Abstract
Sphingolipid precursors, namely, ceramide and long-chain base phosphates (LCBPs), are important growth regulators with often opposite effects on mammalian cells. A set of enzymes that regulate the levels of these precursors, referred to as a ceramide/LCBP rheostat, is conserved in all eukaryotes. In order to gain further insight into the function of the rheostat in Saccharomyces cerevisiae, we searched for mutants that are synthetically lethal with a deletion of the LCB3 gene encoding LCBP phosphatase. In addition to acquiring expected mutants lacking the LCBP lyase, the screen revealed elo3 (sur4) mutants that were defective in fatty acid elongation and cka2 mutants lacking the α′ subunit of the protein kinase CK2 (casein kinase). Both mutations affected the in vivo activity of the acyl coenzyme A (acyl-CoA)-dependent and fumonisin B1-sensitive ceramide synthase (CS). The Elo3 protein is necessary for synthesis of C26-CoA, which in wild-type yeast is a source of C26 fatty acyls found in the ceramide moieties of all sphingolipids. In the in vitro assay, CS had a strong preference for acyl-CoAs containing longer acyl chains. This finding suggests that a block in the formation of C26-CoA in yeast may cause a reduction in the conversion of LCBs into ceramides and lead to an overaccumulation of LCBPs that is lethal in strains lacking the Lcb3 phosphatase. In fact, elo3 mutants were found to accumulate high levels of LCBs and LCBPs. The cka2 mutants, on the other hand, exhibited only 25 to 30% of the in vitro CS activity found in wild-type membranes, indicating that the α′ subunit of CK2 kinase is necessary for full activation of CS. The cka2 mutants also accumulated high levels of LCBs and had elevated levels of LCBPs. In addition, both the elo3 and cka2 mutants showed increased sensitivity to the CS inhibitors australifungin and fumonisin B1. Together, our data demonstrate that the levels of LCBPs in yeast are regulated by the rate of ceramide synthesis, which depends on CK2 kinase activity and is also strongly affected by the supply of C26-CoA. This is the first evidence indicating the involvement of protein kinase in the regulation of de novo sphingolipid synthesis in any organism.
Cellular membranes are complex structures whose lipid compositions change depending on their subcellular localization, extracellular signals, and environmental conditions. Mechanisms of biogenesis and maintenance of these dynamic structures and the role of individual lipid species in cell growth and survival are not fully understood and have been a subject of increased interest in recent years. Saccharomyces cerevisiae, nearly all of whose genes involved in lipid metabolism have been identified (many of them in recent years [6]), provides a convenient model for such studies. It has long been recognized that yeast cells can tolerate the complete lack of some membrane lipids, e.g., phosphatidylserine (1), but that proper concentrations of others are absolutely required for viability, e.g., sterols. In fact, the sensitivity of fungi to membrane sterol imbalance is the mechanism underlying most successful antifungal agents (10).
Sphingolipids, similar to sterols, are important constituent membrane lipids that are critical for the normal function and viability of both yeast and mammalian cells. These essential molecules comprise a diverse group of ceramide-containing lipids that are predominantly localized in the outer leaflets of the plasma membranes of eukaryotic cells, where they play important structural roles in cell recognition and adhesion (14) and in the formation of functional lipid microdomains (44). In addition, short-lived sphingolipid precursors, namely, long-chain bases (LCBs), long-chain base phosphates (LCBPs), and ceramides (N-acyl-LCBs), have been shown to affect cell growth, differentiation, and death (16). Ceramides play prominent roles in stress responses and programmed cell death (15, 21) and are often viewed as growth-inhibitory mediators. In contrast, the major mammalian LCBP sphingosine-1-phosphate has been shown to protect cells from ceramide-mediated apoptosis (5) and has been implicated as a growth-promoting second messenger (46). It has been proposed that the dynamic balance between ceramide and sphingosine-1-phosphate is important for determining cell fate (47). This balance is maintained by a group of sphingolipid metabolic enzymes, which can be referred to as the ceramide/LCBP rheostat (42, 47).
The ceramide/LCBP rheostat is conserved in yeast, despite the fact that there are significant differences between the major yeast and mammalian sphingolipids. For example, the inositolphosphorylceramides (IPCs), mannosyl-IPCs, and inositolphosphoryl-mannosyl-IPCs, the only sphingolipids found in yeast, are not present in mammalian cells (7). However, yeast genes encoding enzymes that are involved in the metabolism of sphingolipid precursors, including LCB kinase, LCBP phosphatase, LCBP lyase, and ceramidase, and recently discovered genes operating ceramide synthesis, LAC1 and LAG1, were all found to have homologues in mammalian cells (12, 19, 28, 29, 30, 32, 33, 36, 41, 43, 49). Changes in the levels of LCBPs and ceramides also accompany stress responses and affect cell growth in yeast. Mild heat stress causes transient increases in LCBs and LCBPs and a sustained increase in free ceramides (8, 17, 45). Mutant cells that accumulate moderate levels of LCBPs are more resistant to heat stress (29, 31, 45); however, the overaccumulation of LCBPs (up to a 400-fold increase over the basal level) observed in mutants defective in both LCBP phosphatase and LCBP lyase is lethal (18, 48). As with phosphates, the levels of free ceramide have to be carefully regulated. Free-ceramide levels in cells that are defective in the IPC synthase or treated with the selective IPC synthase inhibitor aureobasidin A are approximately eightfold increased subsequent to cell death (35). It is possible that an increase in free ceramide plays a role in cell death because cells that are defective in ceramide formation can survive a blockage of IPC synthase activity (35, 43). In order to gain further insight into the function of the ceramide/LCBP rheostat in yeast, we set up a genetic screen for mutants that require the Lcb3 (Ysr2/Lbp1 [29, 33, 40]) LCBP phosphatase for growth. We found that, apart from previously documented defects in the Dpl1 LCBP lyase, mutations in ELO3 (SUR4) and CKA2 impede the proper function of the rheostat in yeast by impairing the in vivo activity of the acyl coenzyme A (acyl-CoA)-dependent ceramide synthase.
MATERIALS AND METHODS
Strains, media, and general growth conditions.
The S. cerevisiae strains used in this study are listed in Table 1. The screening strain Y388-2 was obtained from strain Y388 (2) by PCR-based deletion of the LCB3 gene. Briefly, a G418 resistance marker was PCR amplified from plasmid DNA (13) and homologous recombination was directed by incorporating 40 bp of complementing LCB3 genomic sequences at the 5′ ends of the primers. Standard yeast genetic procedures were used to obtain and verify gene deletions (13). Strain Y388-DD was obtained by deletion of the DPL1 gene in strain Y388-2. The DPL1 knockout cassette carrying the LEU2 marker was PCR amplified from the DPL1 deletion strain MSS204 (45). In order to facilitate this deletion, Y388-2 was first transformed with plasmid L3A3-1, grown in synthetic complete medium minus uracil (SC−ura), and recovered for one doubling in yeast extract-peptone-dextrose (YEPD) prior to transformation with the PCR deletion cassette. The chromosomal deletions in strain Y388-DD were confirmed by restriction enzyme analysis of the PCR products. All strains were grown at 30°C in either a rich YEPD medium (2% dextrose) or, alternatively, in a synthetically defined medium containing yeast nitrogen base, 2% dextrose, and the appropriate amino acid supplements (i.e., SC) as indicated. Additionally, 5-fluoroorotic acid (5-FOA; 1 g/liter) was used as an addition to SC to select for Ura− strains.
TABLE 1.
Genotypes of strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| Y388 | MATaade2 ade3 ura3 leu2 lys2 | 2 |
| Y382 | MATα ade2 ade3 ura3 leu2 trp1 | 2 |
| Y388-2 | MATalcb3::KAN ade2 ura3 leu2 lys2 | This study |
| Y382-2 | MATα lcb3::KAN ade2 ade3 ura3 leu2 trp1 | This study |
| Y388-DD | MATalcb3::KAN dpl1::LEU2 ade2 ura3 leu2 lys2 | This study |
| Y388-2p | MATalcb3::KAN ade2 ura3 leu2 lys2 [URA3 LCB3 ADE3] | This study |
| MSS204 | MATaleu3 rme1 trp1 his4 dpl1::LEU2 | 45 |
| BY4741 | MATahis3 leu2 met15 ura3 | Research Genetics |
| BY4742 | MATα his3 leu2 lys2 ura3 | Research Genetics |
| BY4741elo2 | MATaelo2::KAN his3 leu2 met15 ura3 | Research Genetics |
| BY4741elo3 | MATaelo3::KAN his3 leu2 met15 ura3 | Research Genetics |
| BY4741cka2 | MATacka2::KAN his3 leu2 met15 ura3 | Research Genetics |
| BY4741lcka1 | MATacka1::KAN his3 leu2 met15 ura3 | Research Genetics |
| BY4741ckb1 | MATackb1::KAN his3 leu2 met15 ura3 | Research Genetics |
| BY4741ckb2 | MATackb2::KAN his3 leu2 met15 ura3 | Research Genetics |
| BY4741dpl1 | MATadpl1::KAN his3 leu2 met15 ura3 | Research Genetics |
| BY4742elo3 | MATα elo3::KAN his3 leu2 lys2 ura3 | Research Genetics |
Australifungin fermentation and isolation.
Australifungin was purified from Sporormiella australis ATCC 74157 according to the methods of Mandala et al. (27). Briefly, S. australis was grown on solid cracked-corn-based fermentation medium and extracted after 14 days of fermentation with ethyl acetate. Australifungin was purified by use of both gel column and high-speed countercurrent chromatography. Purity was approximately 80%.
Plasmid construction.
The screening plasmid L3A3-1 (LCB3 ADE3 URA3) was constructed from a pRS416 backbone containing the URA3 marker and a CEN replicon. LCB3 was subcloned from the genomic library plasmid pJAB15-1 (40) and inserted between the NotI and SacI restriction sites of similarly digested pRS416. The ADE3 gene was amplified from S. cerevisiae genomic DNA, digested with SmaI and SacII, and cloned into the LCB3-containing plasmid. The integrity of the LCB3 gene was confirmed by sequencing, and ADE3 was confirmed by the complementing colored-phenotype method.
Synthetic-lethality screen and colony-sectoring assay.
The colony-sectoring assay is based on colony color selection with adenine mutants. S. cerevisiae ade2 strains accumulate a red pigment, whereas ade2 ade3 strains do not and phenotypically result in a normal white colony. The colony-sectoring assay was performed by transforming the Y388-2 screening strain with plasmid L3A3-1. The resulting strain, Y388-2p, was grown under selective pressure in SC−ura in order to retain the plasmid. The screening strain Y388-2p is functionally ade2 and should appear as a red colony. However, frequent plasmid loss in yeast occurs at the approximate rate of 1% per cell per generation and thus results in a colony with a red- and white-sectored phenotype when it is grown under nonselective conditions (i.e., on YEPD agar).
Synthetically lethal LCB3 mutants were generated by mutation of strain Y388-2p by either UV irradiation or treatment with ethane methyl sulfonate (EMS). To obtain UV-induced mutants, strain Y388-2p cells were grown in SC−ura, washed with 50 mM potassium phosphate buffer (pH 7.0), plated on YEPD, and exposed to 1 mJ of UV radiation in a Stratalinker 2400 (Stratagene, La Jolla, Calif.) to obtain a killing efficiency of 90%. EMS treatment yielded a killing efficiency of 50% (22). The surviving cells from both treatments were grown on YEPD agar for 4 days at 30°C and analyzed for color phenotype. Uniformly red colonies were selected and tested for phenotype stability on YEPD agar. 5-FOA plates were used to eliminate non-LCB3-related adenine mutants. The remaining mutants were transformed with the LCB3 plasmid pJAB15-1(LCB3 LEU2), grown in SC−ura−leu, and plated on YEPD agar. At this point, only mutants that sectored were selected and considered to be synthetically lethal with LCB3. In order to determine if the mutations were dominant or recessive, genetic crosses were made with strain Y382-2 by patching the strains onto SC−trp−lys agar. The resulting diploids from these crosses were then analyzed for color phenotype on YEPD agar. Sectoring colonies indicated a recessive mutation, whereas solid-red colonies indicated dominance.
Agar diffusion assay.
Agar diffusion was used to evaluate sensitivity of the screening strain to ceramide synthase inhibitors and other antifungals and to identify antifungal sensitivity differences among the synthetically lethal mutants. For this assay, cells grown in SC−ura were washed one time in sterile phosphate-buffered saline. Cells (106) were added to 4 ml of 0.8% agarose in YEPD and plated on 10-cm-diameter petri dishes. Compounds were spotted on the preinoculated agar plates at the indicated volumes and concentrations. Growth and color changes were noted after 4 days.
Mutant complementation with a genomic library.
Mutants E37, U48, U4, and U33 were transformed with a yeast genomic library based on the p366 CEN vector (ATCC 77162) by selecting for leucine prototrophs. The transformants (20,000 to 50,000) were pooled and plated on SC plates containing 1 mg of 5-FOA per ml. Plasmid DNA was recovered by E. coli transformation from 5-FOA-resistant cells that formed white colonies and retransformed into respective mutants. Genomic DNA inserts in plasmids that conferred colony sectoring and a 5-FOA-resistant phenotype were identified by sequencing both ends of the insert DNA and matching the sequence with the S. cerevisiae genome sequence (4). Plasmids containing the ELO3 (SUR4) gene were recovered by complementing mutants E37 and U48, but only plasmids carrying the LCB3 gene were obtained in complementation experiments involving mutants U4 and U33. To obtain plasmids complementing mutants U4 and U33, pools of transformants were plated on SC plates containing 20 μM fumonisin B1 and prepared with agarose in place of agar. Plasmids pELO3 and pELO2 were used to test the ability of individual genes to complement the phenotype of synthetic-lethality mutants. These plasmids were obtained by ligation of pRS415 vector DNA (Stratagene) cut with BamHI and PstI or BamHI and NotI, with the appropriately cut PCR product containing the ELO3 or ELO2 gene amplified from the genomic DNA obtained from the wild-type strain BY4741. The integrity of each plasmid insert was verified by DNA sequencing. To test the ability of individual genes present on library plasmids to complement mutants U4 and U33, in vivo recombination in yeast cells was utilized to construct plasmids (26). Briefly, each gene was amplified by using primers containing 40 nucleotide sequences complementary to regions flanking the BamHI site in vector p366 and the mutant cells were cotransformed with the PCR product and BamHI-linearized vector to select for Leu+ transformants.
Determination of the MIC.
Susceptibilities of various strains to ceramide synthase inhibitors were measured by using the broth microdilution method. For australifungin, cells were inoculated at an initial density of 105 cells/ml in a 96-well plate containing 200 μl of YEPD medium per well. Drug was added from a 10 mM stock solution in dimethyl sulfoxide. For fumonisin B1 (Sigma) experiments, 95 μl of SC inoculated with 105 cells/ml was mixed with 5 μl of an appropriate fumonisin B1 dilution prepared from a 10 mM stock solution in phosphate-buffered saline. Plates were incubated for 48 h at 30°C, and the optical density at 600 nm (OD600) was measured in a Spectramax Plus spectrometer (Molecular Devices) after the cells were completely suspended in the medium.
Membrane preparations.
Ceramide synthase enzyme activity was measured in crude membrane preparations from each strain. Briefly, the membranes were isolated as follows: about 50 to 100 OD units of cells grown to late logarithmic phase in YEPD medium were collected and resuspended in 1 ml of breaking buffer, consisting of 50 mM potassium phosphate buffer (pH 7.0), 5 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, and a mixture of protease inhibitors (Complete, EDTA free; Boehringer Mannheim). About 500 μl of 500-μm-diameter glass beads (G8772; Sigma) was added to the tube, and cells were disrupted by vortexing them five times for 30 s each time. The supernatant of the low-speed centrifugation (5 min at 1,000 × g) was subjected to high-speed centrifugation (15 min at 100,000 × g), and the resulting pellet was briefly washed with the breaking buffer, resuspended in about 500 μl of the breaking buffer supplemented with 25% glycerol, and stored at −80°C for subsequent analysis.
Ceramide synthase assay.
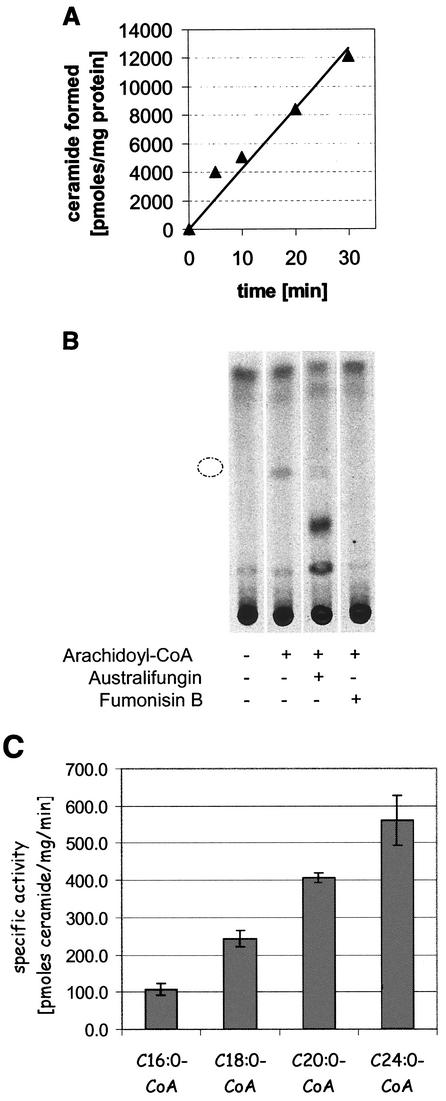
For measuring ceramide synthase activity, a 100-μl reaction mixture comprised of 30 μM [3H]dihydrosphingosine (DHS; 2.5 Ci/mmol), 100 mM Tricine (pH 8.1), 10 μM arachidoyl-CoA (Sigma), and 0.02% Tergitol NP-40 was prepared in a glass tube and sonicated for 1 min in an ultrasonic water bath. Alternatively, arachidoyl-CoA (C20:0-CoA) was replaced with palmitoyl-CoA (C16:0-CoA), stearoyl-CoA (C18:0-CoA), or lignoceroyl-CoA (C24:0-CoA) (all from Sigma) as indicated in Results. Methanolic solutions of tritiated DHS (catalog number ART-460; American Radiolabeled Chemicals) and cold d-erythro-DHS (catalog number 1831; Matreya) were added first to the tube and dried under a stream of N2. The reaction was initiated by the addition of 4 μg of membrane protein, which wasincubated for 30 min at 30°C with gentle shaking, and it was stopped by the addition of 640 μl of a chloroform-methanol mixture (1:1). The mixture was centrifuged at 1,000 × g for 5 min, and 100 μl of the stopped-reaction mixture along with 10 mmol of cold lignoceroyl DHS (Sigma) was spotted on an LK5 silica gel thin-layer chromatography (TLC) plate (Whatman). Chromatography was developed by using a chloroform-methanol solvent (19:1.5), and the plates were quantified with an AR-2000 imaging scanner (Bioscan). Alternatively, plates were exposed to tritium storage screens (Kodak) and read with a PhosphorImager (Molecular Dynamics). The bands representing product and substrate were quantified with ImageQuant software (Molecular Dynamics). The plates were sprayed with a 10% solution of CuSO4 · 5H2O in 8% H3PO4 and charred at 160°C to visualize the N-lignoceroyl DHS standard.
LCB and LCBP analyses.
Measurements of intracellular LCB and LCBP levels were carried out by a method of Lester and Dickson (23), modified such that samples were analyzed by TLC. Briefly, yeast cell culture samples (10 to 30 OD600 units) were killed by adding trichloroacetic acid to a final concentration of 5% followed by 10 min of incubation on ice and were washed once with cold 5% trichloroacetic acid and three times with water. The cell pellet was resuspended in 75 μl of water and extracted by the addition of 425 μl of 70.6 mM triethylamine in ethanol followed by a 2-min treatment in a sonic bath and 30 min of incubation with occasional mixing at 65°C. Cell debris was removed by low-speed centrifugation, and 100 μl of the extract was incubated with 20 μl of 10.5 mM 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate in acetonitrile (AQC reagent; Waters Corporation) for 1 to 2 h at room temperature. Phospholipids were degraded by the addition of 15 μl of 1.5 N KOH in methanol and a 30-min incubation at 37°C. The sample was neutralized with 15 μl of 1.74 N acetic acid in methanol. For LCB analysis, a 10-μl sample of AQC-reacted extract was resolved on LHPKD TLC plates (Whatman) in solvent I (chloroform-methanol [19:1.5]) by using AQC-derivatized phytosphingosine (PHS) and DHS as standards. For LCBP analysis, 50 μl of AQC-reacted extracts was loaded on 500-mg C18 columns (Varian) that had been preequilibrated with 1 ml of a methanol-water mixture (1:1). The column was washed with 1.65 ml of methanol-water (1:1), and lipids were eluted with 1.5 ml of an acetonitrile-water mixture (8:1). Samples were dried under a stream of nitrogen, resuspended in 20 ml of methanol-water (9:1), and developed on LHPKD plates with solvent II (chloroform-methanol-1.26 N ammonia [76:34:6]), with AQC-derivatized DHS-1-phosphate (DHS-1-P) and PHS-1-P being used as standards. Fluorescently tagged lipids were visualized by UV light.
RESULTS
Genetic link between ceramide and LCBPs.
LCBs formed in the endoplasmic reticulum can be used for synthesis of both ceramide and LCBPs (Fig. 1). Both genetic and biochemical evidence indicates that the overaccumulation of LCBPs is lethal in yeast cells (18, 45, 48). Under normal growth conditions, inactivation of either the LCBP phosphatase or the LCBP lyase (i.e., deletion of the LCB3 or DPL1 gene, respectively) has no effect on yeast cell growth. However, mutants with both genes deleted or dpl1 mutants challenged with high concentrations of sphingosine (41) are unable to grow. In both cases, viability is restored by an additional deletion of the major LCB kinase encoded by the LCB4 gene (18, 36, 48). These observations indicate that scenarios leading to gross accumulation of LCBPs are lethal and that LCB phosphates rather than free LCBs are responsible for cell death in these situations. Additionally, mutants defective in the LCB3 gene and therefore lacking major LCBP phosphatase are supersensitive to the ceramide synthase inhibitor australifungin (29), thus indicating that decreased consumption of LCBs for ceramide formation is reflected by the increased synthesis of LCBPs. In order to further explore this apparent association between the Lcb3 phosphatase and ceramide synthesis, we undertook a search for mutants that require an intact LCB3 gene for growth.
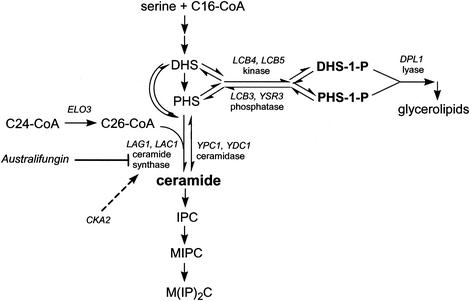
FIG. 1.
Metabolism of sphingoid LCBs in S. cerevisiae. The LCBs DHS and PHS are either N acylated by ceramide synthase or phosphorylated by Lcb4 (major) and Lcb5 (minor) kinases. LCBPs can be hydrolyzed by Dpl1 lyase, forming ethanolamine-1-phosphate and palmitaldehyde, or they can be dephosphorylated by Lcb3 (DHS-1-P selective) and Ysr3 (PHS-1-P selective) lipid phosphatases. C26-CoA, a preferred substrate for ceramide synthase, is formed during the elongation cycle that requires the ELO3 gene. Ceramide synthase activity is sensitive to the natural product inhibitors australifungin and fumonisin B1. As demonstrated by the results presented here, the activity of the α′ isoform of CK2 kinase encoded by CKA2 is necessary for the development of full ceramide synthase activity. MIPC, mannosyl-IPC; M(IP)2C, inositolphosphoryl-mannosyl-IPC.
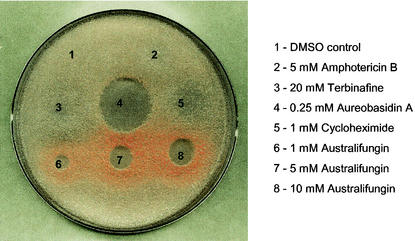
First, we constructed a strain that would allow the application of a classic colony-sectoring assay (2) to screen for mutants synthetically lethal with an LCB3 gene deletion. The LCB3 gene was deleted from the chromosome of strain Y388, and the resulting strain, Y388-2, was confirmed to have an elevated steady-state level of long-chain phosphates similar to that of previously described lcb3 mutants (data not shown; 18, 45). The Y388-2 strain, which carries additional chromosomal mutations in the ade2, ade3, ura3, and leu2 genes, was transformed with the centromere-based plasmid containing intact copies of the ADE3, URA3, and LCB3 genes. The cells in this strain form red colonies when they contain the plasmid and white colonies when they lack the plasmid. Since deletion of the LCB3 gene does not affect growth on YPD medium, the strain undergoes high-frequency plasmid loss under nonselective conditions and forms sectored red-white or white colonies (Fig. 2). We validated the utility of this strain for identification of mutants that are lethal in the lcb3 background by showing that the introduction of a dpl1 chromosomal deletion in the screening strain causes the expected LCB3 synthetic lethality and thus forms solid-red colonies on YEPD plates. Furthermore, we found that in an agar diffusion format, cells of the screening strain treated with the ceramide synthase inhibitor australifungin formed a distinct ring of red colonies surrounding the zone of the growth inhibition. This effect was selective for australifungin and was not caused by antifungal compounds with unrelated modes of action (Fig. 3). Similar effects were observed in experiments when cells were treated with various concentrations of drugs in a broth microdilution format of the growth assay. Cells in wells containing subinhibitory concentrations of australifungin and fumonisin B1 had a distinct red color, whereas the absence of drug or the presence of subinhibitory concentrations of cycloheximide, voriconazole, and amphotericin B did not result in a red color. Cells exposed to sublethal concentrations of ceramide synthase inhibitors turn red because they are forced to retain the plasmid that provides a functional Lcb3 phosphatase, thus indicating that the inhibition of ceramide synthesis leads to an elevation of LCBP above the level that can be tolerated by an lcb3 mutant. This result provides for the direct phenotypic manifestation of a genetic link between ceramide synthesis and LCBP levels. As expected, the Y388-2p strain not only was an indicator of drugs that inhibit ceramide synthesis but also turned red upon being treated with sphingosine, DHS, and to a lesser extent PHS and an IPC synthase inhibitor, aureobasidin A (data not shown).
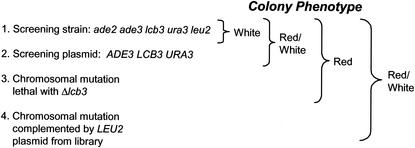
FIG. 2.
Schematic representation of the results of a Δlcb3 synthetic-lethality screen. A Δlcb3 screening strain constructed for the purpose of the synthetic-lethality screen forms red colonies when it contains the LCB3 plasmid and white colonies when it lacks the plasmid. Under nonselective growth conditions, this strain undergoes frequent plasmid loss, resulting in formation of sectored red and white colonies. Mutations that give advantage to cells that retain the plasmid will result in solid-red colonies.
FIG. 3.
The screening strain is a selective indicator of ceramide synthase inhibition. Cells were grown in SC−ura broth, plated on YEPD, and spotted with various antifungal agents. Assay conditions are described in Materials and Methods. DMSO, dimethyl sulfoxide.
Identification of mutants synthetically lethal with a deletion of lcb3.
Cells of the screening strain were mutagenized by either UV or EMS treatment and plated on YEPD plates, and red colonies were selected for further analysis. One hundred fifty-eight red colonies were found in the pool of 200,000 colonies that survived mutagenesis. Thirty-six of these had a stable color phenotype and were sensitive to 5-FOA, which is consistent with their inability to lose the URA3 plasmid. Fourteen mutants changed their color phenotype from red to either red-white or white upon transformation with the LEU2 plasmid carrying the LCB3 gene. These transformants also exhibited 5-FOA resistance. The purpose of this plasmid-shuffling experiment was to eliminate mutants whose red color phenotype and 5-FOA sensitivity resulted from either gene conversion or plasmid integration events. Ten out of the remaining 14 Δlcb3 synthetically lethal mutants were ascribed to result from mutations in the DPL1 gene due to the reversion of the phenotype upon transformation with a LEU2- and DPL1-containing plasmid. The four remaining mutants were subjected to further analysis. All four mutants were found to carry recessive mutations based on the reversal of the red colony phenotype in the diploid strain obtained by mating the mutant with the Y382-2 strain.
Non-DPL1 mutants are hypersensitive to australifungin.
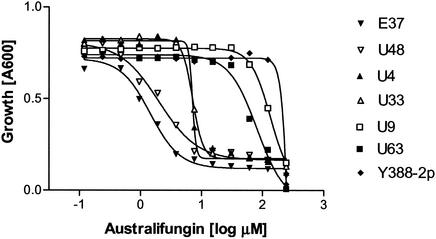
The growth-inhibitory activity of the ceramide synthase inhibitor australifungin is potentiated by a deletion of the LCB3 gene, which likely results in the toxic accumulation of LCBP species (29). One possible outcome of the synthetic-lethality screen was to obtain mutations leading to a deficiency in ceramide formation. Such mutants would potentially be more susceptible to australifungin. To address this possibility, we tested the MICs of australifungin for the non-DPL1 mutants E37, U48, U4, and U33 (Fig. 4). All four mutants were supersensitive to the drug. However, the mutants split into two distinct susceptibility patterns. E37 and U48 were approximately 200-fold more sensitive to australifungin than the parent strain, whereas U4 and U33 showed a 30-fold increase in sensitivity. U9 and U63 are DPL1 mutants and showed only slight differences from the control (Fig. 4). None of the mutants displayed a substantial increase in sensitivity to amphotericin B or aureobasidin A (data not shown).
FIG. 4.
Effect of australifungin on the growth of LCB3 synthetically lethal mutants. LCB3 synthetically lethal mutants E37, U48, U4, U33, U63, and U9 were tested for australifungin sensitivity in a broth microdilution assay and compared to the screening strain Y388-2p. The OD600 was observed after 48 h of growth in the presence of drug.
ELO3 (SUR4) and CKA2 are required for normal growth of LCB3 deletion mutants.
In order to identify mutations that resulted in LCB3 plasmid retention and increased sensitivity to australifungin in the four mutants, we transformed them with a yeast genomic library and assayed for a red colony color phenotype reversal. This approach was successful for mutants E37 and U48. Sequence analysis of plasmids extracted from independent clones revealed that both E37 and U48 were complemented by library plasmids that carry ELO3 (SUR4), a gene which is responsible for the formation of C26 very-long-chain fatty acids (VLCFA) in yeast (37). In order to confirm that ELO3 and not one of the adjacent genes present in the library plasmid complemented the phenotype of E37 and U48 mutants, we constructed a plasmid (pELO3) that carries the wild-type ELO3 gene on the single-copy vector and showed a reversal in colony color phenotype in the E37 and U48 mutants. This plasmid was unable to reverse the colony color phenotype of the U4 and U33 mutants. A similar plasmid that carried the ELO2 gene was unable to complement any of the four mutants. Furthermore, the ELO3 locus was amplified from each mutant and sequence analysis of two independent amplicons obtained from each strain revealed the presence of nonsense mutations in codons 236 and 200 of the E37 and U48 alleles, respectively, and the wild-type ELO3 sequence in mutants U4 and U33. These results indicate that, in strain Y388, inactivation of both the LCB3 and ELO3 genes results in a loss of viability.
Genes defective in mutants U4 and U33 were identified by complementation of the drug supersensitivity phenotype. In the complementation experiments, the ceramide synthase inhibitor fumonisin B1 was used in place of australifungin. For each mutant, a complementing plasmid carried a genomic DNA insert that contained the entire CKA2 gene encoding the α′ catalytic subunit of CK2 (casein) kinase. A plasmid containing a CKA2 gene not only reversed a fumonisin B1 supersensitivity phenotype but also caused a change in colony color from red to a sectored red-white in both the U4 and U33 mutants. Sequence analysis of the CKA2 locus amplified from both mutants revealed the presence of a nonsense mutation in codon 227 of mutant U4 and a frameshift mutation in codon 80 of mutant U33.
In order to verify that both the lcb3 elo3 double deletion and the lcb3 cka2 double deletion cause growth defects, we analyzed genetic crosses among deletion strains BY4742lcb3, BY4741elo3, and BY4741cka2. Twenty-one out of 23 spores that came from 5 nonparental ditype and 13 tetratype tetrads and that were deduced to be double lcb3 cka2 mutants formed distinctly small colonies. In the case of elo3 mutants, 14 out of 16 double-deletion spores either formed extremely small colonies (11 spores) or were inviable (3 spores). This result clearly shows that a strong negative synthetic interaction exists between the lcb3 and elo3 as well as the lcb3 and cka2 deletions; however, in most cases, the interaction results in a poor-growth phenotype rather than lethality.
Crude membranes from cka2 mutants have reduced ceramide synthase activity.
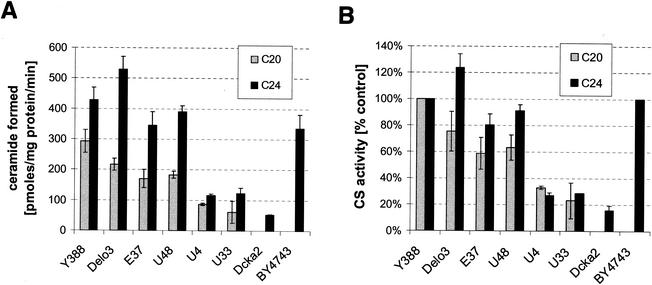
In order to further characterize the nature of australifungin and fumonisin B1 sensitivity, we directly measured the activity of ceramide synthase in crude membrane preparations obtained from elo3 and cka2 mutants and the parental strains. Ceramide formation in this assay was linear with time (Fig. 5A), was dependent on the addition of long-chain acyl-CoA, and was completely inhibited by 100 μM fumonisin B1 and 10 μM australifungin (Fig. 5B). In a separate experiment, we determined that fumonisin B1 was a submicromolar inhibitor of yeast ceramide synthase activity (50% inhibitory concentration, 0.2 μM). We also found that the rate of the reaction increased with increasing lengths of acyl chains in the acyl-CoA substrate; e.g., there was fivefold more product formed in reaction mixtures that contained C24:0-CoA than in those that contained C16:0-CoA (Fig. 5C). Ceramide was not formed when free long-chain fatty acid was added to the reaction mixture, thus indicating that the measured activity was due to the acyl-CoA-dependent ceramide synthase and not due to the reverse activity of ceramidase (30). The specific activity of acyl-CoA-dependent ceramide synthase in the crude membrane preparations of the four non-DPL1 mutants obtained in the synthetic-lethality screen was consistently reduced compared to that of the parental strain Y-388. The activity in membranes from elo3 (E37 and U48) mutants was reduced by 30 to 40% when C20:0-CoA was used as a substrate. This difference, however, was significantly diminished when C24:0-CoA was used as a substrate in the assay (Fig. 6). Similar to the E37 and U48 elo3 mutants, the ELO3 deletion strain also had a slightly reduced ability to incorporate C20:0-CoA into ceramide in the in vitro enzyme assay. This defect was not present when C24:0-CoA instead of C20:0-CoA was used in the assay (Fig. 6).
FIG. 5.
Acyl-CoA-dependent ceramide synthase has a strong preference for longer-chain fatty acyls. Ceramide synthase activity was measured by the incubation of crude membranes with [3H]DHS in the presence of acyl-CoA as described in Materials and Methods. (A) The reaction was linear for at least 30 min. (B) The formation of the product that comigrated with the ceramide standard, indicated by a broken circle, upon silica gel TLC was dependent on the addition of acyl-CoA and was inhibited by 10 μM australifungin and 100 μM fumonisin B1. (C) Crude membranes from the screening strain Y388-2 were assayed for ceramide formation after incubation with 10 μM acyl-CoAs containing saturated fatty acyls of various lengths.
FIG. 6.
Crude membrane fractions from australifungin-sensitive mutants have reduced ceramide synthase activity. Membranes isolated from LCB3 synthetically lethal mutants E37, U48, U4, and U33 from parental strain Y388 and from the elo3 deletion mutant Y388elo3 were assayed for acyl-CoA-dependent ceramide synthase activity with arachidonyl-CoA (grey bars) and lignoceroyl-CoA (black bars). Ceramide synthase activities in membranes isolated from the Δcka2 strain and from its parental strain, BY4741, were assayed with lignoceryl-CoA as the substrate. Reaction mixtures were extracted, separated by silica gel TLC, and quantitated as described in Materials and Methods. The activity of the enzyme was expressed as specific activity (A) or as a percentage of the activity found in the control parental strain (B).
The activity in membranes from cka2 mutants (U4 and U33) was reproducibly reduced by 70 to 75% regardless of the chain length of the acyl-CoA used in the assay (Fig. 6). The independently obtained cka2 deletion strain also had severely reduced ceramide synthase activity (Fig. 6).
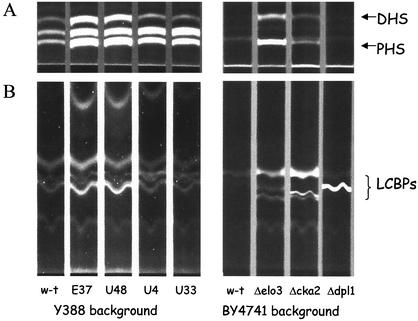
The elo3 and cka2 mutants have elevated levels of LCBs and LCBPs.
As mentioned previously, the combination of lcb3 and dpl1, two mutations that individually result in modest elevation of LCBPs, leads to a large accumulation of LCBPs and results in cell death (18, 45, 48). We expected that other mutations that cause poor growth in the lcb3 deletion background would also have elevated levels of LCBPs. We measured the levels of both free LCBs and free LCBPs in four mutants obtained in the screen as well as in the Δcka2, Δelo3, and Δdpl1 deletion strains and the respective parental strains (Fig. 7). Both the elo3 mutants E37 and U48 and the elo3 deletion strain accumulated high levels of free DHS and PHS and also had highly elevated levels of LCBPs compared to those of parental wild-type strains. Similarly, the cka2 mutants U4 and U33 and the Δcka2 strain had highly elevated levels of PHS and elevated levels of DHS and LCBPs. The Δelo3 and Δcka2 mutants, like the Δdpl1 strain, accumulated high levels of LCBPs, but each of the mutants appeared to accumulate different species of LCBPs. Both the Δelo3 and Δcka2 mutants, consistent with the block in ceramide synthesis, accumulated both LCBs and LCBPs, whereas the Δdpl1 mutant, which lacks LCBP lyase, accumulated mostly LCBPs.
FIG. 7.
The Δlcb3 synthetically lethal mutants accumulate LCBs and LCBPs. LCBs and LCBPs were extracted from 12 OD units of Y388 background cells and from 30 OD units of BY4741 background cells, derivatized with AQC, and treated with KOH to deacylate glycerophospholipids as described in Materials and Methods. (A) Ten microliters of each extract was resolved by TLC in solvent I under conditions separating AQC-LCBs. (B) Fifty microliters of each extract was absorbed on a C18 column, and AQC-LCBPs that eluted from the column were separated by TLC in solvent II. The migration of AQC-derivatized standards is indicated. TLC plates were visualized with UV light. w-t, wild type.
Deletion of elo3 and deletion of cka2 result in hypersensitivity to ceramide synthase inhibitors.
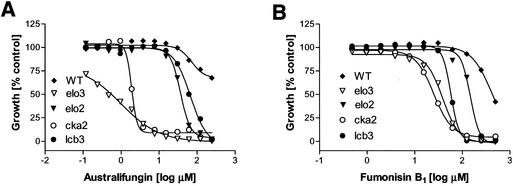
As shown above, the elo3 and cka2 mutants obtained in the genetic screen were supersensitive to drugs inhibiting acyl-CoA-dependent ceramide synthase. In order to confirm this conclusion, we assayed an elo3 deletion and cka2 deletion strains for sensitivity to both australifungin and fumonisin B1. Similar to the truncated elo3 mutants E37 and U48, an elo3 deletion strain, BY4741elo3, was about 200-fold more sensitive to australifungin than the parental strain, BY4741 (Fig. 8). Interestingly, the isogenic elo2 deletion strain was also more sensitive to the drug than the parent; however, it was much less sensitive than the elo3 mutant. Likewise, the deletion of the LCB3 gene had an observable but comparatively smaller effect on australifungin sensitivity than did the deletion of ELO3. The elo3 deletion mutant was also supersensitive to the structurally unrelated inhibitor of the ceramide synthase fumonisin B1, and as in the case of australifungin, a deletion of ELO3 sensitized the cells to fumonisin B1 much more so than a deletion of ELO2. The Δlcb3 strain was also sensitive to fumonisin B1. A Δcka2 strain similar to the cka2 mutants U4 and U33 also showed highly increased sensitivity to australifungin and fumonisin B1 (Fig. 8). Notably, mutants lacking other subunits of the CK2 kinase, namely, the Δcka1, Δckb1, and Δckb2 mutants, did not show any increase in sensitivity to fumonisin B1 (data not shown).
FIG. 8.
Null elo3 and cka2 mutants are supersensitive to ceramide synthase inhibitors. Cells of various deletion mutants were exposed to australifungin (A) or fumonisin B1 (B) in the broth microdilution assay, and the growth was measured after 48 h of incubation. WT, wild type.
DISCUSSION
We have isolated and characterized S. cerevisiae mutants that require the normally dispensable LCB3 gene for growth. Since Lcb3 LCB phosphatase is a part of the system that maintains LCBP-ceramide homeostasis, we expected to uncover mutations in this screen that affect other parts of this system. Out of 14 mutants identified in the screen and confirmed to carry mutations causing synthetic lethality with the LCB3 deletion, 10 were defective in the DPL1 gene encoding the LCBP lyase, 2 were mutated in the ELO3 (SUR4) elongase gene required for formation of the C26 VLCFA, and the remaining 2 were defective in in vitro acyl-CoA-dependent ceramide synthase activity. Mutants defective in the ceramide synthase activity lacked a functional CKA2 gene encoding the α′ catalytic subunit of the CK2 kinase, thus indicating that CK2 kinase is a novel determinant of a fully functional ceramide synthase. Deletion of DPL1 was previously demonstrated to be lethal in strains lacking Lcb3 phosphatase (18, 48). A single LCB3 or DPL1 deletion results in a 5- to 40-fold increase of the steady-state levels of the LCBPs and does not have a measurable effect on cell growth. However, elimination of both the Lcb3 phosphatase and the Dpl1 lyase activities from the cell causes massive accumulation of LCBPs, over 400-fold above the wild-type levels, which in turn leads to cell death. Recent detailed analysis of LCBP accumulation in yeast indicates that DHS-1-P species are more toxic than PHS-1-Ps and that the phosphatase rather than the lyase is involved in the removal of the toxic excess of these intermediates (48). Over two-thirds of the mutants in our screen were classified in the expected DPL1 category, indicating that the screen was reasonably saturated.
Mutants E37 and U48 carry mutations in the ELO3 gene, and their dependence for growth on the LCB3 plasmid was relieved by transformation with an ELO3 plasmid. Additionally, we confirmed by genetic crosses that spores carrying deletions of both the LCB3 and ELO3 genes have poor viability. elo3 mutants are unable to synthesize C26 VLCFA, which are the primary fatty acids linked by the yeast ceramide synthase to PHS or DHS to form the ceramide found in all yeast sphingolipids (7, 37) (Fig. 1). Deletion of ELO3 results in the incorporation of shorter-chain fatty acyls into ceramides, mostly C22 and C24 species that are formed with participation of the Elo2 protein. Additionally, Δelo3 mutants appear to accumulate even shorter fatty acyl ceramides that are not found in wild-type cells (20, 37). ELO3 deletion mutants grow almost normally under standard laboratory conditions, but were shown to have elevated LCB levels (20, 37) (Fig. 8). These observations, together with our findings that elo3 mutations are lethal in cells lacking Lcb3 phosphatase and that elo3 cells accumulate high levels of LCBPs, indicate that a defect in the formation of the C26 VLCFA in yeast causes a backlog in the flux through the sphingolipid pathway at ceramide synthesis, which in turn leads to the elevation of free LCB levels and increased production of the LCBPs. This scenario indicates a significant in vivo preference of the ceramide synthase for the C26-CoA substrate. This view is supported by our observation that the in vitro specific activity of ceramide synthase is nearly 40% higher in assays utilizing C24:0 than in assays utilizing C20:0-CoA as a substrate (Fig. 5c). An even more dramatic preference for C26:0-CoA was observed in the in vitro ceramide synthase assays by Guillas and coworkers (12). Mutants E37 and U48, as well as the elo3 deletion mutants, show extraordinary growth sensitivity to the ceramide synthase inhibitors australifungin and fumonisin B1 (Fig. 4 and 8). Since elo3 mutants have normal levels of in vitro acyl-CoA-dependent ceramide synthase activity, it is not likely that the lack of the Elo3 protein directly impacts the sensitivity of the enzyme to drugs. This sensitivity is more likely caused by the decreased consumption of LCBs in cells lacking C26-CoA and the consequent elevation of LCBP levels, which must be elevated even further in cells treated with normally subinhibitory levels of ceramide synthase inhibitors. Interestingly, elo3 mutants were more sensitive to both fumonisin B1 and australifungin than were the elo2 and lcb3 mutants, which were also more susceptible to both drugs than was the homogenic wild-type strain (Fig. 8). It is possible that in the absence of C26-CoA, a preferred substrate for ceramide synthase, the enzyme becomes significantly more sensitive to both drugs. Taken together, these results highlight the importance of the ELO3 gene and C26-CoA formation for LCB homeostasis in yeast. This is likely a reflection of the extraordinary significance of VLCFA in the formation of functional plasma membranes in single-celled eukaryotes. Most of the C26 VLCFA in yeast are found in sphingolipids, which are located primarily in the plasma membrane (38). Yeast SLC mutants that are able to survive without making any sphingolipids produce compensatory glycerolipids that are based on a C26 VLCFA-containing phosphatidylinositol not found in normal cells (24), further stressing the importance of C26 VLCFA for the survival of yeast cells. Mutants that survive despite their inability to form ceramide also produce novel glycerolipids, and indeed, preliminary experiments showed that they do contain VLCFA (12, 43). The plasma membrane lipid bilayer in yeast, as indicated by the lengths of predicted transmembrane domains in the membrane proteins, is 50% thicker than the lipid bilayers of internal membranes (25). It is possible that VLCFA-containing lipids are indispensable for thickening of the plasma membrane and therefore may be essential for protein sorting and for formation of functional cell envelope proteins in yeast.
The strain constructed for the purpose of our LCB3 genetic screen formed a distinct ring of red colonies surrounding the zone of inhibition due to australifungin treatment, which confirmed that the effect of a block in ceramide formation is exacerbated in lcb3 mutants (Fig. 3). This finding indicated that this genetic screen was capable of yielding mutants defective in ceramide synthase. We assayed acyl-CoA-dependent ceramide synthase activity in crude membranes isolated from four non-dpl1 mutants and found that specific enzyme activity was reduced three- to fourfold in mutants U4 and U33 (Fig. 6). Both mutants were also more sensitive to growth inhibition by australifungin and fumonisin B1 (Fig. 4), and both accumulated free LCBs and had increased levels of LCBPs (Fig. 7). Surprisingly, both mutants carried mutations inactivating the α′ catalytic subunit of the CK2 kinase encoded by the CKA2 gene. An independently obtained deletion of the CKA2 gene led to all phenotypes found in the U4 and U33 mutants, including a defect in in vitro ceramide synthase activity (Fig. 6 to 8). CK2 kinase, formerly known as casein kinase, is a highly conserved, ubiquitous, and pleiotropic protein kinase whose activity is essential for growth in yeast. The enzyme exists in cells as a heterotetrameric holoenzyme consisting of two catalytic subunits, α and α′, and two regulatory subunits, β and β′. A recent study demonstrated differential distributions of the holoenzyme and individual CK2 subunits in mammalian cells and showed that individual CK2 subunits are located on the cytosolic side of the rough endoplasmic reticulum and the smooth endoplasmic reticulum-Golgi complex (9). CK2 kinase has been implicated in numerous fundamental cell processes, including cell proliferation, regulation of polymerase III activity, cell polarity, ion homeostasis, signal transduction, and information storage in neuronal cells, although the cellular function of this kinase is still unclear (3, 11, 39). Our observations that the α′ subunit of CK2 kinase is selectively needed for optimal activity of ceramide synthase and that CKA2 mutants accumulate ceramide synthase substrates define a new important cellular role for CK2 kinase in the activation of ceramide synthase activity. This is the first example of regulation of sphingolipid synthesis in any organism (34). At this point, it is not known if CK2 kinase is directly involved in the activation of ceramide synthase through phosphorylation of the enzyme or if the kinase is required in some indirect way for the formation of the fully active enzyme.
The specific activities of ceramide synthase in membranes from the elo3 mutants E37 and U48, as well as from the elo3 deletion strain, were reduced in comparison to that of wild-type membranes when C20:0-CoA was provided as a substrate. However, the ability to incorporate longer C24 fatty acyl-CoA into ceramide was not impaired in elo3 membranes (Fig. 6). One possible explanation for this observation is that the acyl-CoA in our crude membrane assay may be further elongated in vitro. Thus, given the preference of the enzyme for longer-chain fatty acyl moieties, elo3 membranes that are less efficient in the elongation of the added substrate would make less ceramide.
All of the mutants isolated in the LCB3 screen were affected in elements of the ceramide/LCBP rheostat itself. The fact that we did not find mutants defective in either the LAG1 or LAC1 determinants of the ceramide synthase activity is not surprising given the likelihood that these genes are highly redundant and that single mutants are not defective in the rate of ceramide synthesis. It is worth noting that we did not find mutations affecting genes that encode potential sensors or effectors of altered levels of LCBs or LCBPs, that is, mutations that themselves do not cause elevation of sphingolipid metabolites but rather result in increased sensitivity to the intracellular accumulation of LCBPs. It is possible that such hypothetical effectors have subtle effects on cell growth and therefore would not be expected to emerge from our screen. One other possibility is that these lipids do not have highly selective receptors in yeast and exert their effects through localized changes in the physical properties of membrane bilayers rather than acting as sensu stricto secondary messengers.
Acknowledgments
We thank Bob Dickson and Bob Lester (University of Kentucky) for advice on the ceramide synthase assay and for strains and plasmids, Alan Bender (Indiana University) for strains, Ming-Shang Kuo for a sample of australifungin, Kathleen Schellin and Leanne Seaver for DNA sequencing, and Rolf Kletzien and John Wilks for support and encouragement.
REFERENCES
- 1.Atkinson, K., S. Fogel, and S. A. Henry. 1980. Yeast mutant defective in phosphatidylserine synthesis. J. Biol. Chem. 255:6653-6661. [PubMed] [Google Scholar]
- 2.Bender, A., and J. R. Pringle. 1991. Use of a screen for synthetic lethal and multicopy suppressee mutants to identify two new genes involved in morphogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 11:1295-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanquet, P. R. 2000. Casein kinase 2 as a potentially important enzyme in the nervous system. Prog. Neurobiol. 60:211-246. [DOI] [PubMed] [Google Scholar]
- 4.Cherry, J. M., C. Ball, S. Weng, G. Juvik, R. Schmidt, C. Adler, B. Dunn, S. Dwight, L. Riles, R. K. Mortimer, and D. Botstein 1997. Genetic and physical maps of Saccharomyces cerevisiae. Nature 387:67-73. [PMC free article] [PubMed] [Google Scholar]
- 5.Cuvillier, O., G. Pirianov, B. Kleuser, P. G. Vanek, O. A. Coso, S. Gutkind, and S. Spiegel. 1996. Suppression of ceramide-mediated programmed cell death by sphingosine-1-phosphate. Nature 381:800-803. [DOI] [PubMed] [Google Scholar]
- 6.Daum, G., N. D. Lees, M. Bard, and R. Dickson. 1998. Biochemistry, cell biology and molecular biology of lipids of Saccharomyces cerevisiae. Yeast 14:1471-1510. [DOI] [PubMed] [Google Scholar]
- 7.Dickson, R. C., and R. L. Lester. 1999. Yeast sphingolipids. Biochim. Biophys. Acta 1426:347-357. [DOI] [PubMed] [Google Scholar]
- 8.Dickson, R. C., E. E. Nagiec, M. Skrzypek, P. Tillman, G. B. Wells, and R. L. Lester. 1997. Sphingolipids are potential heat stress signals in Saccharomyces. J. Biol. Chem. 272:30196-30200. [DOI] [PubMed] [Google Scholar]
- 9.Faust, M., M. Jung, J. Gunther, R. Zimmermann, and M. Montenarh. 2001. Localization of individual subunits of protein kinase CK2 to the endoplasmic reticulum and to the Golgi apparatus. Mol. Cell. Biochem. 227:73-80. [PubMed] [Google Scholar]
- 10.Georgopapadakou, N. H., and T. J. Walsh. 1996. Antifungal agents: chemotherapeutic targets and immunologic strategies. Antimicrob. Agents Chemother. 40:279-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glover, C. V., III. 1998. On the physiological role of casein kinase II in Saccharomyces cerevisiae. Prog. Nucleic Acid Res. Mol. Biol. 59:95-133. [DOI] [PubMed] [Google Scholar]
- 12.Guillas, I., P. A. Kirchman, R. Chuard, M. Pfefferli, J. C. Jiang, S. M. Jazwinski, and A. Conzelmann. 2001. C26-CoA-dependent ceramide synthesis of Saccharomyces cerevisiae is operated by Lag1p and Lac1p. EMBO J. 20:2655-2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guldener, U., S. Heck, T. Fielder, J. Beinhauer, and J. H. Hegemann. 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24:2519-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hakomori, S., and Y. Igarashi. 1995. Functional role of glycosphingolipids in cell recognition and signaling. J. Biochem. (Tokyo) 118:1091-1103. [DOI] [PubMed] [Google Scholar]
- 15.Hannun, Y. A., and C. Luberto. 2000. Ceramide in the eukaryotic stress response. Trends Cell Biol. 10:73-80. [DOI] [PubMed] [Google Scholar]
- 16.Hannun, Y. A., C. Luberto, and K. M. Argraves. 2001. Enzymes of sphingolipid metabolism: from modular to integrative signaling. Biochemistry 40:4893-4903. [DOI] [PubMed] [Google Scholar]
- 17.Jenkins, G. M., A. Richards, T. Wahl, C. Mao, L. Obeid, and Y. Hannun. 1997. Involvement of yeast sphingolipids in the heat stress response of Saccharomyces cerevisiae. J. Biol. Chem. 272:32566-32572. [DOI] [PubMed] [Google Scholar]
- 18.Kim, S., H. Fyrst, and J. Saba. 2000. Accumulation of phosphorylated sphingoid long chain bases results in cell growth inhibition in Saccharomyces cerevisiae. Genetics 156:1519-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kohama, T., A. Olivera, L. Edsall, M. M. Nagiec, R. Dickson, and S. Spiegel. 1998. Molecular cloning and functional characterization of murine sphingosine kinase. J. Biol. Chem. 273:23722-23728. [DOI] [PubMed] [Google Scholar]
- 20.Kohlwein, S. D., S. Eder, C.-S. Oh, C. E. Martin, K. Gable, D. Bacikova, and T. Dunn. 2001. Tsc13p is required for fatty acid elongation and localizes to a novel structure at the nuclear-vacuolar interface in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:109-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kolesnick, R., and Y. A. Hannun. 1999. Ceramide and apoptosis. Trends Biochem. Sci. 24:224-225. [DOI] [PubMed] [Google Scholar]
- 22.Lawrence, C. W. 1991. Classical mutagenesis techniques. Methods Enzymol. 194:273-281. [DOI] [PubMed] [Google Scholar]
- 23.Lester, R. L., and R. C. Dickson. 2001. High-performance liquid chromatography analysis of molecular species of sphingolipid-related long chain bases and long chain base phosphates in Saccharomyces cerevisiae after derivatization with 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate. Anal. Biochem. 298:283-292. [DOI] [PubMed] [Google Scholar]
- 24.Lester, R. L., G. B. Wells, G. Oxford, and R. C. Dickson. 1993. Mutant strains of Saccharomyces cerevisiae lacking sphingolipids synthesize novel inositol glycerophospholipids that mimic sphingolipid structures. J. Biol. Chem. 268:845-856. [PubMed] [Google Scholar]
- 25.Levine, T. P., C. A. Wiggins, and S. Munro. 2000. Inositol phosphorylceramide synthase is located in the Golgi apparatus of Saccharomyces cerevisiae. Mol. Biol. Cell 11:2267-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lutfiyya, L. L., V. R. Iyer, J. DeRisi, M. J. DeVit, P. O. Brown, and M. Johnston. 1998. Characterization of three related glucose repressors and genes they regulate in Saccharomyces cerevisiae. Genetics 150:1377-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mandala, S. M., R. A. Thornton, B. R. Frommer, J. E. Curotto, W. Rozdilsky, M. B. Kurtz, R. A. Giacobbe, G. F. Bills, M. A. Cabello, I. Martin, et al. 1995. The discovery of australifungin, a novel inhibitor of sphinganine N-acyltransferase from Sporormiella australis. Producing organism, fermentation, isolation, and biological activity. J. Antibiot. (Tokyo) 48:349-356. [DOI] [PubMed] [Google Scholar]
- 28.Mandala, S. M., R. Thornton, I. Galve-Roperh, S. Poulton, C. Peterson, A. Olivera, J. Bergstrom, M. B. Kurtz, and S. Spiegel. 2000. Molecular cloning and characterization of a lipid phosphohydrolase that degrades sphingosine-1-phosphate and induces cell death. Proc. Natl. Acad. Sci. USA 97:7859-7864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mandala, S. M., R. Thornton, Z. Tu, M. B. Kurtz, J. Nickels, J. Broach, R. Menzeleev, and S. Spiegel. 1998. Sphingoid base 1-phosphate phosphatase: a key regulator of sphingolipid metabolism and stress response. Proc. Natl. Acad. Sci. USA 95:150-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mao, C., R. Xu, A. Bielawska, and L. M. Obeid. 2000. Cloning of an alkaline ceramidase from Saccharomyces cerevisiae. An enzyme with reverse (CoA-independent) ceramide synthase activity. J. Biol. Chem. 275:6876-6884. [DOI] [PubMed] [Google Scholar]
- 31.Mao, C., J. D. Saba, and L. M. Obeid. 1999. The dihydrosphingosine-1-phosphate phosphatases of Saccharomyces cerevisiae are important regulators of cell proliferation and heat stress responses. Biochem. J. 342:667-675. [PMC free article] [PubMed] [Google Scholar]
- 32.Mao, C., R. Xu, Z. M. Szulc, A. Bielawska, S. H. Galadari, and L. M. Obeid. 2001. Cloning and characterization of a novel human alkaline ceramidase. A mammalian enzyme that hydrolyzes phytoceramide. J. Biol. Chem. 276:26577-26588. [DOI] [PubMed] [Google Scholar]
- 33.Mao, C., M. Wadleigh, G. M. Jenkins, Y. A. Hannun, and L. M. Obeid. 1997. Identification and characterization of Saccharomyces cerevisiae dihydrosphingosine-1-phosphate phosphatase. J. Biol. Chem. 272:28690-28694. [DOI] [PubMed] [Google Scholar]
- 34.Merrill, A. H., Jr. 2002. De novo sphingolipid biosynthesis: a necessary, but dangerous, pathway. J. Biol. Chem. 277:25843-25846. [DOI] [PubMed] [Google Scholar]
- 35.Nagiec, M. M., E. E. Nagiec, J. A. Baltisberger, G. B. Wells, R. L. Lester, and R. C. Dickson. 1997. Sphingolipid synthesis as a target for antifungal drugs. Complementation of the inositol phosphorylceramide synthase defect in a mutant strain of Saccharomyces cerevisiae by the AUR1 gene. J. Biol. Chem. 272:9809-9817. [DOI] [PubMed] [Google Scholar]
- 36.Nagiec, M. M., M. Skrzypek, E. E. Nagiec, R. L. Lester, and R. C. Dickson. 1998. The LCB4 (YOR171c) and LCB5 (YLR260w) genes of Saccharomyces encode sphingoid long chain base kinases. J. Biol. Chem. 273:19437-19442. [DOI] [PubMed] [Google Scholar]
- 37.Oh, C. S., D. A. Toke, S. Mandala, and C. E. Martin. 1997. ELO2 and ELO3, homologues of the Saccharomyces cerevisiae ELO1 gene, function in fatty acid elongation and are required for sphingolipid formation. J. Biol. Chem. 272:17376-17384. [DOI] [PubMed] [Google Scholar]
- 38.Patton, J. L., and R. L. Lester. 1991. The phosphoinositol sphingolipids of Saccharomyces cerevisiae are highly localized in the plasma membrane. J. Bacteriol. 173:3101-3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pinna, L. A. 2002. Protein kinase CK2: a challenge to canons. J. Cell Sci. 115:3873-3878. [DOI] [PubMed] [Google Scholar]
- 40.Qie, L., M. M. Nagiec, J. A. Baltisberger, R. L. Lester, and R. C. Dickson. 1997. Identification of a Saccharomyces gene, LCB3, necessary for incorporation of exogenous long chain bases into sphingolipids. J. Biol. Chem. 272:16110-16117. [DOI] [PubMed] [Google Scholar]
- 41.Saba, J. D., F. Nara, A. Bielawska, S. Garrett, and Y. A. Hannun. 1997. The BST1 gene of Saccharomyces cerevisiae is the sphingosine-1-phosphate lyase. J. Biol. Chem. 272:26087-26090. [DOI] [PubMed] [Google Scholar]
- 42.Schneiter, R. 1999. Brave little yeast, please guide us to Thebes: sphingolipid function in S. cerevisiae. Bioessays 21:1004-1010. [DOI] [PubMed] [Google Scholar]
- 43.Schorling, S., B. Vallee, W. P. Barz, H. Riezman, and D. Oesterhelt. 2001. Lag1p and Lac1p are essential for the Acyl-CoA-dependent ceramide synthase reaction in Saccharomyces cerevisae. Mol. Biol. Cell 12:3417-3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simons, K., and E. Ikonen. 1997. Functional rafts in cell membranes. Nature 387:569-572. [DOI] [PubMed] [Google Scholar]
- 45.Skrzypek, M. S., M. M. Nagiec, R. L. Lester, and R. C. Dickson. 1999. Analysis of phosphorylated sphingolipid long-chain bases reveals potential roles in heat stress and growth control in Saccharomyces. J. Bacteriol. 181:1134-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spiegel, S., O. Cuvillier, L. C. Edsall, T. Kohama, R. Menzeleev, Z. Olah, A. Olivera, G. Pirianov, D. M. Thomas, Z. Tu, J. R. Van Brocklyn, and F. Wang. 1998. Sphingosine-1-phosphate in cell growth and cell death. Ann. N. Y. Acad. Sci. 845:11-18. [DOI] [PubMed] [Google Scholar]
- 47.Spiegel, S., and S. Milstien. 2000. Sphingosine-1-phosphate: signaling inside and out. FEBS Lett. 476:55-57. [DOI] [PubMed] [Google Scholar]
- 48.Zhang, X., M. S. Skrzypek, R. L. Lester, and R. C. Dickson. 2001. Elevation of endogenous sphingolipid long-chain base phosphates kills Saccharomyces cerevisiae cells. Curr. Genet. 40:221-233. [DOI] [PubMed] [Google Scholar]
- 49.Zhou, J., and J. D. Saba. 1998. Identification of the first mammalian sphingosine phosphate lyase gene and its functional expression in yeast. Biochem. Biophys. Res. Commun. 242:502-507. [DOI] [PubMed] [Google Scholar]