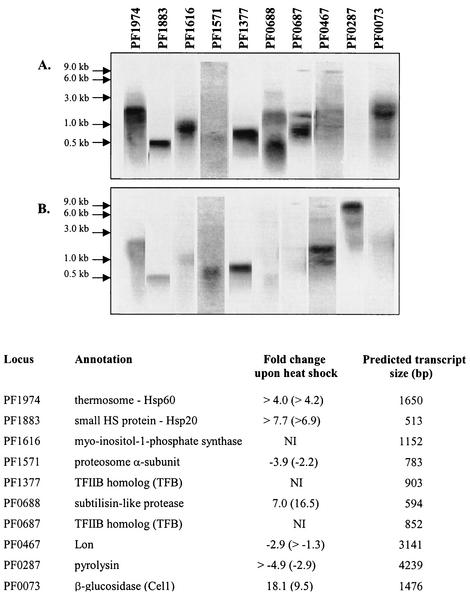
Intensity comparisons from the Northern hybridizations on selected genes (Fig. 3) were consistent with differential expression patterns observed in the microarrays. In addition, the microarray expression levels obtained here compared well with those reported by Schut et al. (35), who used a targeted microarray to study the effect of sulfur on maltose-grown P. furiosus at 95°C (see our website, http://www.che.ncsu.edu/extremophiles/publications/Pfu_heatshock.html, for details).
FIG. 3.
Northern analysis of selected genes included on the P. furiosus targeted cDNA microarray under conditions of 105°C (A) and 90°C (B). Also shown are genes encoding a putative myo-inositol phosphatase (PF1616) and two transcription factors homologs, TFIIB (PF1377 and PF0687), not included on the array (NI). Fold changes for the biological replicate are shown in parentheses.
Comparative genomic analyses predicted conserved archaeal heat shock regulons consisting of genes encoding the small heat shock protein, a thermosome subunit, two VAT homolog CDC48-2 proteins, and two histones in P. furiosus (11). With the exception of the genes encoding the two histones, all of these genes in this experiment showed substantial increased fold changes and high levels of expression under heat shock conditions. While the gene encoding the archaeal histone A1 (PF1831) was not induced, it was expressed at very high levels under both normal and heat shock conditions.
Previous work in Methanosarcina mazeii with the grpE-dnaK-dnaJ chaperone gene cluster suggests that TATA-binding protein and eukaryotic transcription factor IIB homolog (TFB) interact more strongly with stress-gene promoters during heat shock (7). These chaperone-encoding genes are not present in P. furiosus, but this organism's genome contains two different TFB-related genes. The presence of multiple TFB and TATA-binding protein homologs in some species of archaea has led to speculation that these proteins may play a role similar to that of sigma factors in bacteria by recognizing promoters with different sequences (3). Indeed, one of the six TFB-related genes in Haloferax volcanii is heat shock induced (36). Northern analyses in this study reveal that PF0687 is induced while PF1377 expression levels are similar during unstressed growth and heat shock (see Fig. 3).
Whether additional heat shock elements exist in P. furiosus and related hyperthermophilic archaea remains to be seen. Consistent with previous work with the extremely thermoacidophilic archaeon Metallosphaera sedula (14), the heat shock response in P. furiosus appears to involve a much more limited set of genes than is found for most of the less thermophilic prokaryotes examined to date. This is consistent with previous work with Metallosphaera sedula, an extremely thermoacidophilic archaeon (14). Further work is needed to understand heat shock gene regulation in P. furiosus, especially as it relates to less thermophilic organisms. Efforts along these lines have recently been reported (39), and related studies will help elucidate thermal stress response mechanisms among the most thermally active microorganisms known. Finally, even though gene regulation is most often controlled at the level of transcription initiation, additional investigations are needed to confirm that the microarray data presented in this study conform to true biological response. Such efforts are currently under way in our laboratory.
Acknowledgments
This work was supported in part by grants from the Department of Energy (Energy Biosciences Program) and the National Science Foundation (Biotechnology Program). K.R.S. acknowledges support from a Department of Education GAANN Fellowship. S.B.C. acknowledges support from an IGERT Fellowship in Bioinformatics.
The authors thank Bryon Sosinski and Len van Zyl for helpful discussions and Amy Grunden for providing selected proteolytic fermentation PCR products.
REFERENCES
- 1.Alexandre, H., V. Ansanay-Galeote, S. Dequin, and B. Blondin. 2001. Global gene expression during short term ethanol stress in Saccharomyces cerevisiae. FEBS Lett. 498:98-103. [DOI] [PubMed] [Google Scholar]
- 2.Bauer, M. W., S. H. Bauer, and R. M. Kelly. 1997. Purification and characterization of a proteasome from the hyperthermophilic archaeon Pyrococcus furiosus. Appl. Environ. Microbiol. 63:1160-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell, S. D., and S. P. Jackson. 1998. Transcription and translation in archaea: a mosaic of eukaryl and bacterial features. Trends Microbiol. 6:222-228. [DOI] [PubMed] [Google Scholar]
- 4.Bouyoub, A., G. Barbier, J. Querellou, and P. Forterre. 1995. A putative SOS repair gene (dinF-like) in a hyperthermophilic archaeon. Gene 167:147-149. [DOI] [PubMed] [Google Scholar]
- 5.Canovas, D., S. A. Fletcher, M. Hayashi, and L. N. Csonka. 2001. Role of trehalose in growth at high temperature of Salmonella enterica serovar Typhimurium. J. Bacteriol. 183:3365-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chhabra, S. R., K. R. Shockley, D. E. Ward, and R. M. Kelly. 2002. Regulation of endo-acting glycosyl hydrolases in the hyperthermophilic bacterium Thermotoga maritima grown on glucan- and mannan-based polysaccharides. Appl. Environ. Microbiol. 68:545-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Biase, A., A. J. L. Macario, and E. C. de Macario. 2002. Effect of heat stress on promoter binding by transcription factors in the cytosol of the archaeon Methanosarcina mazei. Gene 282:189-197. [DOI] [PubMed] [Google Scholar]
- 8.De Mot, R., I. Nagy, J. Walz, and W. Baumeister. 1999. Proteasomes and other self-compartmentalizing proteases in prokaryotes. Trends Microbiol. 7:88-92. [DOI] [PubMed] [Google Scholar]
- 9.Fiala, G., and K. O. Stetter. 1986. Pyrococcus furiosus sp. nov represents a novel genus of marine heterotrophic archaebacteria growing optimally at 100° C. Arch. Microbiol. 145:56-61. [Google Scholar]
- 10.Fukui, T., T. Egushi, H. Atomi, and T. Imanaka. 2002. A membrane-bound archaeal Lon protease displays ATP-independent proteolytic activity towards unfolded proteins and ATP-dependent activity for folded proteins. J. Bacteriol. 184:3689-3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gelfand, M. S., E. V. Koonin, and A. A. Mirinov. 2000. Prediction of transcription regulatory sites in archaea by a comparative genomic approach. Nucleic Acids Res. 28:695-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Golbik, R., A. N. Lupas, K. K. Koretke, W. Baumeister, and J. Peters. 1999. The janus face of the archaeal Cdc48/p97 homologue VAT: protein folding versus unfolding. Biol. Chem. 380:1049-1062. [DOI] [PubMed] [Google Scholar]
- 13.Gottesman, S. 1996. Proteases and their targets in Escherichia coli. Annu. Rev. Genet. 30:465-506. [DOI] [PubMed] [Google Scholar]
- 14.Han, C. J., S. H. Park, and R. M. Kelly. 1997. Acquired thermotolerance and stressed-phase growth of the extremely thermoacidophilic archaeon Metallospaera sedula in continuous culture. Appl. Environ. Microbiol. 63:2391-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hasseman, J. 2001. TIGR microarray protocols. [Online.] http://www.tigr.org/tdb/microarray/protocolsTIGR.shtml.
- 16.Hedge, P., R. Qi, R. Abernathy, C. Gay, S. Dharap, R. Gaspard, J. Earle-Hughes, E. Snesrud, N. Lee, and J. Quackenbush. 2000. A concise guide to cDNA microarray analysis. BioTechniques 29:548-562. [DOI] [PubMed] [Google Scholar]
- 17.Hottiger, T., C. De Virgilio, M. Hall, T. Boller, and A. Wiemken. 1994. The role of trehalose synthesis for the acquisition of thermotolerance in yeast. II. Physiological concentrations of trehalose increase the thermal stability of proteins in vitro. Eur. J. Biochem. 15:187-193. [DOI] [PubMed] [Google Scholar]
- 18.Kannan, Y., Y. Koga, Y. Inoue, M. Haruki, M. Takagi, T. Imanaka, M. Morikawa, and S. Kanaya. 2001. Active subtilisin-like protease from a hyperthermophilic archaeon in a form with a putative prosequence. Appl. Environ. Microbiol. 67:2445-2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klumpp, M., and W. Baumeister. 1998. The thermosome: archetype of group II chaperonins. FEBS Lett. 430:73-77. [DOI] [PubMed] [Google Scholar]
- 20.Komori, K., T. Miyata, J. DiRuggiero, R. Holley-Shanks, I. Hayashi, I. K. Cann, K. Mayanagi, H. Shinagawa, and Y. Ishino. 2000. Both RadA and RadB are involved in homologous recombination in Pyrococcus furiosus. J. Biol. Chem. 275:33782-33790. [DOI] [PubMed] [Google Scholar]
- 21.Koonin, E. V., Y. I. Wolf, and L. Aravind. 2001. Prediction of the archaeal exosome and its connections with the proteasome and the translation and transcription machineries by a comparative-genomic approach. Genome Res. 11:240-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuckelkorn, U., C. Knuehl, B. Boes-Fabian, I. Drung, and P. M. Kloetzel. 2000. The effect of heat shock on 20S/26S proteasomes. Biol Chem. 381:1017-1023. [DOI] [PubMed] [Google Scholar]
- 23.Laksanalamai, P., D. L. Maeder, and F. T. Robb. 2001. Regulation and mechanism of action of the small heat shock protein from the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 183:5198-5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamosa, P., L. O. Martins, M. S. Da Costa, and H. Santos. 1998. Effects of temperature, salinity, and medium composition on compatible solute accumulation by Thermococcus spp. Appl. Environ. Microbiol. 64:3591-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leroux, M. R., M. Fandrich, D. Klunker, K. Siegers, A. N. Lupas, J. R. Brown, E. Schiebel, C. M. Dobson, and F. U. Hartl. 1999. MtGimC, a novel archaeal chaperone related to the eukaryotic chaperonin cofactor GimC/prefoldin. EMBO J. 18:6730-6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Macario, A. J. L., and E. C. de Macario. 1999. The archaeal molecular chaperone machine: peculiarities and paradoxes. Genetics 152:1277-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Macario, A. J. L., and E. C. de Macario. 2001. The molecular chaperone system and other anti-stress mechanisms in archaea. Front. Biosci. 6:D262-D283. [DOI] [PubMed] [Google Scholar]
- 28.Martins, L. O., and H. Santos. 1995. Accumulation of mannosylglycerate and di-myo-inositol-phosphate by Pyrococcus furious in response to salinity and temperature. Appl. Environ. Microbiol. 61:3299-3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maupin-Furlow, J., S. Kaczowka, M. Ou, and H. Wilson. 2001. Archaeal proteasomes: proteolytic nanocompartments of the cell. Adv. Appl. Microbiol. 50:279-338. [DOI] [PubMed] [Google Scholar]
- 30.Nickells, R. W., and L. W. Browder. 1988. A role for glyceraldehyde-3-phosphate dehydrogenase in the development of thermotolerance in Xenopus laevis embryos. J. Cell Biol. 107:1901-1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruepp, A., C. Eckerskorn, M. Bogyo, and W. Baumeister. 1998. Proteasome function is dispensable under normal but not under heat shock conditions in Thermoplasma acidophilum. FEBS Lett. 425:87-90. [DOI] [PubMed] [Google Scholar]
- 32.Ruepp, A., B. Rockel, I. Gutsche, W. Baumeister, and A. N. Lupas. 2001. The chaperones of the archaeon Thermoplasma acidophilum. J. Struct. Biol. 135:126-138. [DOI] [PubMed] [Google Scholar]
- 33.Santos, H., and M. S. da Costa. 2002. Compatible solutes of organisms that live in hot saline environments. Environ. Microbiol. 4:501-509. [DOI] [PubMed] [Google Scholar]
- 34.Scholz, S., S. Wolff, and R. Hensel. 1998. The biosynthesis pathway of di-myo-inositol-1,1′-phosphate in Pyrococcus woesei. FEMS Microbiol. Lett. 168:37-42. [Google Scholar]
- 35.Schut, G. J., J. Z. Zhou, and M. W. W. Adams. 2001. DNA microarray analysis of the hyperthermophilic archaeon Pyrococcus furiosus: evidence for a new type of sulfur-reducing enzyme complex. J. Bacteriol. 183:7027-7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson, D. K., J. R. Palmer, and A. C. J. Daniels. 1999. Expression and heat-responsive regulation of a TFIIB homolog from the archaeon Haloferax volcanii. Mol. Microbiol. 33:1081-1092. [DOI] [PubMed] [Google Scholar]
- 37.van der Oost, J., G. Schut, S. W. Kengen, W. R. Hagen, M. Thomm, and W. M. de Vos. 1998. The ferredoxin-dependent conversion of glyceraldehyde-3-phosphate in the hyperthermophilic archaeon Pyrococcus furiosus represents a novel site of glycolytic regulation. 273:28149-28154. [DOI] [PubMed] [Google Scholar]
- 38.Vickerman, M. M., N. Mather, P. Minick, and C. Edwards. 2002. Initial characterization of the Streptococcus gordonii htpX gene. Oral Microbiol. Immunol. 17:22-31. [DOI] [PubMed] [Google Scholar]
- 39.Vierke, G., A. Engelmann, C. Hebbeln, and M. Thomm. 2003. A novel archaeal transcriptional regulator of heat shock response. J. Biol. Chem. 278:18-26. [DOI] [PubMed] [Google Scholar]
- 40.Voit, E. O., and T. Radivoyevitch. 2000. Biochemical systems analysis of genome-wide expression data. Bioinformatics 16:1023-1037. [DOI] [PubMed] [Google Scholar]
- 41.Voorhorst, W. G. B., R. I. L. Eggen, A. C. M. Geerling, C. Platteeuw, R. J. Siezen, and W. M. deVos. 1996. Isolation and characterization of the hyperthermostable serine protease, pyrolysin, and its gene from the hyperthermophilic archaeon Pyrococcus furiosus. J. Biol. Chem. 271:20426-20431. [DOI] [PubMed] [Google Scholar]
- 42.Ward, D. E., S. W. M. Kengen, J. van der Oost, and W. M. de Vos. 2000. Purification and characterization of the alanine aminotransferase from the hyperthermophilic archaeon Pyrococcus furiosus and its role in alanine production. J. Bacteriol. 182:2559-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ward, D. E., K. R. Shockley, L. S. Chang, R. D. Levy, J. K. Michel, S. B. Conners, R. M. Kelly. 2002. Proteolysis in hyperthermophilic microorganisms. Archaea 1:63-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xavier, K. B., L. O. Martins, R. Peist, M. Kossmann, W. Boos, and H. Santos. 1996. High-affinity maltose/trehalose transport system in the hyperthermophilic archaeon Thermococcus litoralis. J. Bacteriol. 178:4773-4777. [DOI] [PMC free article] [PubMed] [Google Scholar]