Abstract
The lack of malolactic activity in H+-ATPase-deficient mutants of Oenococcus oeni selected previously was analyzed at the molecular level. Western blot experiments revealed a spot at 60 kDa corresponding to the malolactic enzyme only in the parental strain. Moreover, the mleA transcript encoding the malolactic enzyme was not detected by reverse transcription (RT)-PCR analysis of mutants. These results suggest that the malolactic operon was not transcribed in ATPase-deficient mutants. The mleR gene encoding a LysR-type regulatory protein which should be involved in expression of the malolactic genes was described previously for O. oeni. Results obtained in this study show that the mleR transcript was not detected in the mutants by RT-PCR. No mutation in the nucleotide sequences of the mleR gene and the malolactic operon was found. The effect of a reduction in H+-ATPase activity on l-malate metabolism was then investigated by using other malolactic bacteria. Spontaneous H+-ATPase-deficient mutant strains of Lactococcus lactis and Leuconostoc mesenteroides were isolated by using neomycin resistance. Two mutants were selected. These mutants exhibited ATPase activities that were reduced to 54 and 70% of the activities obtained for the L. lactis and L. mesenteroides parental strains, respectively. These mutants were also acid sensitive. However, in contrast to the ATPase-deficient mutants of O. oeni, activation of l-malate metabolism was observed with the L. lactis and L. mesenteroides mutants under optimal or acidic growth conditions. These data support the suggestion that expression of the genes encoding malolactic enzymes in O. oeni is regulated by the mleR product, as it is in L. lactis. Nevertheless, our results strongly suggest that there is a difference between the regulation of expression of the malolactic locus in O. oeni and the regulation of expression of this locus in less acidophilic lactic acid bacteria.
Lactic acid bacteria are extensively used as starters in food and beverage fermentations, such as dairy fermentations, as well as in wine production. Although lactic acid bacteria are considered acid tolerant, the harsh environment resulting from accumulation of the lactic acid produced during carbohydrate catabolism limits their growth and their survival. On the other hand, an initial high level of acidity of the medium due to a significant concentration of organic acids can prevent bacterial growth. In the case of winemaking, the level of malic acid, which depends directly on grape maturity, affects the pH of the wine.
In order to withstand the low external pH values (pHex) of environments, bacteria have developed different strategies. When the pHex decreases, maintenance of a neutral cytoplasmic pH is essential for survival of the fermentative bacterium Enterococcus faecalis (23). Many acid-tolerant fermentative bacteria use another strategy: the internal pH (pHin) decreases as the pHex decreases in order to maintain a constant transmembrane pH gradient rather than a constant pHin (24).
Whatever the strategy, the most important mechanism by which fermentative bacteria can regulate pHin is the proton-translocating ATPase. For Enterococcus hirae, this enzyme has been shown previously to be similar to the F1-F0 ATPase of aerobic bacteria, which synthesizes ATP coupled with the proton motive force (16, 23). However, the unique function of the H+-ATPase of enterococci is regulation of the cytoplasmic pH (9). This enzyme is also involved in regulation of pHin in other lactic acid bacteria (18). An acid-sensitive variant of Lactobacillus helveticus was isolated as an H+-ATPase-deficient strain (27). More recently, Kullen and Klaenhammer (12) characterized the pH-inducible F1F0-ATPase operon of Lactobacillus acidophilus. An increase in atp mRNA was induced by low pH and was correlated with an increase in the activity of the H+-ATPase in membrane extracts. Genes encoding F1F0-ATPase in Lactococcus lactis have also been cloned and sequenced (11). Results obtained with a mutant strain in which expression of H+-ATPase on the chromosome is under control of the nisA promoter clearly demonstrated a requirement for nisin for growth of the mutant. The H+-ATPase was therefore essential for growth of this bacterium. Moreover, the acid sensitivity of a mutant of L. lactis with reduced membrane-bound H+-ATPase activity confirmed the major role of this enzyme in regulation of the cytoplasmic pH (1, 28).
Other mechanisms involving decarboxylase systems are thought to participate in pHin regulation. Decarboxylation of amino acids may protect bacterial cells against intracellular and extracellular acidification, particularly when the main carbon and energy sources have been consumed (4, 25). Decarboxylation of carboxylic acids also leads to internal consumption of protons. This is the case for Oenococcus oeni, the lactic acid bacterium responsible for malolactic fermentation in wine. Studies of the energetics of l-malic acid metabolism (5, 19, 21) have demonstrated that malolactic fermentation is an energy-producing pathway based on the electrogenic uptake of l-malate, intracellular decarboxylation of this compound by the malolactic enzyme (MLE), and efflux of the decarboxylation product l-lactic acid. Not only does this metabolic pathway result in the generation of a proton motive force sufficient to drive ATP synthesis via the membrane-bound F1F0-ATPase, but the proton consumption during the decarboxylation of l-malate also participates in the regulation of pHin (22). We previously cloned and characterized the mleA and mleP genes encoding the MLE and the malate permease, respectively, in O. oeni (13). These two genes are organized in an mle locus. Upstream of the mleA gene, an open reading frame likely to encode a LysR-type regulatory protein was found (14). The role of this regulatory protein in malolactic gene expression in O. oeni has not been determined yet. On the other hand, it has been shown that activation of the malolactic system in L. lactis is mediated by mleR (20). The protein encoded by mleR is homologous to a class of positive regulatory proteins belonging to the LysR family of proteins.
In our laboratory, we isolated spontaneous ATPase-deficient mutants of O. oeni (26). The acid sensitivity of these mutants suggested a role for H+-ATPase in the acid tolerance of O. oeni, like that demonstrated for other lactic acid bacteria. However, all of the mutants isolated lacked malolactic activity, suggesting that there is linkage between the ATPase and malolactic activities in O. oeni. In order to investigate more precisely the nature of the relationship between these two activities, we analyzed the absence of malolactic activity at the molecular level in ATPase-deficient mutant strains of O. oeni. Moreover, the lack of information about the influence of a deficiency in H+-ATPase activity on l-malic acid metabolism in other malolactic bacteria led us to isolate ATPase-deficient mutant strains of L. lactis and Leuconostoc mesenteroides by using neomycin resistance as a positive marker. The metabolism of l-malic acid in these mutant strains under optimal or acidic growth conditions was also investigated.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The parental strain O. oeni IOB84.13 (Oenological Institute of Bordeaux) and neomycin-resistant strains 9.01.07 and 6.27.06 obtained previously (26) were cultured at 30°C and pH 5.3 in FT80 medium modified (FT80m) as described by Labarre et al. (13). L. mesenteroides subsp. mesenteroides 19D isolated in our laboratory and L. lactis subsp. lactis biovar diacetylactis CNRZ 126 (INRA, Jouy en Josas, France) were used as other malolactic bacteria. L. mesenteroides, L. lactis, and neomycin-resistant strains obtained from both of these strains were cultured at 30°C and pH 6.5 in MRS medium (6) supplemented with 10 g of dl-malate per liter.
Isolation of neomycin-resistant mutants of L. lactis and L. mesenteroides.
Spontaneous neomycin-resistant mutants of L. lactis and L. mesenteroides were isolated by the method of Yokota et al. (28), with some modifications. Bacteria were grown in 40 ml of MRS medium until the exponential phase was reached (optical density at 600 nm [OD600], 0.3). Cells were harvested by centrifugation, concentrated in 2 ml of fresh medium, and spread onto MRS medium plates containing 300 μg of neomycin sulfate ml−1. The plates were then incubated at 30°C for 48 h. Neomycin-resistant colonies were selected and inoculated onto plates containing 5 ml of MRS medium supplemented with 300 μg of neomycin sulfate ml−1. A first screening, based on the bacterial growth characteristics, was conducted with MRS medium at pH 6.5. The specific growth rates were calculated during the exponential phase (i.e., between 10 and 15 h for L. lactis strains and between 5 and 12 h for L. mesenteroides strains). Biomass concentrations were calculated for an initial OD600 of 0.06 after 27 and 38 h of growth for L. lactis and L. mesenteroides, respectively. Cell biomass was deduced based on a preliminary calibration, as follows: 1 OD600 unit = 0.4 mg (dry weight). Specific l-malate consumption was calculated by determining the amount of l-malate metabolized, expressed as millimoles per milligram of biomass at the end of culture.
Preparation of membranes and ATPase activity assay.
Membranes were prepared and ATPase activity assays were performed as described by Tourdot-Maréchal et al. (26). The methods were adapted to L. lactis and L. mesenteroides as follows: the buffer used to break the cells was Tris-HCl buffer (50 mM Tris, 10 mM MgSO4; pH 7.0); and ATPase activities were measured in Tris-HCl buffer (pH 7.0) with 6.25 mM ATP. ATPase activity was expressed as micromoles of Pi produced per minute per milligram of protein.
Western immunoblot analysis.
Protein extraction was performed as described by Labarre et al. (15). Proteins were transferred from sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels to nitrocellulose membranes (PROTEAN; Schleicher and Schuell) by using a trans-blot semidry transfer cell (Bio-Rad, Richmond, Calif.), and immunoblot analysis was performed with a specific rabbit polyclonal antiserum directed against the 60-kDa MLE protein from O. oeni (15).
RNA isolation.
O. oeni cells cultured at pH 5.3 were harvested by centrifugation for10 min at 5,000 × g and resuspended in 1 ml of Tri reagent (Sigma). Cells were mechanically broken with 200 mg of glass beads (diameter, 70 to 110 μm) by using six 40-s periods of homogenization with 30-s intervals between the periods with a FastPrep cell disintegrator (FP120; Instrument Savant; Bio 101). Samples were then treated as recommended by the manufacturer.
RT-PCR.
RNA samples were treated with RNase-free DNase (1 U μl−1) as recommended by the manufacturer (Life Technologies, Gibco BRL). Reverse transcription (RT) was done in a 25-μl mixture containing 4 μl of 5× reverse transcriptase buffer, 2 μl of 0.1 M dithiothreitol, and 4 μl of a preparation containing each deoxynucleoside triphosphate at a concentration of 2.5 mM. The reaction mixture was incubated at 42°C for 2 min. Then 200 U of Escherichia coli SuperScript II Rnase H− reverse transcriptase (Life Technologies, Gibco BRL) was added. Incubation was continued for 50 min at 42°C and then for 15 min at 70°C for enzyme denaturation. Amplification was performed by using a 50-μl (final volume) reaction mixture containing 50 ng of cDNA, 30 pmol of each primer, each deoxynucleoside triphosphate at a concentration of 2 μM, and 2.5 U of Taq DNA polymerase. Specific cDNA was amplified for 40 cycles consisting of denaturation for 45 s at 92°C, annealing for 45 s at 55°C, and elongation for 1 min at 72°C. The PCR products were analyzed on a 1.6% agarose gel. The primers used for this experiment were 23- and 24-mer oligonucleotides for the mleA gene fragment (mleA probes 60440 [5′-ACC AAA ATG GTC GGG TGG ACA GC-3′] and 43406 [5′-GGA AGA TTT TGG CCG TTC GAA TGC-3′]) and 18- and 22-mer oligonucleotides for the mleR-like gene fragment (mleR4 [5′-CGA TAA AAC CAA TCC CGG-3′] and Reg1 [5′-GGT TTG GAA ACA ATT GAA ATC G-3′]). The gene encoding the thioredoxin gene trxA, used as an internal control, was amplified by using 20- and 21-mer oligonucleotides (Trx1 [5′-TTG CCG AAT TTA ACC CTC GA-3′] and Trx5 [5′-AGG AGG AAT TAT ATG GCA AT-3′]).
Other procedures.
Protein concentration was determined by the Bradford method (3) by using a Bio-Rad protein assay kit (Bio-Rad Laboratories, Richmond, Calif.) with bovine serum albumin as the standard. l-Malic acid concentrations were determined enzymatically by using Boehringer Mannheim kits.
RESULTS
Immunoblot analysis of MLE in O. oeni ATPase-deficient strains.
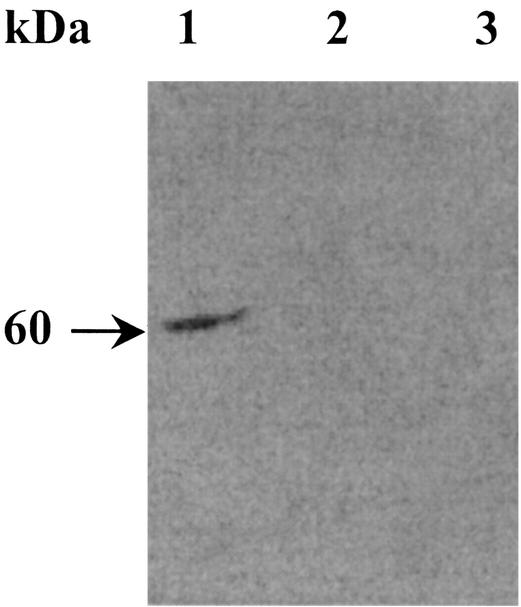
The absence of malolactic activity in both of the H+-ATPase-deficient strains of O. oeni used (26) led us to first determine the amounts of MLE in the mutant strains. Proteins (10 μg) obtained from disrupted cells grown on FT80m medium were loaded on SDS-PAGE gels. An immunoblot experiment was then performed with specific polyclonal antibodies directed against the MLE of O. oeni. The MLE corresponds to a 60-kDa protein (15), and it was detected only in the parental strain (Fig. 1, lane 1). No cross-reaction signals were obtained with protein extracts from the H+-ATPase-deficient mutants (Fig. 1, lanes 1 and 3). This result led us to hypothesize that the MLE was not synthesized in the two mutant strains. However, we cannot exclude the possibility that the enzyme was unstable or there was defective enzyme synthesis, which prevented a cross-reaction with the antiserum. To answer these questions, a transcriptional study of mleA gene expression was carried out.
FIG. 1.
Immunodetection of MLE by Western blotting. Harvested cells in the mid-exponential growth phase were washed in 10 mM of Tri-HCl buffer (pH 8) and disrupted as described by Labarre et al. (15). Protein extracts (10 μg) were analyzed by SDS-PAGE (10.5% acrylamide) and immunoblotting. Lane 1, total proteins of the wild-type strain; lanes 2 and 3, total proteins of strains 9.01.07 and 6.27.06, respectively. The arrow indicates the position of the MLE.
Transcriptional analysis of malolactic gene expression.
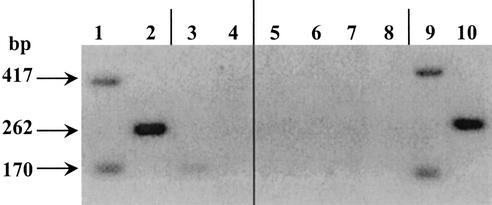
In order to detect mleA mRNA, RT-PCR experiments were performed. The RT-PCR experiments were performed with total RNA from exponentially growing cells of strains IOB84.13 and 9.01.07. Probes for mleA were used to amplify a 417-bp DNA fragment of the MLE gene (Fig. 2). The thioredoxin gene, trxA, was used as a PCR positive control. This gene was detected at a significant level and was estimated to be at roughly the same level during the exponential phase (10). A DNA strand of the expected length (417 bp) was amplified by PCR from the reverse transcriptase product only for the parental strain (Fig. 2, lane 1). No amplification was observed for mutant strain 9.01.07 (Fig. 2, lane 3). The positive results observed for amplification of a 170-bp DNA fragment of the trxA gene (control) from the parental strain and mutant 9.01.07 validated our experiments. A lack of amplification with the mleA probes was also observed with strain 6.27.06 (data not shown). These data suggest that either there was no transcription or the mleA messenger was more unstable in the two mutants.
FIG. 2.
RT-PCR analysis of malolactic gene expression. One microliter of the product was amplified by PCR. Lanes 1 and 3, amplification of mleA and trxA genes of strains IOB 84.13 and 9.01.07, respectively; lanes 2 and 4, amplification of the mleR gene of strains IOB 84.13 and 9.01.07, respectively. Amplification was also carried out with total DNA of the parental strain as a positive control (lanes 9 and10). To check that there was no DNA contamination in the RNA sample, total RNA not treated with reverse transcriptase was amplified (lanes 5 and 6, strain IOB 84.13; lanes 7 and 8, strain 9.01.07). The thioredoxin gene trxA was used as a PCR positive control.
An RT-PCR analysis was conducted in order to detect mleR mRNA in the parental strain and the H+-ATPase-deficient mutants. The RT-PCR experiment was performed with total RNA from exponentially growing cells. Primers mleR4 and Reg1 were used to amplify the 262-bp DNA fragment of the mleR-like gene. A DNA strand of the expected length was amplified only from the parental strain (Fig. 2, lane 2). No amplification was obtained for mutant strain 9.01.07 (Fig. 2, lane 4). The same result was obtained with mutant strain 6.27.06 (data not shown).
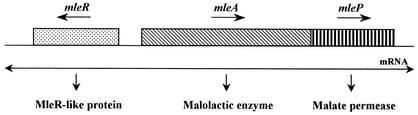
According to Labarre et al. (14), the mleR-like gene of O. oeni is transcribed divergently from the mle operon (Fig. 3), and the promoter regions of the mleR gene and the mle operon certainly overlap. To further investigate the lack of putative mleR gene expression in H+-ATPase-deficient mutants, the mleR-like gene and the hypothetical region of overlapping promoters were sequenced. No change in the nucleotide sequence of the promoter region and the mleR-like gene was observed. Moreover, the complete mle operon was sequenced for both mutant strains. No mutation was observed (data not shown). These data led us to conclude that the malolactic deficiency phenotype is not due to a mutation in the malolactic locus in H+-ATPase-deficient mutants.
FIG. 3.
Genetic organization of the mle genes of O. oeni. The mleA and mleP genes encoding the MLE and the malate permease of O. oeni, respectively, are transcribed in an operon. Upstream of the mle operon, another gene encoding an MleR-like protein is transcribed divergently. This protein is related to the LysR-type regulatory protein family (14).
Isolation of H+-ATPase-deficient mutants of L. lactis and L. mesenteroides.
The effect of a reduction in the H+-ATPase activity on l-malate metabolism in other lactic acid bacteria was also investigated. We used L. lactis and L. mesenteroides for this analysis. In L. lactis, the mleS gene encoding the MLE was cloned and sequenced (2, 7), and activation of the malolactic system was shown to be mediated by mleR (20). Moreover, mutants of L. lactis with reduced membrane-bound ATPase activity were previously obtained by selection for neomycin resistance (1). In contrast, little information is available about genes implicated in malolactic activity in L. mesenteroides. However, this bacterium is phylogenetically closely related to O. oeni (8), and it is one of the heterofermentative cocci of wines (17). To isolate spontaneous neomycin-resistant mutants of L. lactis and L. mesenteroides, two sets of cultures containing 2 × 1010 CFU for each strain were treated with a lethal concentration of neomycin sulfate (300 μg ml−1). A total of 1,682 spontaneous neomycin-resistant mutants were obtained from L. lactis with a frequency of 1.9 × 10−7. A total of 345 mutants were obtained from L. mesenteroides with a frequency of 4.5 × 10−6. Fifty L. lactis colonies and 30 L. mesenteroides colonies were randomly picked and first analyzed to determine their growth profiles under optimal growth conditions. We selected neomycin-resistant mutants of L. lactis and L. mesenteroides whose growth profiles were most affected at pH 6.5. Mutant 5.16 of L. lactis and mutant 5.15 of L. mesenteroides had the lowest final biomass (35 and 44% of the wild-type final biomasses, respectively) (Table 1). The membrane-bound ATPase activities of the mutants were also determined. As shown in Table 1, the membrane-bound ATPase activities of mutant strain 5.16 (L. lactis) and mutant strain 5.15 (L. mesenteroides) were reduced to 54 and 70%, respectively, of the activities of the parental strains.
TABLE 1.
Growth parameters and membrane-bound ATPase activities for wild-type strains of L. lactis and L. mesenteroides and neomycin-resistant mutants cultured in MRS medium (pH 6.5)
| Strain | Specific growth rate (h−1)a | Final biomass concn (g [dry wt] liter−1)b | Membrane-bound ATPase sp act (μmol of P1 min−1 mg of protein−1) |
|---|---|---|---|
| L. lactis CNRZ 126 (wild type) | 0.46 | 1.04 | 1.27 |
| Mutant 5.16 | 0.16 | 0.36 | 0.59 |
| L. mesenteroides 19D (wild type) | 0.45 | 1.26 | 0.33 |
| Mutant 5.15 | 0.35 | 0.56 | 0.10 |
The specific growth rates are the averages for two independent cultures.
Calculated for an initial OD600 of 0.06, after 27 h of growth for L. lactis and after 38 h of growth for L. mesenteroides.
Growth of ATPase-deficient mutants of L. lactis and L. mesenteroides under acidic conditions.
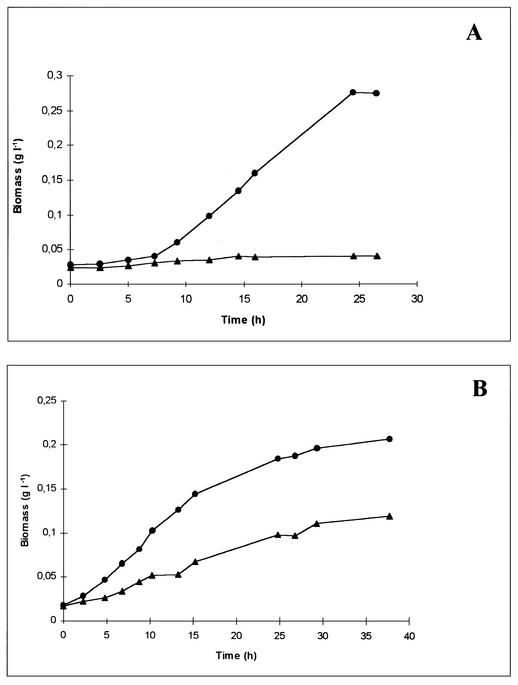
The parental strains, the L. lactis mutant, and the L. mesenteroides mutant were cultured in MRS medium with an initial pH of 5.0 for L. lactis and an initial pH of 4.5 for L. mesenteroides. Under these conditions, the specific growth rates of the L. lactis and L. mesenteroides parental strains were 0.16 and 0.12 h−1, respectively, compared with specific growth rates of 0.46 and 0.45 h−1, respectively, at pH 6.5 (Fig. 4). The 5.16 mutant strain of L. lactis did not grow at pH 5.0. Mutant strain 5.15 of L. mesenteroides had an 85% lower specific growth rate than the parental strain when the organisms were cultured at pH 4.5. A significant decrease (43%) in the final biomass obtained after 38 h of growth was also observed (Fig. 4B).
FIG. 4.
Growth profiles of H+-ATPase-deficient mutants of L. lactis (strain 5.16) and L. mesenteroides (strain 5.15) under acidic conditions. MRS medium at pH 5.0 was used for L. lactis (A), and MRS medium at pH 4.5 was used for L. mesenteroides (B). Symbols: •, parental strain; ▴, mutant strain.
l-Malate metabolism in H+-ATPase-deficient mutants of L. lactis and L. mesenteroides.
The residual malate was assayed enzymatically after growth on MRS medium under optimal growth conditions or under acidic conditions (Table 2). At pH 6.5, the ATPase-deficient mutants of L. lactis and L. mesenteroides and the wild-type strains used approximately 90% of the 37 mM l-malic acid initially present in MRS medium. Moreover, the H+-ATPase-deficient mutants exhibited greater consumption of l-malate (expressed as millimoles of l-malate consumed per milligram of final biomass) than the parental strains exhibited. The L. lactis and L. mesenteroides mutants consumed three and two times more l-malate than the wild-type strains consumed, respectively.
TABLE 2.
Effect of the growth medium pH on the l-malate concentration and l-malate consumption in wild-type strains of L. lactis and L. mesenteroides and H+-ATPase-deficient mutant strains cultured in MRS medium
| Strain | Residual l-malate concn (mM)a
|
l-malate consumption (mol of l-malate g of final biomass−1)
|
||
|---|---|---|---|---|
| pH 6.5 | Acidic conditionsb | pH 6.5 | Acidic conditionsb | |
| L. lactis CNRZ 126 (wild type) | 2.9 | 2.6 | 33 | 126 |
| Mutant 5.16 | 3.4 | 19.2 | 95 | 451 |
| L. mesenteroides 19D (wild type) | 3.2 | 5.6 | 27 | 153 |
| Mutant 5.15 | 3.7 | 10.7 | 60 | 221 |
Concentration after 27 h of growth for L. lactis and after 38 h of growth for L. mesenteroides.
The initial pH values were 5.0 for L. lactis and 4.5 for L. mesenteroides. The results are means for two independent experiments.
Similar measurements were obtained after growth under acidic conditions (Table 2). At pH 5.0, the wild-type strain of L. lactis metabolized 90% of the l-malate, whereas for the ATPase-deficient mutant, in spite of an absence of significant growth, 50% of the l-malate was still present in MRS medium after 27 h of culture. At pH 4.5, the wild-type strain of L. mesenteroides metabolized 90% of the l-malate, compared with 70% for the ATPase-deficient mutant, after 38 h of growth. Compared with the results obtained at pH 6.5, the consumption of l-malate increased four- and fivefold for the L. lactis and L. mesenteroides wild-type strains, respectively. The same results were obtained with the ATPase-deficient mutant strains. Moreover, for the mutant strains of L. lactis and L. mesenteroides, consumption of l-malate increased 3.5- and 1.5-fold, respectively, compared with consumption by the wild-type strains when the organisms were cultured under acidic conditions.
These results show that the ATPase-deficient mutants of L. lactis and L. mesenteroides metabolized l-malic acid, in contrast to the ATPase-deficient mutant of O. oeni. Moreover, the consumption of l-malate was greater than that calculated for wild-type strains. In the same way, under acidic growth conditions the l-malate consumption by the parental strains and also by the ATPase-deficient mutants clearly increased.
DISCUSSION
In this study the lack of malolactic activity previously observed with H+-ATPase-deficient mutants of O. oeni (26) was analyzed at a molecular level to clarify the link between the malolactic system and the membrane-bound H+-ATPase. The 60-kDa MLE was not detected in H+-ATPase-deficient mutants. We can exclude the possibility that the enzyme was not detected by immunoblotting due to weak synthesis by the mutants. Indeed, significant quantities of total proteins (10 μg) were loaded on the gel, and the polyclonal antibodies against MLE prepared by Labarre et al. (15) are highly specific. Moreover, the malolactic activity of O. oeni was previously detected during the log phase of growth with or without l-malate added to the growth medium (15). These findings eliminate the possibility that there was not induction of MLE in ATPase-deficient mutant strains under our experimental conditions. The lack of detection of MLE in mutants was due to either a lack of gene expression or instability of the enzyme. Another possibility is synthesis of a defective MLE that did not cross-react with the antiserum. To examine these hypotheses, a transcriptional study of the malolactic operon was carried out. In a previous report it was shown that a structure in the operon harbors the mleA and mleP genes, which encode the MLE and the malate permease of O. oeni, respectively (13). The mleA transcript was not detected by RT-PCR analysis of two H+-ATPase-deficient mutants when they were compared with the O. oeni parental strain. This result indicates that the mle operon does not seem to be transcribed in ATPase-deficient mutants. However, we cannot exclude the possibility that the malolactic mRNA is unstable, preventing detection by RT-PCR.
It has been proven that in L. lactis induction of the genes necessary to perform malolactic fermentation occurs only in bacteria with a functional copy of mleR. This gene encodes a LysR-type regulatory protein that acts as a positive regulator of the expression of the mleA gene (20). The mleR gene of L. lactis is not clustered with the mleA gene. In contrast, in O. oeni, an mleR-like gene was previously found upstream of the mle operon. This gene is transcribed divergently from the operon (14). The translation initiation sites of mleR and mleA are very close to each other, and the promoter region of the mleR gene and the mle operon certainly overlap (14). The role of mleR in malolactic gene expression in O. oeni remains unknown. Results obtained in this study show that the mleR-like transcript was not detected by RT-PCR in H+-ATPase-deficient mutants. Moreover, sequencing data proved that there is no mutation in the mleR-like gene and promoter region or in the mle operon in the H+-ATPase-deficient mutants. Taken together, these results are in accordance with positive regulation of mle operon expression by the mleR-like gene in O. oeni, like the situation in L. lactis. Further genetics experiments are needed to confirm the role of the mleR gene in mle operon regulation. Disruption of the mleR gene in O. oeni would be of great interest and would certainly allow us to understand regulation of malolactic gene expression. Unfortunately, DNA transfer techniques are only at the experimental stage at this time for this type of genetic approach with O. oeni. Mainly for this reason we chose to investigate l-malate metabolism in H+-ATPase-deficient mutants in another lactic acid bacterium, L. lactis. The MLE of this bacterium and the MLE of O. oeni are encoded by similar genes. Spontaneous neomycin-resistant mutants of L. lactis with reduced membrane-bound ATPase activity were obtained previously (1, 28). We likewise chose L. mesenteroides. This bacterium, in contrast to L. lactis, is one of the heterofermentative cocci of wines (17), and it is phylogenetically closely related to O. oeni.
First, investigations were carried out to select neomycin-resistant mutants of L. lactis and L. mesenteroides on the basis of their growth profiles under optimal conditions. Selected mutants were found to be acid sensitive and had ATPase activities that were significantly reduced. Similar results were obtained previously with spontaneous neomycin-resistant mutants of O. oeni that were acid sensitive and exhibited twofold-reduced H+-ATPase activity (26). The acid sensitivity of the ATPase-deficient mutants of L. lactis and L. mesenteroides confirms the suggestion that in lactic acid bacteria, the major role of the H+-ATPase is maintenance of pHin (1, 27).
Previous data indicated that there is a relationship between a reduction in ATPase activity and a lack of malolactic activity in O. oeni (26). This study showed that there were unexpected increases in l-malate consumption in H+-ATPase-deficient mutants of L. lactis and L. mesenteroides under both optimal and acidic growth conditions compared to the metabolism of the parental strains. In contrast to ATPase-deficient mutants of O. oeni, activation of l-malate metabolism for pHin homeostasis could be a compensatory consequence of the H+-ATPase deficiency in these mutants.
In conclusion, the relationship between the H+-ATPase deficiency and the lack of expression of the malolactic system seems to be a characteristic of mutants of O. oeni. It has been shown previously that expression of the MLE in O. oeni is not effectively modified by the presence of l-malate in the medium (14). In contrast, the malolactic activity in L. lactis, like that in L. mesenteroides, is inducible by l-malate (15). It appears that in O. oeni regulation of the expression of the malolactic operon may involve another regulatory factor that is presumably linked to the metabolic energy. Determination of the amount of ATP in relation to the level of transcription of malolactic genes should certainly help us elucidate the link between the ATPase and malolactic activities in O. oeni.
Acknowledgments
This study was supported by the Ministère de la Recherche et de l'Enseignement (France) and by the Conseil Régional de Bourgogne.
REFERENCES
- 1.Amachi, S., K. Ishikawa, S. Toyoda, Y. Kagawa, A. Yokota, and F. Tomita. 1998. Characterization of a mutant of Lactococcus lactis with reduced membrane-bound ATPase activity under acidic conditions. Biosci. Biotechnol. Biochem. 62:1574-1580. [DOI] [PubMed] [Google Scholar]
- 2.Ansanay, V., S. Dequin, B. Blondin, and P. Barre. 1993. Cloning, sequence and expression of the gene encoding the malolactic enzyme from Lactococcus lactis. FEBS Lett. 332:74-80. [DOI] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Castanie-Cornet, M. P., T. A. Penfound, D. Smith, J. F. Elliott, and J. W. Foster. 1999. Control of acid resistance in Escherichia coli. J. Bacteriol. 181:3525-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cox, D. J., and T. Henick-Kling. 1989. Proton-motive force and ATP generation during malolactic fermentation. Am. J. Enol. Vitic. 46:319-323. [Google Scholar]
- 6.De Mann, J. C., M. Rogosa, and M. E. Sharpe. 1960. A medium for the cultivation of lactobacilli. J. Appl. Bacteriol. 23:130-135. [Google Scholar]
- 7.Denayrolles, M., M. Aigle, and A. Lonvaud-Funel. 1994. Cloning and sequence analysis of the gene encoding Lactococcus lactis malolactic enzyme: relationships with malic enzymes. FEMS Microbiol. Lett. 116:79-86. [DOI] [PubMed] [Google Scholar]
- 8.Dicks, L. M., F. Dellaglio, and M. D. Collins. 1995. Proposal to reclassify Leuconostoc oenos as Oenococcus oeni [corrig.] gen. nov., comb. nov. Int. J. Syst. Bacteriol. 45:395-397. [DOI] [PubMed] [Google Scholar]
- 9.Heefner, D. L., H. Kobayashi, and F. M. Harold. 1980. ATP-linked sodium transport in Streptococcus faecalis. II. Energy coupling in everted membrane vesicles. J. Biol. Chem. 255:11403-11407. [PubMed] [Google Scholar]
- 10.Jobin, M. P., D. Garmyn, C. Diviès, and J. Guzzo. 1999. Expression of the Oenococcus oeni trxA gene is induced by hydrogen peroxide and heat shock. Microbiology 145:1245-1251. [DOI] [PubMed] [Google Scholar]
- 11.Koebmann, B. J., D. Nilsson, O. P. Kuipers, and P. R. Jensen. 2000. The membrane-bound H+-ATPase complex is essential for growth of Lactococcus lactis. J. Bacteriol. 182:4738-4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kullen, M., and T. R. Klaenhammer. 1999. Identification of the pH-inducible, proton-translocating F1F0-ATPase (atpBEFHAGDC) operon of Lactobacillus acidophilus by differential display: gene structure, cloning and characterization. Mol. Microbiol. 33:1152-1161. [DOI] [PubMed] [Google Scholar]
- 13.Labarre, C., J. Guzzo, J. F. Cavin, and C. Diviès. 1996. Cloning and characterization of the genes encoding the malolactic enzyme and the malate permease of Leuconostoc oenos. Appl. Environ. Microbiol. 62:1274-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Labarre, C., C. Diviès, and J. Guzzo. 1996. Genetic organization of the mle locus and identification of a mleR-like gene from Leuconostoc oenos. Appl. Environ. Microbiol. 62:4493-4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Labarre, C., J. F. Cavin, C. Diviès, and J. Guzzo. 1998. Using specific polyclonal antibodies to study the malolactic enzyme from Leuconostoc oenos and other lactic acid bacteria. Lett. Appl. Microbiol. 26:293-296. [DOI] [PubMed] [Google Scholar]
- 16.Leimgruber, R. M., C. Jensen, and A. Abrams. 1981. Purification and characterization of the membrane adenosine triphosphatase complex from the wild-type and N,N′-dicyclohexylcarbodiimide-resistant strains of Streptococcus faecalis. J. Bacteriol. 147:363-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lonvaud-Funel, A. 1999. Lactic acid bacteria in the quality improvement and depreciation of wine. Antonie Leeuwenhoek 76:317-331. [PubMed] [Google Scholar]
- 18.Nannen, N. L., and R. W. Hutkins. 1991. Proton-translocating adenosine triphosphatase activity in lactic acid bacteria. J. Dairy Sci. 74:747-751. [Google Scholar]
- 19.Olsen, E. B., J. B. Russell, and T. Henick-Kling. 1991. Electrogenic l-malate transport by Lactobacillus plantarum: a basis for energy derivation from malolactic fermentation. J. Bacteriol. 173:6199-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Renault, P., C. Gaillardin, and H. Heslot. 1989. Product of the Lactococcus lactis gene required for malolactic fermentation is homologous to a family of positive regulators. J. Bacteriol. 171:3108-3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salema, M., B. Poolman, J. S. Lolkema, M. C. Loureiro Dias, and W. N. Konings. 1994. Uniport of monoanionic l-malate in membrane vesicles from Leuconostoc oenos. FEBS Eur. J. Biochem. 124:1-7. [DOI] [PubMed] [Google Scholar]
- 22.Salema, M., J. S. Lolkema, M. V. San Romao, and M. C. Loureiro Dias. 1996. The proton-motive force generated in Leuconostoc oenos by l-malate fermentation. J. Bacteriol. 178:3127-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shibata, C., T. Ehara, K. Tomura, K. Igarashi, and H. Kobayashi. 1992. Gene structure of Enterococcus hirae (Streptococcus faecalis) F1F0-ATPase which functions as a regulator of cytoplasmic pH. J. Bacteriol. 174:6117-6124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siegumfeldt, H., K. B. Rechinger, and M. Jakobsen. 2000. Dynamic changes of intracellular pH in individual lactic acid bacterium cells in response to a rapid drop in extracellular pH. Appl. Environ. Microbiol. 66:2330-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ten Brink, B., C. Damink, H. M. L. J. Joosten, and J. Huis in't Veld. 1990. Occurrence and formation of biologically active amines in foods. Int. J. Food Microbiol. 11:3-84. [DOI] [PubMed] [Google Scholar]
- 26.Tourdot-Maréchal, R., L.-C. Fortier, J. Guzzo, L. Byong, and C. Diviès. 1999. Acid sensitivity of neomycin-resistant mutants of Oenococcus oeni: a relationship between reduction of ATPase activity and lack of malolactic activity. FEMS Microbiol. Lett. 178:319-326. [DOI] [PubMed] [Google Scholar]
- 27.Yamamoto, N., Y. Masujima, and T. Takano. 1996. Reduction of membrane-bound ATPase activity in a Lactobacillus helveticus strain with slower growth at low pH. FEMS Microbiol. Lett. 138:179-184. [Google Scholar]
- 28.Yokota, A., S. Amachi, S. Ishii, and S. Tomita. 1995. Acid sensitivity of a mutant of Lactococcus lactis subsp. lactis C2 with reduced membrane-bound ATPase activity. Biosci. Biotechnol. Biochem. 59:2004-2007. [DOI] [PubMed] [Google Scholar]